Submitted:
20 September 2023
Posted:
22 September 2023
You are already at the latest version
Abstract
Keywords:
1. Overview
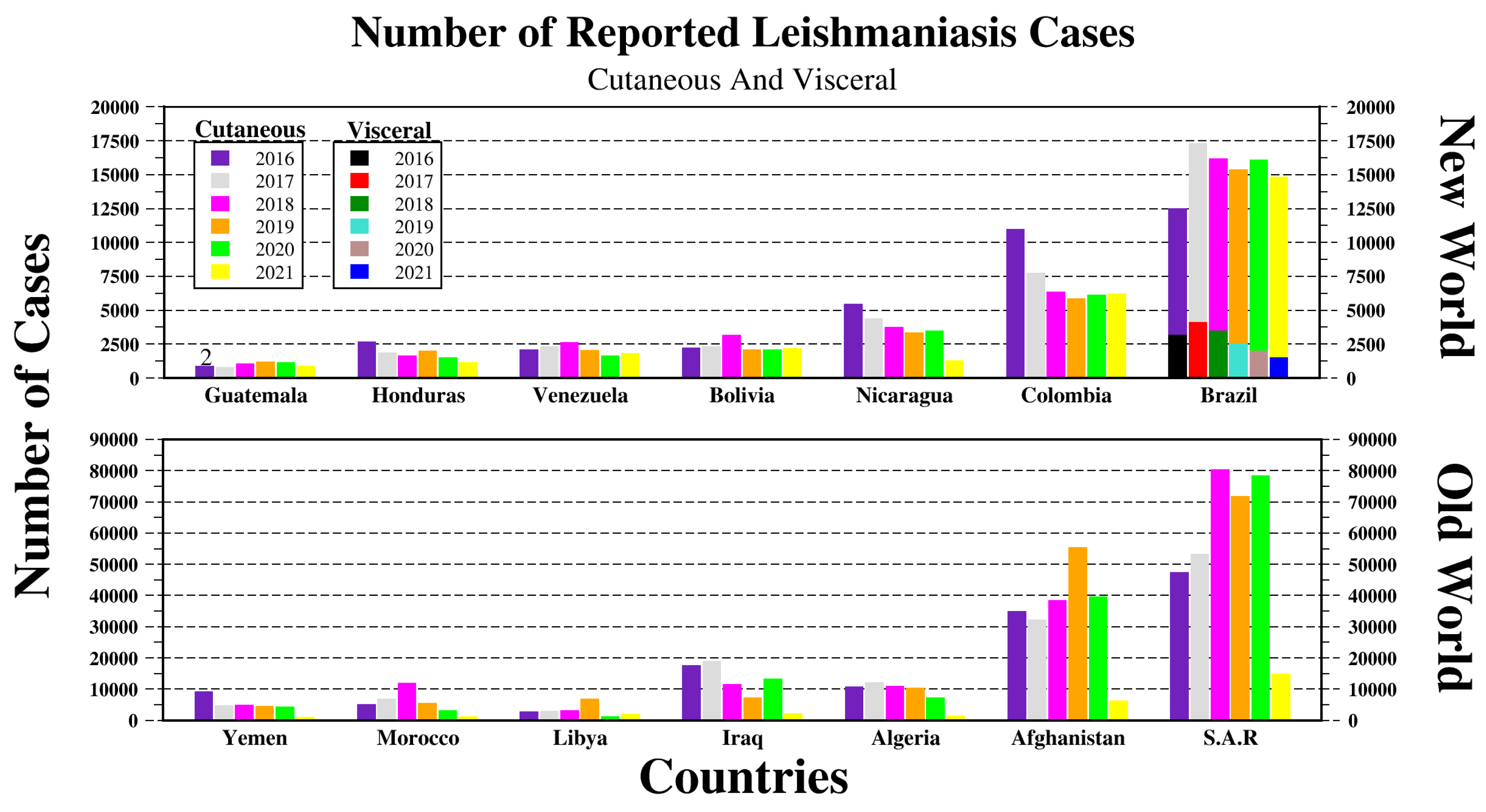
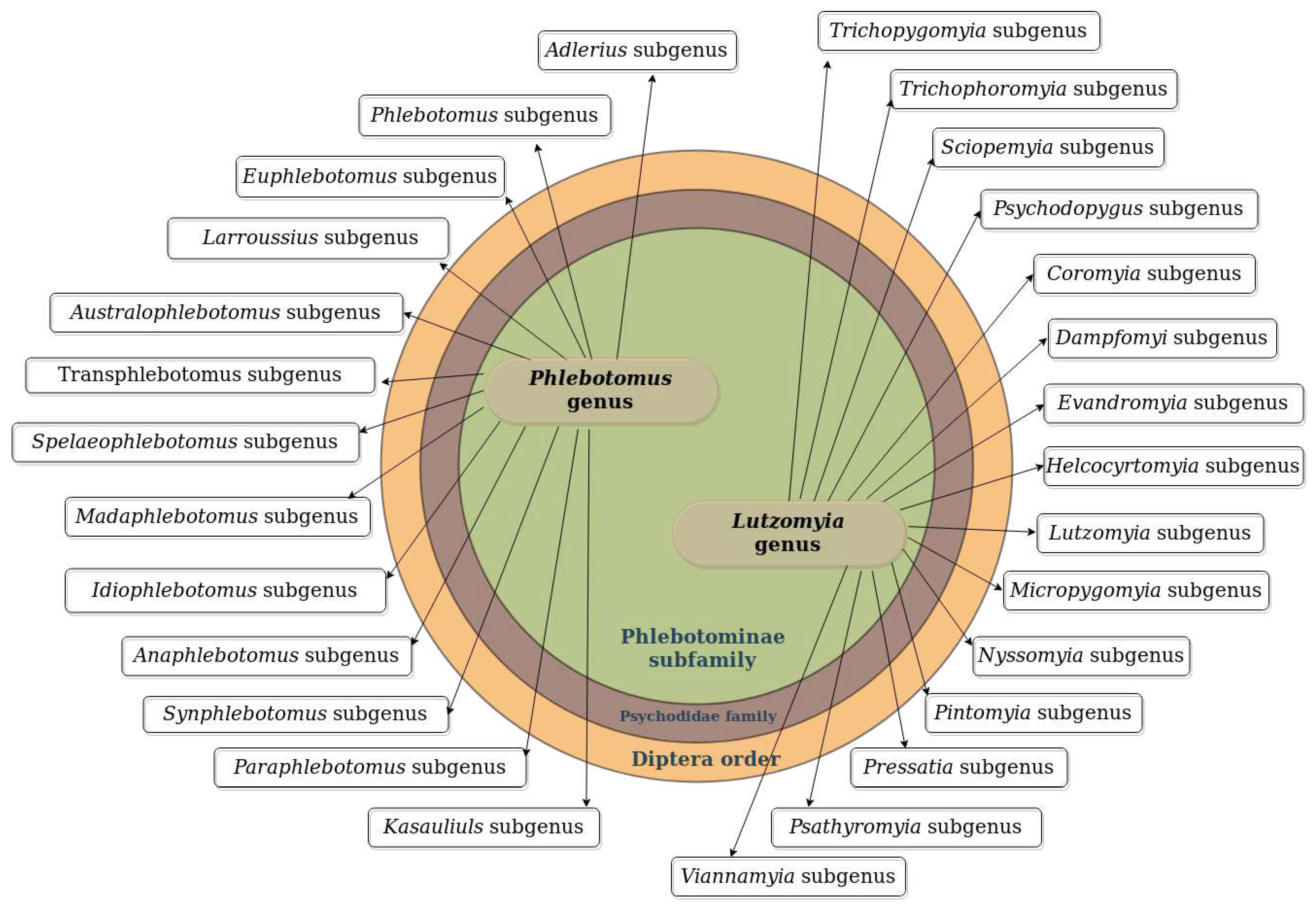
2. Leishmania Vectors
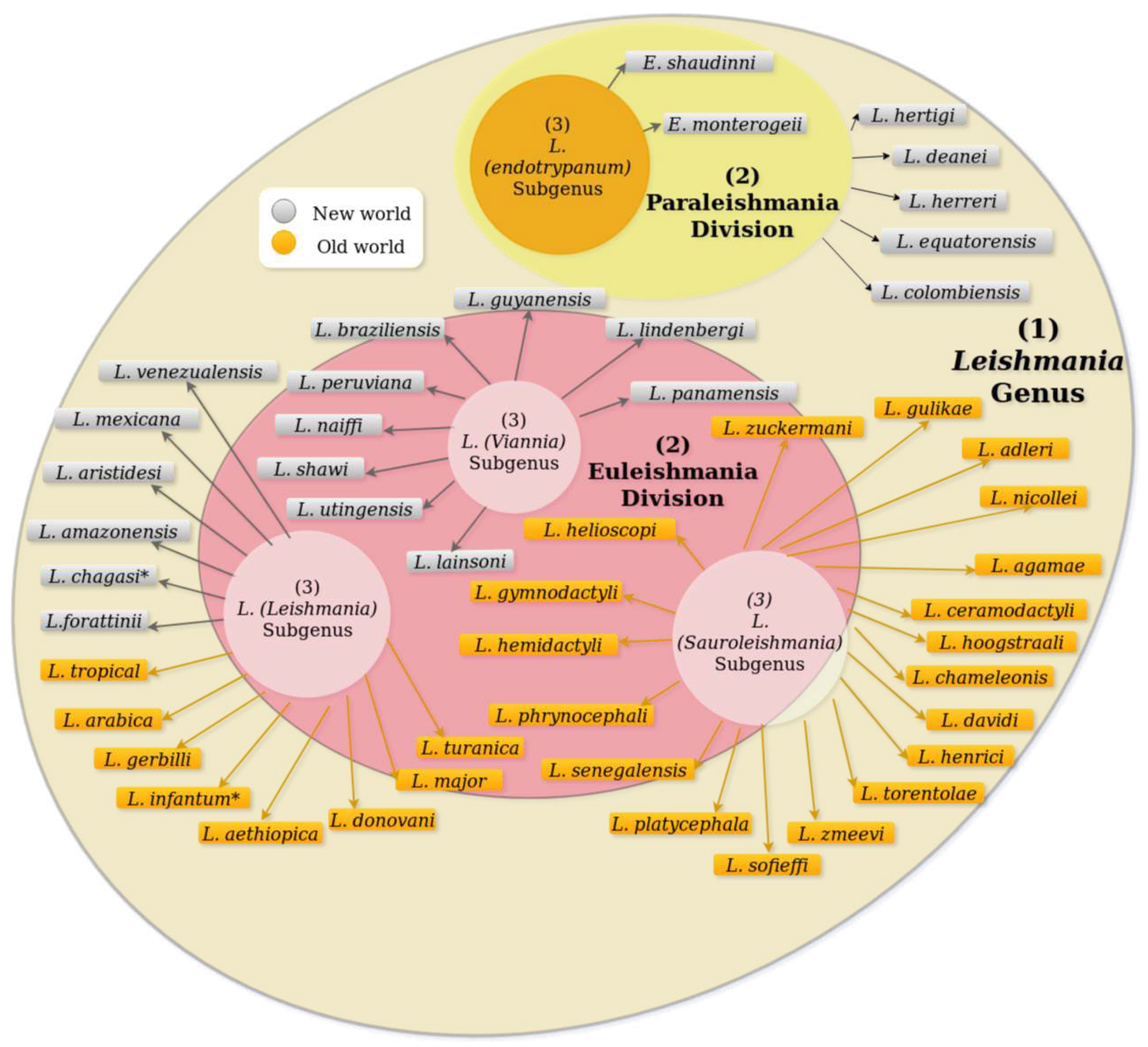
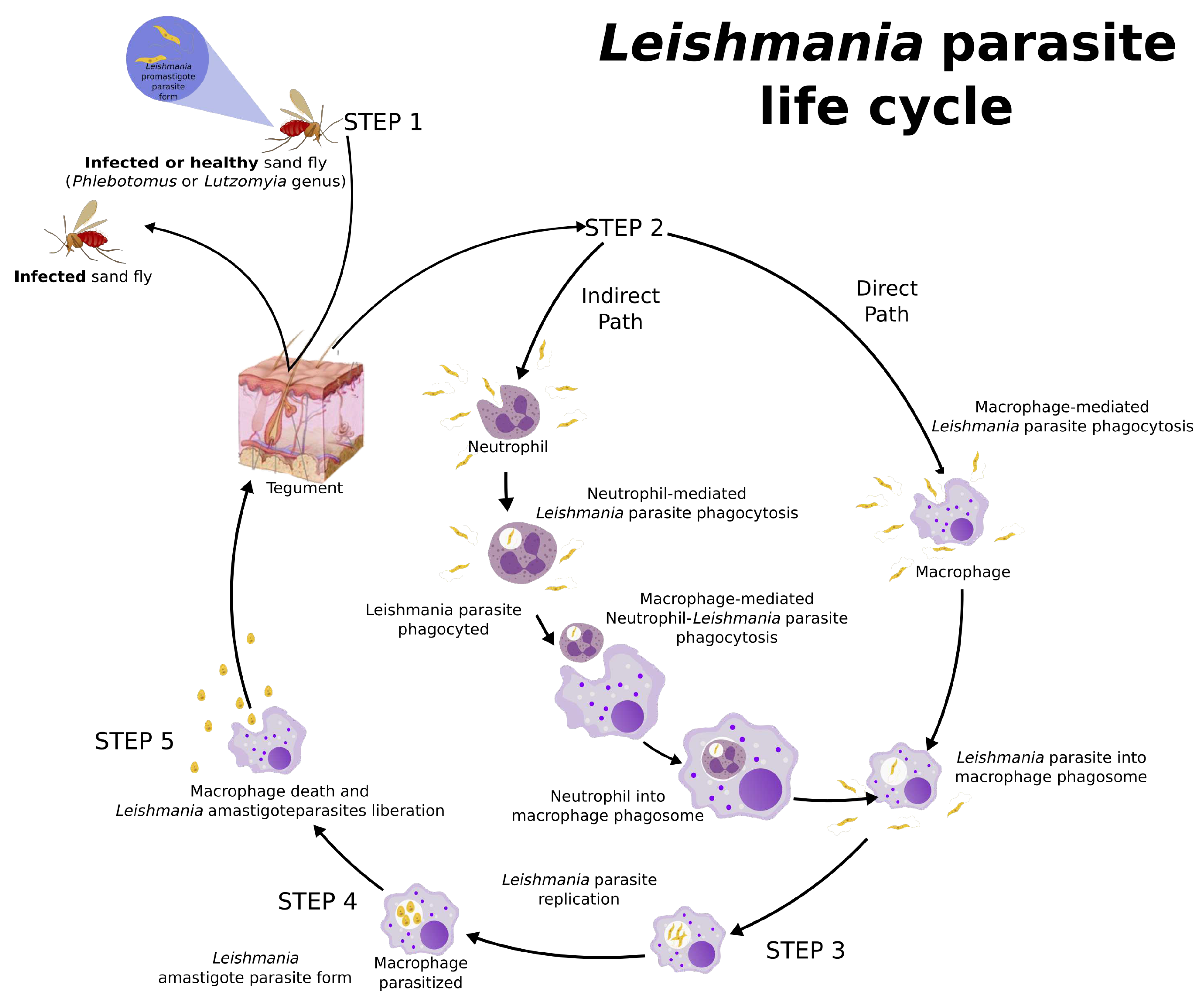
3. Leishmania Life Cycle and Host Immune Response
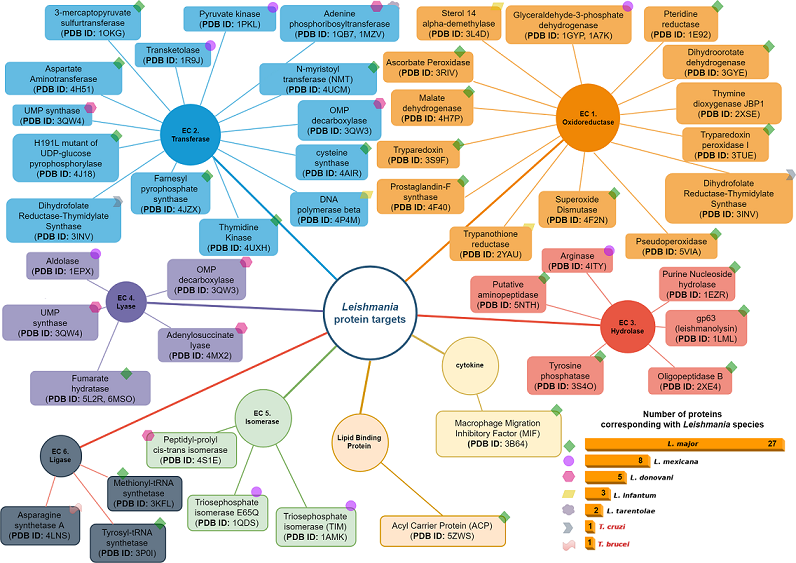
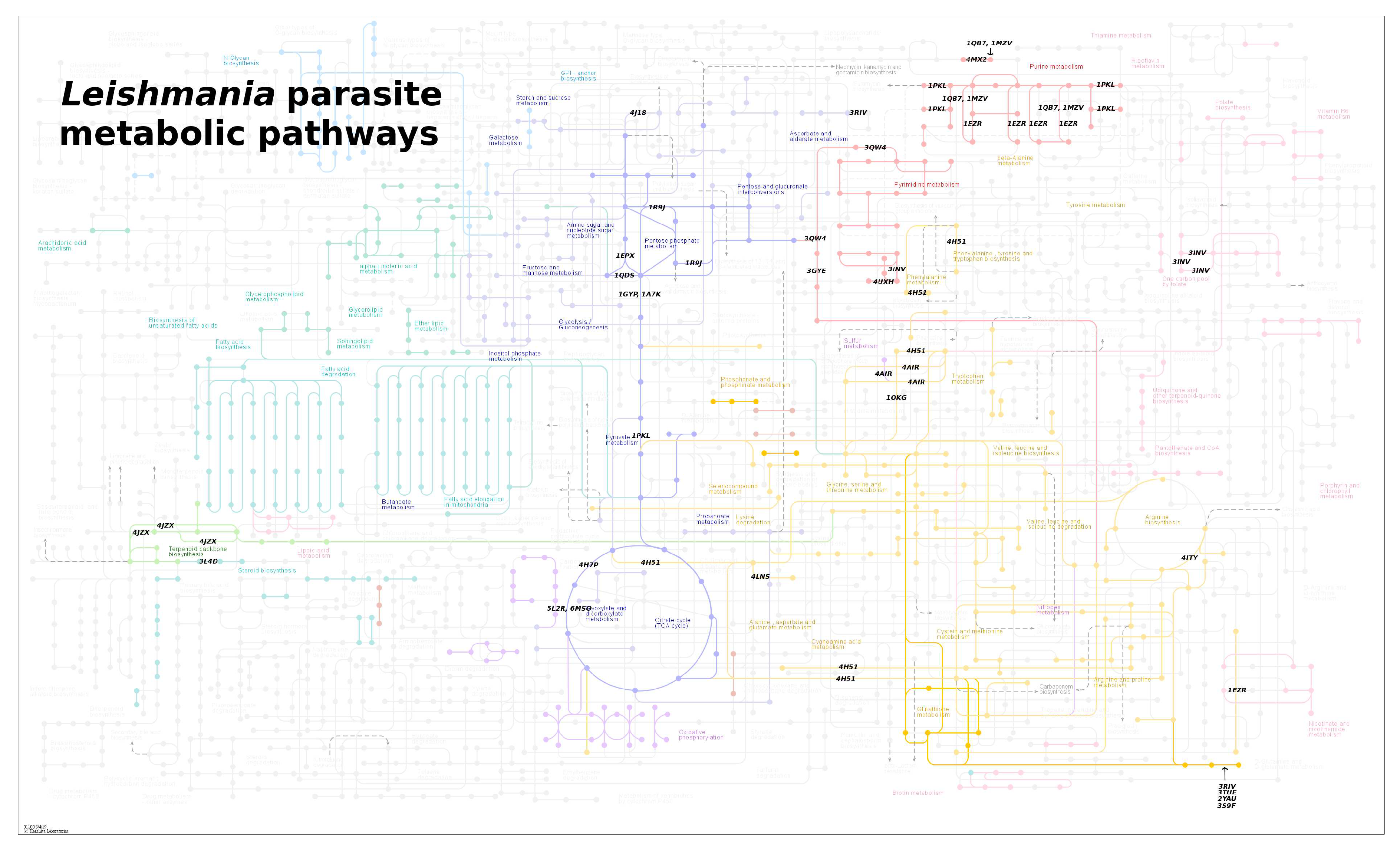
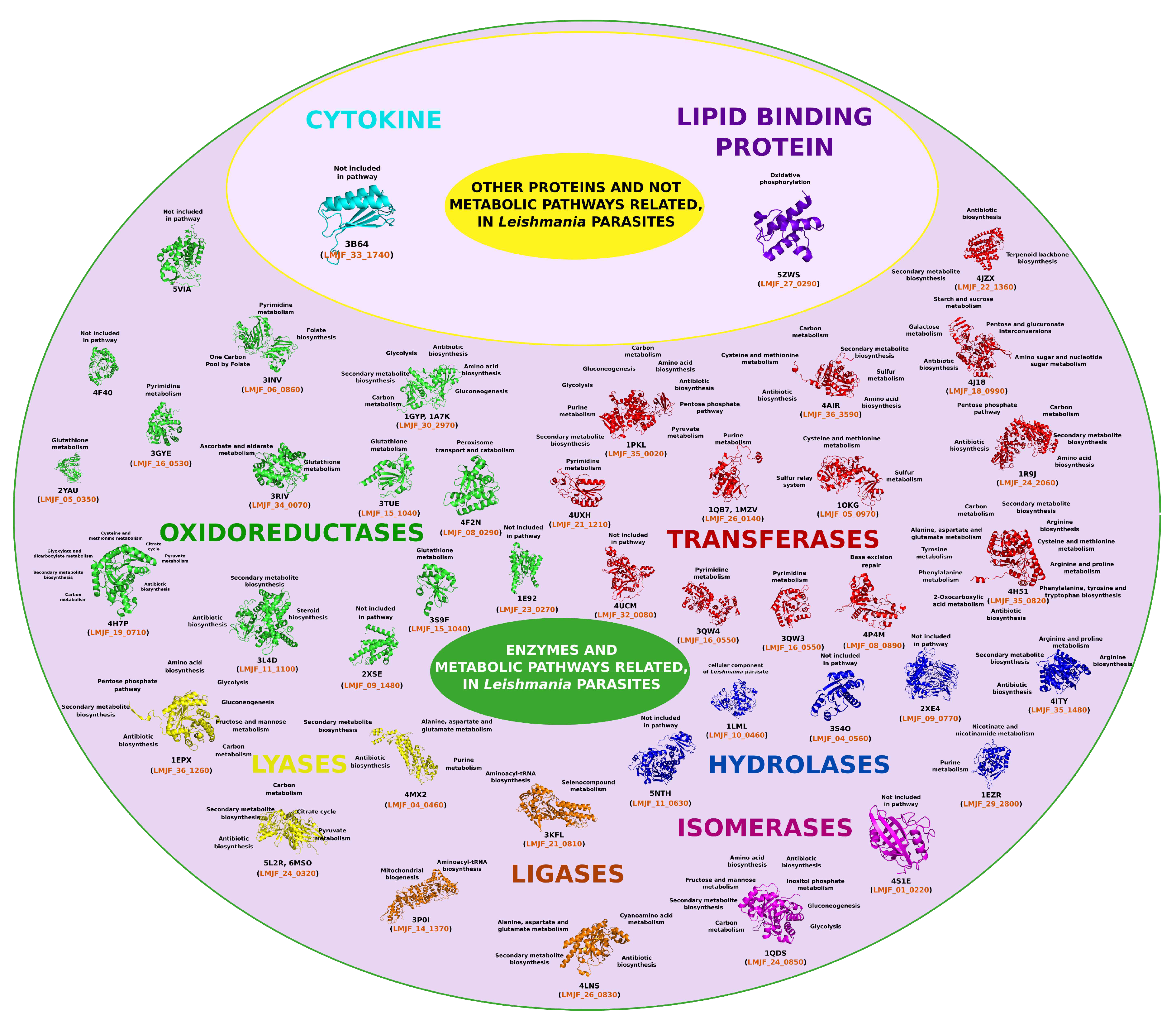
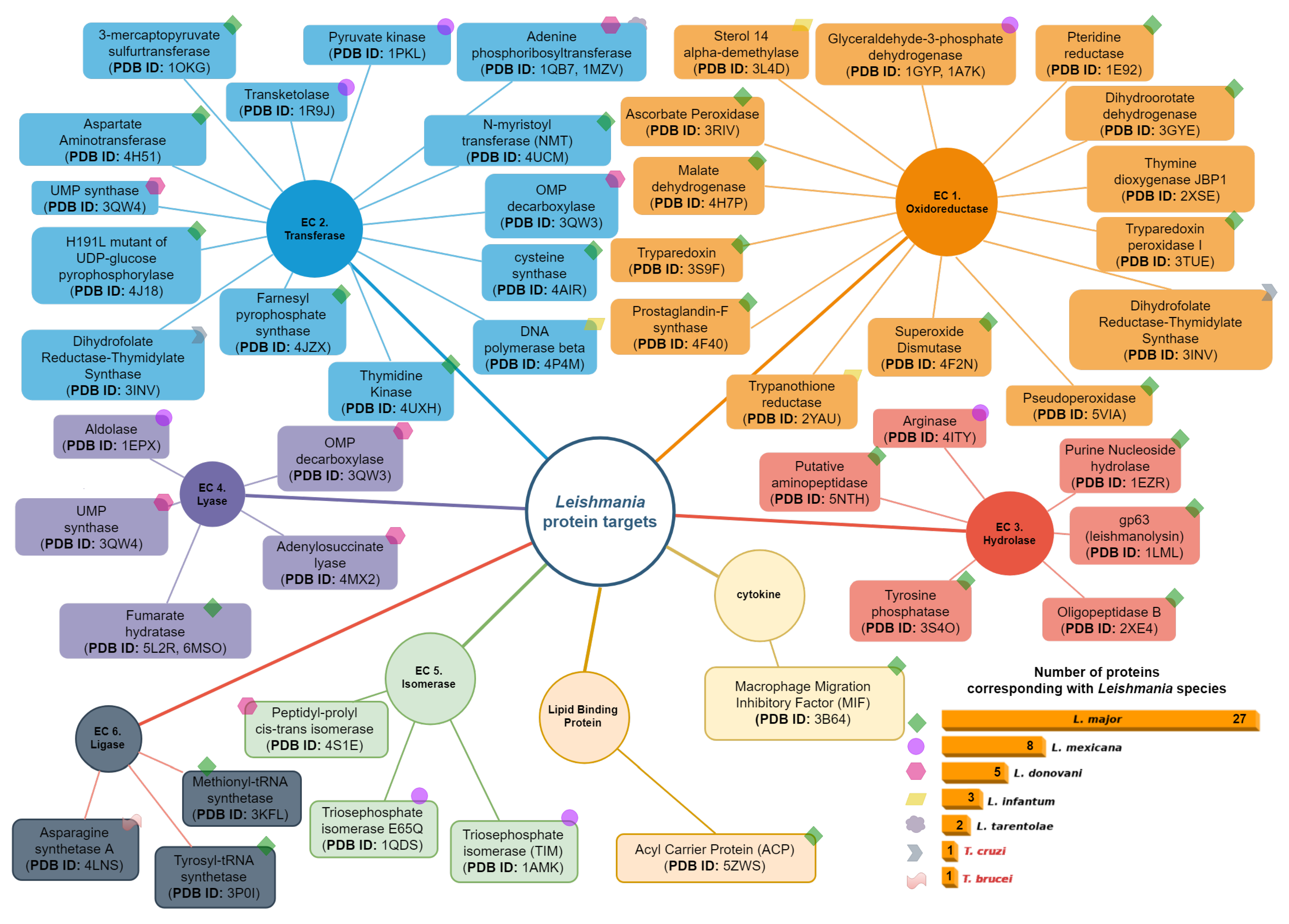
4. Leishmania Proteomics and Metabolomics Analysis
4.1. Oxidoreductases (EC 1)
4.2. Transferases Group (EC. 2)
4.3. Hydrolases Group (EC. 3)
4.4. Lyase Group (EC. 4)
4.5. Isomerases Group (EC. 5)
4.6. Ligases Group (EC. 6)
4.7. Cytokines Group
4.8. Lipid Binding Protein Group
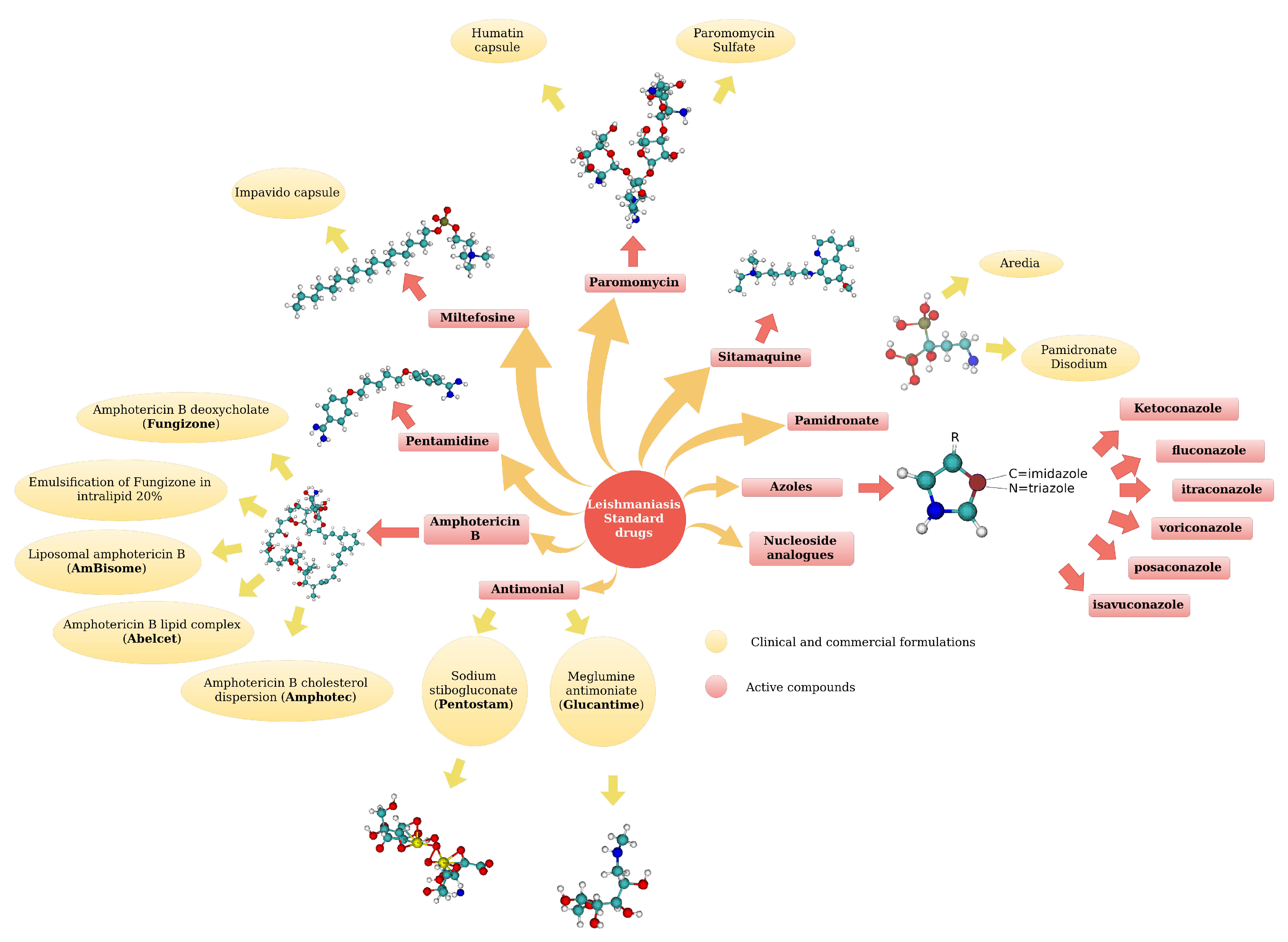
5. Leishmaniasis Drugs
5.1. Standard Drugs
5.2. Metabolomics: Non Standard Drugs
5.2.1. From Venom to Medicine: Phospholipases A2 (PLA2) as Potential Antiparasitic Agents against Leishmania Parasites
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ornellas-Garcia, U.; Cuervo, P.; Ribeiro-Gomes, F.L. Malaria and leishmaniasis: Updates on co-infection. Frontiers in Immunology 2023, 14, 1122411. [Google Scholar] [CrossRef] [PubMed]
- Awab, G.R. Leishmaniasis Epidemiology and Psychosocial Aspect; IntechOpen, 2023. [Google Scholar] [CrossRef]
- Grifferty, G.; Shirley, H.; McGloin, J.; Kahn, J.; Orriols, A.; Wamai, R. Vulnerabilities to and the Socioeconomic and Psychosocial Impacts of the Leishmaniases: A Review. Research and Reports in Tropical Medicine 2021, Volume 12, 135–151. [Google Scholar] [CrossRef]
- Maheshwari, K.K.; Bandyopadhyay, D. Heterocycles in the Treatment of Neglected Tropical Diseases. Current Medicinal Chemistry 2021, 28, 472–495. [Google Scholar] [CrossRef] [PubMed]
- Morales-Yuste, M.; Martín-Sánchez, J.; Corpas-Lopez, V. Canine Leishmaniasis: Update on Epidemiology, Diagnosis, Treatment, and Prevention. Veterinary Sciences 2023, 9, 387. [Google Scholar] [CrossRef]
- Gao, C.h.; Wang, J.y.; Zhang, S.; Yang, Y.t.; Wang, Y. Survey of Wild and Domestic Mammals for Infection with Leishmania infantum following an Outbreak of Desert Zoonotic Visceral Leishmaniasis in Jiashi, People’s Republic of China. PLOS ONE 2023, 10, e0132493. [Google Scholar] [CrossRef] [PubMed]
- Alcover, M.M.; Riera, M.C.; Fisa, R. Leishmaniosis in Rodents Caused by Leishmania infantum: A Review of Studies in the Mediterranean Area. Frontiers in Veterinary Science 2021, 8, 702687. [Google Scholar] [CrossRef] [PubMed]
- Rezaei, Z.; Pourabbas, B.; Asaei, S.; Sepehrpour, S.; Ahmadnia Motlagh, S.; Pourabbas, P.; Abdolahi Khasibi, S.; Alborzi, A. Livestock infected with Leishmania spp. in southern Iran. Parasites & Vectors 2023, 15, 215. [Google Scholar] [CrossRef]
- Ratzlaff, F.R.; Osmari, V.; da Silva, D.; de Paula Vasconcellos, J.S.; Pötter, L.; Fernandes, F.D.; de Mello Filho, J.A.; de Avila Botton, S.; Vogel, F.S.F.; Sangioni, L.A. Identification of infection by Leishmania spp. in wild and domestic animals in Brazil: a systematic review with meta-analysis (2001–2021). Parasitology Research 2023, 122, 1605–1619. [Google Scholar] [CrossRef]
- J, B.; M, B.M.; Chanda, K. An Overview on the Therapeutics of Neglected Infectious Diseases—Leishmaniasis and Chagas Diseases. Frontiers in Chemistry 2021, 9. [Google Scholar] [CrossRef]
- Pandian, S.R.K.; Panneerselvam, T.; Pavadai, P.; Govindaraj, S.; Ravishankar, V.; Palanisamy, P.; Sampath, M.; Sankaranarayanan, M.; Kunjiappan, S. Nano Based Approach for the Treatment of Neglected Tropical Diseases. Frontiers in Nanotechnology 2021, 3. [Google Scholar] [CrossRef]
- J, B.; M, B.M.; Chanda, K. An Overview on the Therapeutics of Neglected Infectious Diseases—Leishmaniasis and Chagas Diseases. Frontiers in Chemistry, 9.
- Choi, H.L.; Jain, S.; Postigo, J.A.R.; Borisch, B.; Dagne, D.A. The global procurement landscape of leishmaniasis medicines. PLOS Neglected Tropical Diseases, P: Publisher; 15, e0009181. [CrossRef]
- Zulfiqar, B.; Shelper, T.B.; Avery, V.M. Leishmaniasis drug discovery: recent progress and challenges in assay development. Drug Discovery Today 2017, 22, 1516–1531. [Google Scholar] [CrossRef] [PubMed]
- Reimão, J.Q.; Coser, E.M.; Lee, M.R.; Coelho, A.C. Laboratory Diagnosis of Cutaneous and Visceral Leishmaniasis: Current and Future Methods. Microorganisms, 8, 1632. [CrossRef]
- OPS/OMS, Organización Panamericana de la Salud. Leishmaniasis: Informe Epidemiológico de las Américas. Technical report, OPS, Organización Panamericana de la Salud, 2021.
- Cupolillo, E.; Medina-Acosta, E.; Noyes, H.; Momen, H.; Grimaldi, G. A revised classification for Leishmania and Endotrypanum. Parasitology Today 2000, 16, 142–144. [Google Scholar] [CrossRef] [PubMed]
- Momen, H.; Cupolillo, E. Speculations on the Origin and Evolution of the Genus Leishmania. Revista da Sociedade Brasileira de Medicina Tropical 2000, 95, 583–588. [Google Scholar] [CrossRef]
- Bruschi, F.; Gradoni, L. (Eds.) The leishmaniases: Old neglected tropical diseases; Springer International Publishing: Cham, 2018; pp. 1–245. [Google Scholar] [CrossRef]
- Kostygov, A.Y.; Yurchenko, V. Revised classification of the subfamily Leishmaniinae (Trypanosomatidae). Folia Parasitologica 2017, 64, 1–5. [Google Scholar] [CrossRef] [PubMed]
- (a)Lainson, R.; Shaw, J. Evolution, classification and geographical distribution. In The Leishmaniases in Biology and Epidemiology; Peters, W.; Killick-Kendrick, R., Eds.; Academic Press, 1987; Vol. 1, chapter 1, pp. 1 – 120.
- Lainson, R.; Shaw, J. The Role of Animals in the Epidemiology of South American Leishmaniasis. In Biology of the Kinetoplastida, 2 ed.; In WHR Lumsden, D.E., Ed.; Academic Press: London and New York, 1979; p. 116. [Google Scholar]
- Corrêa, J.R.; Brazil, R.P.; Soares, M.J. Leishmania (Viannia) lainsoni (Kinetoplastida: Trypanosomatidae), a divergent Leishmania of the Viannia subgenus - A mini review. Memorias do Instituto Oswaldo Cruz 2005, 100, 587–592. [Google Scholar] [CrossRef] [PubMed]
- Franco, A.M.; Grimaldi, G. Characterization of Endotrypanum (Kinetoplastida: Trypanosomatidae), a Unique Parasite Infecting the Neotropical Tree Sloths (Edentata). Memorias do Instituto Oswaldo Cruz 1999, 94, 261–268. [Google Scholar] [CrossRef]
- Akhoundi, M.; Kuhls, K.; Cannet, A.; Votýpka, J.; Marty, P.; Delaunay, P.; Sereno, D. A Historical Overview of the Classification, Evolution, and Dispersion of Leishmania Parasites and Sandflies. PLoS Neglected Tropical Diseases 2016, 10, 1–40. [Google Scholar] [CrossRef]
- Mesnil F, B.E. Sur un hematozoaire nouveau (Endotrypanum n.gen.) d’un edente de la Guyane. C R Soc Biol 1908, 65, 587. [Google Scholar]
- JJ, S. The Haemoflagellates of Sloths H.K. Lewis and Co. Ltd., London, In London School of Hygiene and Tropical Medicine Memoir, 13 ed.; H. K. Lewis (1969): London, 1969; p. 132. [Google Scholar]
- WHO/PAHO. Leishmaniases: Epidemiological Report in the Americas. Technical report, WHO and PAHO, 2018.
- Universidad del Rosario. Colombia, el país con más especies de parásitos de Leishmania, 2017.
- Kmetiuk, L.B.; Tirado, T.C.; Biondo, L.M.; Biondo, A.W.; Figueiredo, F.B. Leishmania spp. in indigenous populations: A mini-review. Frontiers in Public Health, 10.
- Herrera, G.; Barragán, N.; Luna, N.; Martínez, D.; De Martino, F.; Medina, J.; Niño, S.; Páez, L.; Ramírez, A.; Vega, L.; Velandia, V.; Vera, M.; Zúñiga, M.F.; Bottin, M.J.; Ramírez, J.D. An interactive database of Leishmania species distribution in the Americas. Scientific Data, 7, 110. [CrossRef]
- Frézard, F.; Aguiar, M.M.G.; Ferreira, L.A.M.; Ramos, G.S.; Santos, T.T.; Borges, G.S.M.; Vallejos, V.M.R.; De Morais, H.L.O. Liposomal Amphotericin B for Treatment of Leishmaniasis: From the Identification of Critical Physicochemical Attributes to the Design of Effective Topical and Oral Formulations. Pharmaceutics, 15, 99. [CrossRef]
- Musa, A.M.; Mbui, J.; Mohammed, R.; Olobo, J.; Ritmeijer, K.; Alcoba, G.; Muthoni Ouattara, G.; Egondi, T.; Nakanwagi, P.; Omollo, T.; Wasunna, M.; Verrest, L.; Dorlo, T.P.C.; Musa Younis, B.; Nour, A.; Taha Ahmed Elmukashfi, E.; Ismail Omer Haroun, A.; Khalil, E.A.G.; Njenga, S.; Fikre, H.; Mekonnen, T.; Mersha, D.; Sisay, K.; Sagaki, P.; Alvar, J.; Solomos, A.; Alves, F. Paromomycin and Miltefosine Combination as an Alternative to Treat Patients With Visceral Leishmaniasis in Eastern Africa: A Randomized, Controlled, Multicountry Trial. Clinical Infectious Diseases: An Official Publication of the Infectious Diseases Society of America, 76, e1177–e1185. [CrossRef]
- Román-Álamo, L.; Allaw, M.; Avalos-Padilla, Y.; Manca, M.L.; Manconi, M.; Fulgheri, F.; Fernández-Lajo, J.; Rivas, L.; Vázquez, J.A.; Peris, J.E.; Roca-Geronès, X.; Poonlaphdecha, S.; Alcover, M.M.; Fisa, R.; Riera, C.; Fernàndez-Busquets, X. In Vitro Evaluation of Aerosol Therapy with Pentamidine-Loaded Liposomes Coated with Chondroitin Sulfate or Heparin for the Treatment of Leishmaniasis. Pharmaceutics, 15, 1163. [CrossRef]
- Rojas Cabrera, E.; Verduguez-Orellana, A.; Tordoya-Titichoca, I.J.; Sejas, C.; Ledezma, R.; Álvarez, I.; Limachi-Choque, J.; Ortuño-Gutiérrez, N.; Córdova Rojas, M.; Guzman-Rivero, M. Intralesional Meglumine Antimoniate: Safe, Feasible and Effective Therapy for Cutaneous Leishmaniasis in Bolivia. Tropical Medicine and Infectious Disease, 7, 286. [CrossRef]
- Yadagiri, G.; Singh, A.; Arora, K.; Mudavath, S.L. Immunotherapy and immunochemotherapy in combating visceral leishmaniasis. Frontiers in Medicine, 10.
- Feng, M.; Jin, Y.; Yang, S.; Joachim, A.M.; Ning, Y.; Mori-Quiroz, L.M.; Fromm, J.; Perera, C.; Zhang, K.; Werbovetz, K.A.; Wang, M.Z. Sterol profiling of Leishmania parasites using a new HPLC-tandem mass spectrometry-based method and antifungal azoles as chemical probes reveals a key intermediate sterol that supports a branched ergosterol biosynthetic pathway. International Journal for Parasitology. Drugs and Drug Resistance, 20, 27–42. [CrossRef]
- Hofer, A. Targeting the nucleotide metabolism of Trypanosoma brucei and other trypanosomatids. FEMS Microbiology Reviews, 47, fuad020. [CrossRef]
- Croft, S.L.; Sundar, S.; Fairlamb, A.H. Drug Resistance in Leishmaniasis. Society 2006, 19, 111–126. [Google Scholar] [CrossRef]
- Kumar, A. Leishmania and Leishmaniasis; Vol. 3, SpringerBriefs in Immunology, Springer, 2013. [CrossRef]
- Ghorbani, M.; Farhoudi, R. Leishmaniasis in humans: Drug or vaccine therapy? Drug Design, Development and Therapy 2018, 12, 25–40. [Google Scholar] [CrossRef]
- Tsirigotakis, N.; Pavlou, C.; Christodoulou, V.; Dokianakis, E.; Kourouniotis, C.; Alten, B.; Antoniou, M. Phlebotomine sand flies (Diptera: Psychodidae) in the Greek Aegean Islands: ecological approaches. Parasites & Vectors 2018, 11, 97. [Google Scholar] [CrossRef]
- Bates, P.A. Transmission of Leishmania metacyclic promastigotes by phlebotomine sand flies. International Journal for Parasitology 2007, 37, 1097–1106. [Google Scholar] [CrossRef] [PubMed]
- ESTRADA, L.G.; APONTE, O.A.; BEJARANO, E.E. REGISTROS NUEVOS DE ESPECIES DE Lutzomyia (DIPTERA: PSYCHODIDAE) EN EL DEPARTAMENTO DE CESAR, COLOMBIA. Acta Biológica Colombiana 2015, 20, 225–228. [Google Scholar] [CrossRef]
- Yuko, E.; Sang, R.; Owino, E.A.; Ingonga, J.; Matoke-Muhia, D.; Hassaballa, I.B.; Junglen, S.; Tchouassi, D.P. Sandfly Blood-Feeding Habits and Competence in Transmitting Ntepes Virus, a Recently Discovered Member of the Genus Phlebovirus. BioMed Research International 2022, 2022, 4231978. [Google Scholar] [CrossRef] [PubMed]
- Amaro, F.; Zé-Zé, L.; Alves, M.J. Sandfly-Borne Phleboviruses in Portugal: Four and Still Counting. Viruses, M: Number: 8 Publisher; 14, 1768. [CrossRef]
- Alemayehu, B.; Alemayehu, M. Leishmaniasis: A Review on Parasite, Vector and Reservoir Host. Health Science Journal 2017, 11, 1–6. [Google Scholar] [CrossRef]
- Pérez-doria, A.; Hernández-oviedo, E.; Elías-Bejarano, E. Brumptomyia hamata ( Psychodidae ), A New Addition To The Phlebotomine Fauna Of The Colombian Caribbean. Acta Biológica Colombiana 2009, 14, 135–140. [Google Scholar]
- Ready, P.D. Biology of Phlebotomine Sand Flies as Vectors of Disease Agents. Annual Review of Entomology 2013, 58, 227–250. [Google Scholar] [CrossRef]
- Raj, S.; Sasidharan, S.; Balaji, S.N.; Dubey, V.K.; Saudagar, P. Review on natural products as an alternative to contemporary anti-leishmanial therapeutics. Journal of Proteins and Proteomics 11, 1768–158. [CrossRef]
- de Salud y Protección Social de Colombia, M. Boletín Epidemiológico Semanal, 2022.
- Mann, S.; Frasca, K.; Scherrer, S.; Henao-Martínez, A.F.; Newman, S.; Ramanan, P.; Suarez, J.A. A Review of Leishmaniasis: Current Knowledge and Future Directions. Current Tropical Medicine Reports, 8, 121–132. [CrossRef]
- Catta-Preta, C.M.C.; Mottram, J.C. Drug candidate and target for leishmaniasis. Nature 2018, 560, 171–172. [Google Scholar] [CrossRef]
- Chappuis, F.; Sundar, S.; Hailu, A.; Ghalib, H.; Rijal, S.; Peeling, R.W.; Alvar, J.; Boelaert, M. Visceral leishmaniasis: What are the needs for diagnosis, treatment and control? Nature Reviews Microbiology 2007, 5, 873–882. [Google Scholar] [CrossRef]
- Rittig, M.G.; Bogdan, C. Leishmania-host-cell interaction: Complexities and alternative views. Parasitology Today 2000, 16, 292–297. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, C.; Estaquier, J.; Silvestre, R. Immune-metabolic interactions between Leishmania and macrophage host. Current Opinion in Microbiology 2021, 63, 231–237. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, A.C. Parasitic Adaptive Mechanisms in Infection by Leishmania. Experimental and Molecular Pathology 2002, 72, 132–141. [Google Scholar] [CrossRef] [PubMed]
- Nathan, C. Neutrophils and immunity: challenges and opportunities. Nature Reviews Immunology 2006, 6, 173–182. [Google Scholar] [CrossRef] [PubMed]
- Torres-Vélez, C. ; María Cynthia Fuentes-Lacouture.; Yurany Duarte-Torres.; Myriam Beatriz AmayaBernal.; Jair Figueroa-Emiliani. Visceral Leishmaniasis in A Patient with Multiple Myeloma: Casuality or Causality? Journal of Medical Case Reports and Case Series, 3. [CrossRef]
- Charmoy, M.; Auderset, F.; Allenbach, C.; Tacchini-Cottier, F. The Prominent Role of Neutrophils during the Initial Phase of Infection by Leishmania Parasites. Journal of Biomedicine and Biotechnology 2010, 2010, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Laskay, T.; Zandbergen, G.v.; Solbach, W. Neutrophil granulocytes as host cells and transport vehicles for intracellular pathogens : Apoptosis as infection-promoting factor. Immunology 2008, 213, 183–191. [Google Scholar] [CrossRef]
- Naderer, T.; McConville, M.J. The Leishmania-macrophage interaction: a metabolic perspective. Cellular Microbiology 2008, 10, 301–308. [Google Scholar] [CrossRef]
- Naderer, T.; Vince, J.; McConville, M. Surface Determinants of Leishmania Parasites and their Role in Infectivity in the Mammalian Host. Current Molecular Medicine 2004, 4, 649–665. [Google Scholar] [CrossRef]
- de León Fraga, J. (Ed.) INMUNOLOGÍA by Thomas J. Kindt, Richard A. Goldsby, and Barbara A. Osborne, 6 ed.; McGraw-Hill, Interamericana, 2007.
- Parham, P. The Immune System, Fourth Edition, 4 ed.; Taylor and Francis Group, 2014; p. 624.
- Kima, P.E. Leishmania molecules that mediate intracellular pathogenesis. Microbes and Infection 2014, 16, 721–726. [Google Scholar] [CrossRef]
- McConville, M.J.; Naderer, T. Metabolic Pathways Required for the Intracellular Survival of Leishmania. Annual Review of Microbiology 2011, 65, 543–561. [Google Scholar] [CrossRef]
- Kanehisa, M. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Research 2006, 34, D354–D357. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M. The Protein Data Bank. Nucleic Acids Research 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Cornish-Bowden, A. Current IUBMB recommendations on enzyme nomenclature and kinetics. Perspectives in Science 2014, 1, 74–87. [Google Scholar] [CrossRef]
- De Rycker, M.; Baragaña, B.; Duce, S.L.; Gilbert, I.H. Challenges and recent progress in drug discovery for tropical diseases. Nature, 559, 498–506. [CrossRef]
- Rodriguez-Contreras, D.; Hamilton, N. Gluconeogenesis in Leishmania mexicana. Journal of Biological Chemistry 2014, 289, 32989–33000. [Google Scholar] [CrossRef] [PubMed]
- Seidler, N.W. GAPDH: Biological Properties and Diversity; Vol. 985, Advances in Experimental Medicine and Biology, Springer Netherlands: Dordrecht, 2013; pp. 1–291. [Google Scholar] [CrossRef]
- Kim, H.; Feil, I.K.; Verlinde, C.L.M.J.; Petra, P.H.; Hol, W.G.J. Crystal Structure of Glycosomal Glyceraldehyde-3-phosphate Dehydrogenase from Leishmania mexicana: Implications for Structure-Based Drug Design and a New Position for the Inorganic Phosphate Binding Site. Biochemistry, 34, 14975–14986. [CrossRef]
- Pelley, J.W. Gluconeogenesis and Glycogen Metabolism. In Elsevier’s Integrated Biochemistry; Elsevier, 2007; pp. 65–71. [CrossRef]
- Minárik, P.; Tomásková, N.; Kollárová, M.; Antalík, M. Malate dehydrogenases–structure and function. General physiology and biophysics 2002, 21, 257–65. [Google Scholar]
- Gourley, D.G.; Schüttelkopf, A.W.; Leonard, G.A.; Luba, J.; Hardy, L.W.; Beverley, S.M.; Hunter, W.N. Pteridine reductase mechanism correlates pterin metabolism with drug resistance in trypanosomatid parasites. Nature Publishing Group 2001, 8, 521–525. [Google Scholar] [CrossRef]
- Vadloori, B.; Sharath, A.K.; Prabhu, N.P.; Maurya, R. Homology modelling, molecular docking, and molecular dynamics simulations reveal the inhibition of Leishmania donovani dihydrofolate reductase-thymidylate synthase enzyme by Withaferin-A. BMC Research Notes 2018, 11, 1–7. [Google Scholar] [CrossRef]
- Chang, C.f.; Papadopoulou, B.; Wang, J.; Bray, T.; Whiteley, J.M.; Lin, S.x.; Ouellette, M.; Al, W.E.T. Pterin and Folate Reduction by the Leishmania tarentolae H Locus Short-Chain Dehydrogenase / Reductase PTR1. Archives of Biochemistry and Biophysics 1997, 342, 197–202. [Google Scholar]
- Feliciano, P.R.; Cordeiro, A.T.; Costa-Filho, A.J.; Nonato, M.C. Cloning, expression, purification, and characterization of Leishmania major dihydroorotate dehydrogenase. Protein Expression and Purification 2006, 48, 98–103. [Google Scholar] [CrossRef]
- Cordeiro, A.T.; Feliciano, P.R.; Pinheiro, M.P.; Nonato, M.C. Crystal structure of dihydroorotate dehydrogenase from Leishmania major. Biochimie 2012, 94, 1739–1748. [Google Scholar] [CrossRef]
- Gommers-Ampt, J.H.; Van Leeuwen, F.; de Beer, A.L.; Vliegenthart, J.F.; Dizdaroglu, M.; Kowalak, J.A.; Crain, P.F.; Borst, P. β-d-glucosyl-hydroxymethyluracil: A novel modified base present in the DNA of the parasitic protozoan T. brucei. Cell 1993, 75, 1129–1136. [Google Scholar] [CrossRef] [PubMed]
- van Leeuwen, F.; Taylor, M.C.; Mondragon, A.; Moreau, H.; Gibson, W.; Kieft, R.; Borst, P. -D-Glucosyl-hydroxymethyluracil is a conserved DNA modification in kinetoplastid protozoans and is abundant in their telomeres. Proceedings of the National Academy of Sciences 2002, 95, 2366–2371. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Genest, P.A.; ter Riet, B.; Sweeney, K.; DiPaolo, C.; Kieft, R.; Christodoulou, E.; Perrakis, A.; Simmons, J.M.; Hausinger, R.P.; van Luenen, H.G.; Rigden, D.J.; Sabatini, R.; Borst, P. The protein that binds to DNA base J in trypanosomatids has features of a thymidine hydroxylase. Nucleic Acids Research 2007, 35, 2107–2115. [Google Scholar] [CrossRef] [PubMed]
- Heidebrecht, T.; Christodoulou, E.; Chalmers, M.J.; Jan, S.; Ter Riet, B.; Grover, R.K.; Joosten, R.P.; Littler, D.; Van Luenen, H.; Griffin, P.R.; Wentworth, P.; Borst, P.; Perrakis, A. The structural basis for recognition of base J containing DNA by a novel DNA binding domain in JBP1, 2011. [CrossRef]
- Kryshtafovych, A.; Moult, J.; Bartual, S.G.; Bazan, J.F.; Berman, H.; Casteel, D.E.; Christodoulou, E.; Everett, J.K.; Hausmann, J.; Heidebrecht, T.; Hills, T.; Hui, R.; Hunt, J.F.; Seetharaman, J.; Joachimiak, A.; Kennedy, M.A.; Kim, C.; Lingel, A.; Michalska, K.; Montelione, G.T.; Otero, J.M.; Perrakis, A.; Pizarro, J.C.; van Raaij, M.J.; Ramelot, T.A.; Rousseau, F.; Tong, L.; Wernimont, A.K.; Young, J.; Schwede, T. Target highlights in CASP9: Experimental target structures for the critical assessment of techniques for protein structure prediction. Proteins: Structure, Function, and Bioinformatics 2011, 79, 6–20. [Google Scholar] [CrossRef]
- Barr, S.D.; Gedamu, L. Role of Peroxidoxins in Leishmania chagasi Survival. Journal of Biological Chemistry 2003, 278, 10816–10823. [Google Scholar] [CrossRef]
- Murray, H.W.; Nathan, C.F. Macrophage microbicidal mechanisms in vivo: reactive nitrogen versus oxygen intermediates in the killing of intracellular visceral Leishmania donovani. The Journal of experimental medicine 1999, 189, 741–6. [Google Scholar] [CrossRef]
- Nathan, C.; Shiloh, M.U. Reactive oxygen and nitrogen intermediates in the relationship between mammalian hosts and microbial pathogens. Proceedings of the National Academy of Sciences of the United States of America 2000, 97, 8841–8848. [Google Scholar] [CrossRef]
- Mutlu, O. In silico molecular modeling and docking studies on the leishmanial tryparedoxin peroxidase. Brazilian Archives of Biology and Technology 2014, 57, 244–252. [Google Scholar] [CrossRef]
- Fiorillo, A.; Colotti, G.; Boffi, A.; Baiocco, P.; Ilari, A. The Crystal Structures of the Tryparedoxin-Tryparedoxin Peroxidase Couple Unveil the Structural Determinants of Leishmania Detoxification Pathway. PLoS Neglected Tropical Diseases 2012, 6, e1781. [Google Scholar] [CrossRef]
- Bose, M.; Saha, R.; Sen Santara, S.; Mukherjee, S.; Roy, J.; Adak, S. Protection against peroxynitrite by pseudoperoxidase from Leishmania major. Free Radical Biology and Medicine 2012, 53, 1819–1828. [Google Scholar] [CrossRef]
- Chreifi, G.; Dejam, D.; Poulos, T.L. Crystal structure and functional analysis of Leishmania major pseudoperoxidase. Journal of Biological Inorganic Chemistry 2017. [Google Scholar] [CrossRef]
- Phan, I.Q.H.; Davies, D.R.; Moretti, N.S.; Shanmugam, D.; Cestari, I.; Anupama, A.; Fairman, J.W.; Edwards, T.E.; Stuart, K.; Schenkman, S.; Myler, P.J. Iron superoxide dismutases in eukaryotic pathogens: new insights from Apicomplexa and Trypanosoma structures. Acta Crystallographica Section F Structural Biology Communications 2015, 71, 615–621. [Google Scholar] [CrossRef]
- GHOSH, S.; GOSWAMI, S.; ADHYA, S. Role of superoxide dismutase in survival of Leishmania within the macrophage. Biochemical Journal 2003, 369, 447–452. [Google Scholar] [CrossRef] [PubMed]
- Slauch, J.M. How does the oxidative burst of macrophages kill bacteria? Still an open question. Molecular Microbiology 2011, 80, 580–583. [Google Scholar] [CrossRef] [PubMed]
- Ilari, A.; Baiocco, P.; Messori, L.; Fiorillo, A.; Boffi, A.; Gramiccia, M.; Di Muccio, T.; Colotti, G. A gold-containing drug against parasitic polyamine metabolism: the X-ray structure of trypanothione reductase from Leishmania infantum in complex with auranofin reveals a dual mechanism of enzyme inhibition. Amino Acids 2012, 42, 803–811. [Google Scholar] [CrossRef]
- Fairlamb, A.H.; Cerami, A. Metabolism and Functions of Trypanothione in the Kinetoplastida. Annual Review of Microbiology 1992, 46, 695–729. [Google Scholar] [CrossRef] [PubMed]
- Bernardes, L.; Zani, C.; Carvalho, I. Trypanosomatidae Diseases: From the Current Therapy to the Efficacious Role of Trypanothione Reductase in Drug Discovery. Current Medicinal Chemistry 2013, 20, 2673–2696. [Google Scholar] [CrossRef] [PubMed]
- Kabututu, Z.; Martin, S.K.; Nozaki, T.; Kawazu, S.i.; Okada, T.; Munday, C.J.; Duszenko, M.; Lazarus, M.; Thuita, L.W.; Urade, Y.; Kubata, B.K. Prostaglandin production from arachidonic acid and evidence for a 9,11-endoperoxide prostaglandin H2 reductase in Leishmania. International Journal for Parasitology 2002, 32, 1693–1700. [Google Scholar] [CrossRef]
- Moen, S.O.; Fairman, J.W.; Barnes, S.R.; Sullivan, A.; Nakazawa-Hewitt, S.; Van Voorhis, W.C.; Staker, B.L.; Lorimer, D.D.; Myler, P.J.; Edwards, T.E. Structures of prostaglandin F synthase from the protozoa Leishmania major and Trypanosoma cruzi with NADP. Acta Crystallographica Section F Structural Biology Communications 2015, 71, 609–614. [Google Scholar] [CrossRef]
- Hargrove, T.Y.; Wawrzak, Z.; Liu, J.; Nes, W.D.; Waterman, M.R.; Lepesheva, G.I. Substrate preferences and catalytic parameters determined by structural characteristics of sterol 14$α$-demethylase (CYP51) from Leishmania infantum. Journal of Biological Chemistry 2011. [Google Scholar] [CrossRef]
- Manhas, R.; Tripathi, P.; Khan, S.; Sethu Lakshmi, B.; Lal, S.K.; Gowri, V.S.; Sharma, A.; Madhubala, R. Identification and Functional Characterization of a Novel Bacterial Type Asparagine Synthetase A. Journal of Biological Chemistry 2014, 289, 12096–12108. [Google Scholar] [CrossRef]
- Führing, J.; Cramer, J.T.; Routier, F.H.; Lamerz, A.C.; Baruch, P.; Gerardy-Schahn, R.; Fedorov, R. Catalytic Mechanism and Allosteric Regulation of UDP-Glucose Pyrophosphorylase from Leishmania major. ACS Catalysis 2013, 3, 2976–2985. [Google Scholar] [CrossRef]
- Martin, J.L.; Yates, P.A.; Boitz, J.M.; Koop, D.R.; Fulwiler, A.L.; Cassera, M.B.; Ullman, B.; Carter, N.S. A role for adenine nucleotides in the sensing mechanism to purine starvation in Leishmania donovani. Molecular Microbiology 2016, 101, 299–313. [Google Scholar] [CrossRef]
- Boitz, J.M.; Strasser, R.; Yates, P.A.; Jardim, A.; Ullman, B. Adenylosuccinate synthetase and adenylosuccinate lyase deficiencies trigger growth and infectivity deficits in Leishmania donovani. Journal of Biological Chemistry 2013, 288, 8977–8990. [Google Scholar] [CrossRef] [PubMed]
- Silva, M.; Silva, C.H.; Iulek, J.; Oliva, G.; Thiemann, O.H. Crystal structure of adenine phosphoribosyltransferase from Leishmania tarentolae: Potential implications for APRT catalytic mechanism. Biochimica et Biophysica Acta - Proteins and Proteomics, 2004. [Google Scholar] [CrossRef]
- Phillips, C.L.; Ullman, B.; Brennan, R.G.; Hill, C.P. Crystal structures of adenine phosphoribosyltransferase from Leishmania donovani. EMBO Journal 1999, 18, 3533–3545. [Google Scholar] [CrossRef] [PubMed]
- Resh, M.D. Fatty acylation of proteins: new insights into membrane targeting of myristoylated and palmitoylated proteins. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research 1999, 1451, 1–16. [Google Scholar] [CrossRef]
- Robinson, D.A.; Wyatt, P.G. Identification and structure solution of fragment hits against kinetoplastid N -myristoyltransferase. Acta Crystallographica Section F Structural Biology Communications 2015, 71, 586–593. [Google Scholar] [CrossRef]
- Selvakumar, P.; Kumar, S.; Dimmock, J.R.; Sharma, R.K. NMT1 (N-myristoyltransferase 1). Atlas of genetics and cytogenetics in oncology and haematology, 15, 570–575. [CrossRef]
- Price, H.P.; Menon, M.R.; Panethymitaki, C.; Goulding, D.; McKean, P.G.; Smith, D.F. Myristoyl-CoA:Protein N -Myristoyltransferase, an Essential Enzyme and Potential Drug Target in Kinetoplastid Parasites. Journal of Biological Chemistry 2003, 278, 7206–7214. [Google Scholar] [CrossRef]
- French, J.B.; Yates, P.A.; Soysa, D.R.; Boitz, J.M.; Carter, N.S.; Chang, B.; Ullman, B.; Ealick, S.E. The Leishmania donovani UMP synthase is essential for promastigote viability and has an unusual tetrameric structure that exhibits substrate-controlled oligomerization. Journal of Biological Chemistry 2011. [Google Scholar] [CrossRef]
- Jones, M.E. Pyrimidine Nucleotide Biosynthesis in Animals: Genes, Enzymes, and Regulation of UMP Biosynthesis. Annual Review of Biochemistry 1980, 49, 253–279. [Google Scholar] [CrossRef] [PubMed]
- Williams, R.A.M.; Westrop, G.D.; Coombs, G.H. Two pathways for cysteine biosynthesis in Leishmania major. Biochemical Journal 2009, 420, 451–462. [Google Scholar] [CrossRef]
- Oza, S.L.; Shaw, M.P.; Wyllie, S.; Fairlamb, A.H. Trypanothione biosynthesis in Leishmania major. Molecular and Biochemical Parasitology 2005, 139, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Krauth-Siegel, R.L.; Comini, M.A. Redox control in trypanosomatids, parasitic protozoa with trypanothione-based thiol metabolism. Biochimica et Biophysica Acta (BBA) - General Subjects 2008, 1780, 1236–1248. [Google Scholar] [CrossRef] [PubMed]
- Fyfe, P.K.; Westrop, G.D.; Ramos, T.; Müller, S.; Coombs, G.H.; Hunter, W.N. Structure of Leishmania major cysteine synthase. Acta Crystallographica Section F Structural Biology and Crystallization Communications 2012, 68, 738–743. [Google Scholar] [CrossRef]
- McConville, M.J.; de Souza, D.; Saunders, E.; Likic, V.A.; Naderer, T. Living in a phagolysosome; metabolism of Leishmania amastigotes. Trends in Parasitology 2007, 23, 368–375. [Google Scholar] [CrossRef]
- Mejia, E.; Burak, M.; Alonso, A.; Larraga, V.; Kunkel, T.A.; Bebenek, K.; Garcia-Diaz, M. Structures of the Leishmania infantum polymerase beta. DNA Repair 2014, 18, 1–9. [Google Scholar] [CrossRef]
- Hammond, D.J.; Gutteridge, W.E. Purine and pyrimidine metabolism in the trypanosomatidae. Molecular and Biochemical Parasitology 1984, 13, 243–261. [Google Scholar] [CrossRef] [PubMed]
- Al-Madhoun, A. The Role of Thymidine Kinases in the Activation of Pyrimidine Nucleoside Analogues. Mini Reviews in Medicinal Chemistry 2004, 4, 341–350. [Google Scholar] [CrossRef]
- Timm, J.; Bosch-Navarrete, C.; Recio, E.; Nettleship, J.E.; Rada, H.; González-Pacanowska, D.; Wilson, K.S. Structural and Kinetic Characterization of Thymidine Kinase from Leishmania major. PLOS Neglected Tropical Diseases 2015, 9, e0003781. [Google Scholar] [CrossRef]
- Aripirala, S.; Gonzalez-Pacanowska, D.; Oldfield, E.; Kaiser, M.; Amzel, L.M.; Gabelli, S.B. Structural and thermodynamic basis of the inhibition of Leishmania major farnesyl diphosphate synthase by nitrogen-containing bisphosphonates. Acta Crystallographica Section D: Biological Crystallography, 2014. [Google Scholar] [CrossRef]
- de Mattos Oliveira, L.; Araújo, J.S.C.; Bacelar Costa Junior, D.; Santana, I.B.; Duarte, A.A.; Leite, F.H.A.; Benevides, R.G.; Coelho dos Santos Junior, M. Pharmacophore modeling, docking and molecular dynamics to identify Leishmania major farnesyl pyrophosphate synthase inhibitors. Journal of Molecular Modeling 2018, 24, 314. [Google Scholar] [CrossRef]
- Schomburg, I.; Jeske, L.; Ulbrich, M.; Placzek, S.; Chang, A.; Schomburg, D. The BRENDA enzyme information system–From a database to an expert system. Journal of Biotechnology 2017, 261, 194–206. [Google Scholar] [CrossRef] [PubMed]
- Abendroth, J.; Choi, R.; Wall, A.; Clifton, M.C.; Lukacs, C.M.; Staker, B.L.; Van Voorhis, W.; Myler, P.; Lorimer, D.D.; Edwards, T.E. Structures of aspartate aminotransferases from Trypanosoma brucei, Leishmania major and Giardia lamblia. Acta Crystallographica Section F:Structural Biology Communications, 2015. [Google Scholar] [CrossRef]
- Alphey, M.S.; Williams, R.A.; Mottram, J.C.; Coombs, G.H.; Hunter, W.N. The crystal structure of leishmania major 3-mercaptopyruvate sulfurtransferase. A three-domain architecture with a serine protease-like triad at the active site. Journal of Biological Chemistry 2003, 278, 48219–48227. [Google Scholar] [CrossRef] [PubMed]
- Williams, R.A.M.; Kelly, S.M.; Mottram, J.C.; Coombs, G.H. 3-Mercaptopyruvate Sulfurtransferase of Leishmania Contains an Unusual C-terminal Extension and Is Involved in Thioredoxin and Antioxidant Metabolism. Journal of Biological Chemistry 2003, 278, 1480–1486. [Google Scholar] [CrossRef] [PubMed]
- VEITCH, N.J.; MAUGERI, D.A.; CAZZULO, J.J.; LINDQVIST, Y.; BARRETT, M.P. Transketolase from Leishmania mexicana has a dual subcellular localization. Biochemical Journal 2004, 382, 759–767. [Google Scholar] [CrossRef]
- Rigden, D.J.; Phillips, S.E.; Michels, P.A.; Fothergill-Gilmore, L.A. The structure of pyruvate kinase from Leishmania mexicana reveals details of the allosteric transition and unusual effector specificity 1 1Edited by I. A. Wilson. Journal of Molecular Biology 1999, 291, 615–635. [Google Scholar] [CrossRef]
- Shi, W.; Schramm, V.L.; Almo, S.C. Nucleoside Hydrolase from Leishmania major. The Journal of Biological Chemistry 1999, 274, 21114–21120. [Google Scholar] [CrossRef]
- Schlagenhauf, E.; Etges, R.; Metcalf, P. The crystal structure of the Leishmania major surface proteinase leishmanolysin (gp63). Structure 1998, 6, 1035–1046. [Google Scholar] [CrossRef]
- McKerrow, J.H.; Caffrey, C.; Kelly, B.; Loke, P.; Sajid, M. PROTEASES IN PARASITIC DISEASES. Annual Review of Pathology: Mechanisms of Disease 2006, 1, 497–536. [Google Scholar] [CrossRef]
- McLuskey, K.; Paterson, N.G.; Bland, N.D.; Isaacs, N.W.; Mottram, J.C. Crystal Structure of Leishmania major Oligopeptidase B Gives Insight into the Enzymatic Properties of a Trypanosomatid Virulence Factor. Journal of Biological Chemistry 2010, 285, 39249–39259. [Google Scholar] [CrossRef]
- Teixeira, P.C.; Velasquez, L.G.; Lepique, A.P.; de Rezende, E.; Bonatto, J.M.C.; Barcinski, M.A.; Cunha-Neto, E.; Stolf, B.S. Regulation of Leishmania (L.) amazonensis Protein Expression by Host T Cell Dependent Responses: Differential Expression of Oligopeptidase B, Tryparedoxin Peroxidase and HSP70 Isoforms in Amastigotes Isolated from BALB/c and BALB/c Nude Mice. PLOS Neglected Tropical Diseases 2015, 9, e0003411. [Google Scholar] [CrossRef]
- Timm, J.; Valente, M.; García-Caballero, D.; Wilson, K.S.; González-Pacanowska, D. Structural Characterization of Acidic M17 Leucine Aminopeptidases from the TriTryps and Evaluation of Their Role in Nutrient Starvation in Trypanosoma brucei. mSphere 2017, 2. [Google Scholar] [CrossRef]
- Leitherer, S.; Clos, J.; Liebler-Tenorio, E.M.; Schleicher, U.; Bogdan, C.; Soulat, D. Characterization of the protein tyrosine phosphatase LmPRL-1 secreted by Leishmania major via the exosome pathway. Infection and Immunity 2017. [Google Scholar] [CrossRef] [PubMed]
- D’Antonio, E.L.; Ullman, B.; Roberts, S.C.; Dixit, U.G.; Wilson, M.E.; Hai, Y.; Christianson, D.W. Crystal structure of arginase from Leishmania mexicana and implications for the inhibition of polyamine biosynthesis in parasitic infections. Archives of Biochemistry and Biophysics 2013, 535, 163–176. [Google Scholar] [CrossRef] [PubMed]
- Feliciano, P.R.; Drennan, C.L.; Nonato, M.C. Crystal structure of an Fe-S cluster-containing fumarate hydratase enzyme from Leishmania major reveals a unique protein fold. Proceedings of the National Academy of Sciences 2016, 113, 9804–9809. [Google Scholar] [CrossRef]
- Chudzik, D.M.; Michels, P.A.; De Walque, S.; Hol, W.G. Structures of type 2 peroxisomal targeting signals in two trypanosomatid aldolases. Journal of Molecular Biology 2000, 300, 697–707. [Google Scholar] [CrossRef]
- Williams, J.C.; Zeelen, J.P.; Neubauer, G.; Vriend, G.; Backmann, J.; Michels, P.a.; Lambeir, a.M.; Wierenga, R.K. Structural and mutagenesis studies of leishmania triosephosphate isomerase: a point mutation can convert a mesophilic enzyme into a superstable enzyme without losing catalytic power. Protein engineering 1999, 12, 243–250. [Google Scholar] [CrossRef] [PubMed]
- Lambeir, A.M.; Backmann, J.; Ruiz-Sanz, J.; Filimonov, V.; Nielsen, J.E.; Kursula, I.; Norledge, B.V.; Wierenga, R.K. The ionization of a buried glutamic acid is thermodynamically linked to the stability of Leishmania mexicana triose phosphate isomerase. European Journal of Biochemistry 2000, 267, 2516–2524. [Google Scholar] [CrossRef] [PubMed]
- Bateman, A.; Martin, M.J.; O’Donovan, C.; Magrane, M.; Alpi, E.; Antunes, R.; Bely, B.; Bingley, M.; Bonilla, C.; Britto, R.; Bursteinas, B.; Bye-AJee, H.; Cowley, A.; Da Silva, A.; De Giorgi, M.; Dogan, T.; Fazzini, F.; Castro, L.G.; Figueira, L.; Garmiri, P.; Georghiou, G.; Gonzalez, D.; Hatton-Ellis, E.; Li, W.; Liu, W.; Lopez, R.; Luo, J.; Lussi, Y.; MacDougall, A.; Nightingale, A.; Palka, B.; Pichler, K.; Poggioli, D.; Pundir, S.; Pureza, L.; Qi, G.; Rosanoff, S.; Saidi, R.; Sawford, T.; Shypitsyna, A.; Speretta, E.; Turner, E.; Tyagi, N.; Volynkin, V.; Wardell, T.; Warner, K.; Watkins, X.; Zaru, R.; Zellner, H.; Xenarios, I.; Bougueleret, L.; Bridge, A.; Poux, S.; Redaschi, N.; Aimo, L.; ArgoudPuy, G.; Auchincloss, A.; Axelsen, K.; Bansal, P.; Baratin, D.; Blatter, M.C.; Boeckmann, B.; Bolleman, J.; Boutet, E.; Breuza, L.; Casal-Casas, C.; De Castro, E.; Coudert, E.; Cuche, B.; Doche, M.; Dornevil, D.; Duvaud, S.; Estreicher, A.; Famiglietti, L.; Feuermann, M.; Gasteiger, E.; Gehant, S.; Gerritsen, V.; Gos, A.; Gruaz-Gumowski, N.; Hinz, U.; Hulo, C.; Jungo, F.; Keller, G.; Lara, V.; Lemercier, P.; Lieberherr, D.; Lombardot, T.; Martin, X.; Masson, P.; Morgat, A.; Neto, T.; Nouspikel, N.; Paesano, S.; Pedruzzi, I.; Pilbout, S.; Pozzato, M.; Pruess, M.; Rivoire, C.; Roechert, B.; Schneider, M.; Sigrist, C.; Sonesson, K.; Staehli, S.; Stutz, A.; Sundaram, S.; Tognolli, M.; Verbregue, L.; Veuthey, A.L.; Wu, C.H.; Arighi, C.N.; Arminski, L.; Chen, C.; Chen, Y.; Garavelli, J.S.; Huang, H.; Laiho, K.; McGarvey, P.; Natale, D.A.; Ross, K.; Vinayaka, C.R.; Wang, Q.; Wang, Y.; Yeh, L.S.; Zhang, J. UniProt: the universal protein knowledgebase. Nucleic Acids Research 2017, 45, D158–D169. [Google Scholar] [CrossRef]
- Larson, E.T.; Kim, J.E.; Zucker, F.H.; Kelley, A.; Mueller, N.; Napuli, A.J.; Verlinde, C.L.; Fan, E.; Buckner, F.S.; Van Voorhis, W.C.; Merritt, E.A.; Hol, W.G. Structure of Leishmania major methionyl-tRNA synthetase in complex with intermediate products methionyladenylate and pyrophosphate. Biochimie 2011, 93, 570–582. [Google Scholar] [CrossRef]
- Larson, E.T.; Kim, J.E.; Castaneda, L.J.; Napuli, A.J.; Zhang, Z.; Fan, E.; Zucker, F.H.; Verlinde, C.L.M.J.; Buckner, F.S.; Van Voorhis, W.C.; Hol, W.G.J.; Merritt, E.A. The double-length tyrosyl-tRNA synthetase from the eukaryote leishmania major forms an intrinsically asymmetric pseudo-dimer. Journal of Molecular Biology 2011. [Google Scholar] [CrossRef]
- Kamir, D.; Zierow, S.; Leng, L.; Cho, Y.; Diaz, Y.; Griffith, J.; McDonald, C.; Merk, M.; Mitchell, R.A.; Trent, J.; Chen, Y.; Kwong, Y.K.A.; Xiong, H.; Vermeire, J.; Cappello, M.; McMahon-Pratt, D.; Walker, J.; Bernhagen, J.; Lolis, E.; Bucala, R. A Leishmania Ortholog of Macrophage Migration Inhibitory Factor Modulates Host Macrophage Responses. The Journal of Immunology 2008, 180, 8250–8261. [Google Scholar] [CrossRef] [PubMed]
- Arya, R.; Sharma, B.; Dhembla, C.; Pal, R.K.; Patel, A.K.; Sundd, M.; Ghosh, B.; Makde, R.D.; Kundu, S. A conformational switch from a closed apo- to an open holo-form equips the acyl carrier protein for acyl chain accommodation. Biochimica et Biophysica Acta - Proteins and Proteomics 2019, 1867, 163–174. [Google Scholar] [CrossRef] [PubMed]
- Chawla, B.; Madhubala, R. Drug targets in Leishmania. Journal of Parasitic Diseases 2010, 34, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Soto, J.; Soto, P. Miltefosina oral para el tratamiento de la leishmaniasis. Biomédica 2012, 26, 207. [Google Scholar] [CrossRef]
- Ramón Azanza, J.; García-Quetglas, E.; Sádaba, B. Farmacología de los azoles. Rev Iberoam Micol 2007, 24, 223–227. [Google Scholar] [CrossRef] [PubMed]
- Tiuman, T.S.; Santos, A.O.; Ueda-Nakamura, T.; Filho, B.P.D.; Nakamura, C.V. Recent advances in leishmaniasis treatment. International Journal of Infectious Diseases 2011, 15. [Google Scholar] [CrossRef]
- Sen, R.; Chatterjee, M. Plant derived therapeutics for the treatment of Leishmaniasis. Phytomedicine 2011, 18, 1056–1069. [Google Scholar] [CrossRef]
- Gutiérrez-Rebolledo, G.A.; Drier-Jonas, S.; Jiménez-Arellanes, M.A. Natural compounds and extracts from Mexican medicinal plants with anti-leishmaniasis activity: An update. Asian Pacific Journal of Tropical Medicine 2017, 10, 1105–1110. [Google Scholar] [CrossRef]
- Zahedifard, F.; Rafati, S. Prospects for antimicrobial peptide-based immunotherapy approaches in <i>Leishmania</i> control. Expert Review of Anti-infective Therapy 2018, 16, 461–469. [Google Scholar] [CrossRef]
- Patiño-Márquez, I.A.; Manrique-Moreno, M.; Patiño-González, E.; Jemioła-Rzemińska, M.; Strzałka, K. Effect of antimicrobial peptides from Galleria mellonella on molecular models of Leishmania membrane. Thermotropic and fluorescence anisotropy study. The Journal of Antibiotics 2018, 71, 642–652. [Google Scholar] [CrossRef]
- Balboa, M.A.; Balsinde, J. Phospholipases: From Structure to Biological Function. Biomolecules, 11, 428. [CrossRef]
- Saddhe, A.A.; Potocký, M. Comparative phylogenomic and structural analysis of canonical secretory PLA2 and novel PLA2-like family in plants. Frontiers in Plant Science, 14.
- Tonello, F.; Rigoni, M. Cellular Mechanisms of Action of Snake Phospholipase A2 Toxins. In Snake Venoms; Inagaki, H.; Vogel, C.W.; Mukherjee, A.K.; Rahmy, T.R.; Gopalakrishnakone, P., Eds.; Toxinology, Springer Netherlands, 2017; pp. 49–65. [CrossRef]
- Jaiswal, A.; Rehman, R.; Dutta, J.; Singh, S.; Ray, A.; Shridhar, M.; Jaisankar, J.; Bhatt, M.; Khandelwal, D.; Sahoo, B.; Ram, A.; Mabalirajan, U. Cellular Distribution of Secreted Phospholipase A2 in Lungs of IPF Patients and Its Inhibition in Bleomycin-Induced Pulmonary Fibrosis in Mice. Cells, 12, 1044. [CrossRef]
- Castro-Amorim, J.; Novo de Oliveira, A.; Da Silva, S.L.; Soares, A.M.; Mukherjee, A.K.; Ramos, M.J.; Fernandes, P.A. Catalytically Active Snake Venom PLA2 Enzymes: An Overview of Its Elusive Mechanisms of Reaction. Journal of Medicinal Chemistry, 66, 5364–5376. [CrossRef]
- Moreira, V.; Leiguez, E.; Janovits, P.M.; Maia-Marques, R.; Fernandes, C.M.; Teixeira, C. Inflammatory Effects of Bothrops Phospholipases A2: Mechanisms Involved in Biosynthesis of Lipid Mediators and Lipid Accumulation. Toxins, 13, 868. [CrossRef]
- Lee, K.S.; Kim, B.Y.; Park, M.J.; Deng, Y.; Kim, J.M.; Kim, Y.H.; Heo, E.J.; Yoon, H.J.; Lee, K.Y.; Choi, Y.S.; Jin, B.R. Bee Venom Induces Acute Inflammation through a H2O2-Mediated System That Utilizes Superoxide Dismutase. Toxins, 14, 558. [CrossRef]
- Ma, X.; Liu, H.; Liu, Z.; Wang, Y.; Zhong, Z.; Peng, G.; Gu, Y. Trichosporon asahii PLA2 Gene Enhances Drug Resistance to Azoles by Improving Drug Efflux and Biofilm Formation. International Journal of Molecular Sciences, 24, 8855. [CrossRef]
- Soltan-Alinejad, P.; Alipour, H.; Meharabani, D.; Azizi, K. Therapeutic Potential of Bee and Scorpion Venom Phospholipase A2 (PLA2): A Narrative Review. Iranian Journal of Medical Sciences, 47, 300–313. [CrossRef]
- Rojas-Pirela, M.; Kemmerling, U.; Quiñones, W.; Michels, P.A.M.; Rojas, V. Antimicrobial Peptides (AMPs): Potential Therapeutic Strategy against Trypanosomiases? Biomolecules, M: Number: 4 Publisher; 13, 599. [CrossRef]
- Borges, I.P.; Castanheira, L.E.; Barbosa, B.F.; de Souza, D.L.N.; da Silva, R.J.; Mineo, J.R.; Tudini, K.A.Y.; Rodrigues, R.S.; Ferro, E.A.V.; de Melo Rodrigues, V. Anti-parasitic effect on Toxoplasma gondii induced by BnSP-7, a Lys49-phospholipase A2 homologue from Bothrops pauloensis venom. Toxicon: Official Journal of the International Society on Toxinology, 119, 84–91. [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).




