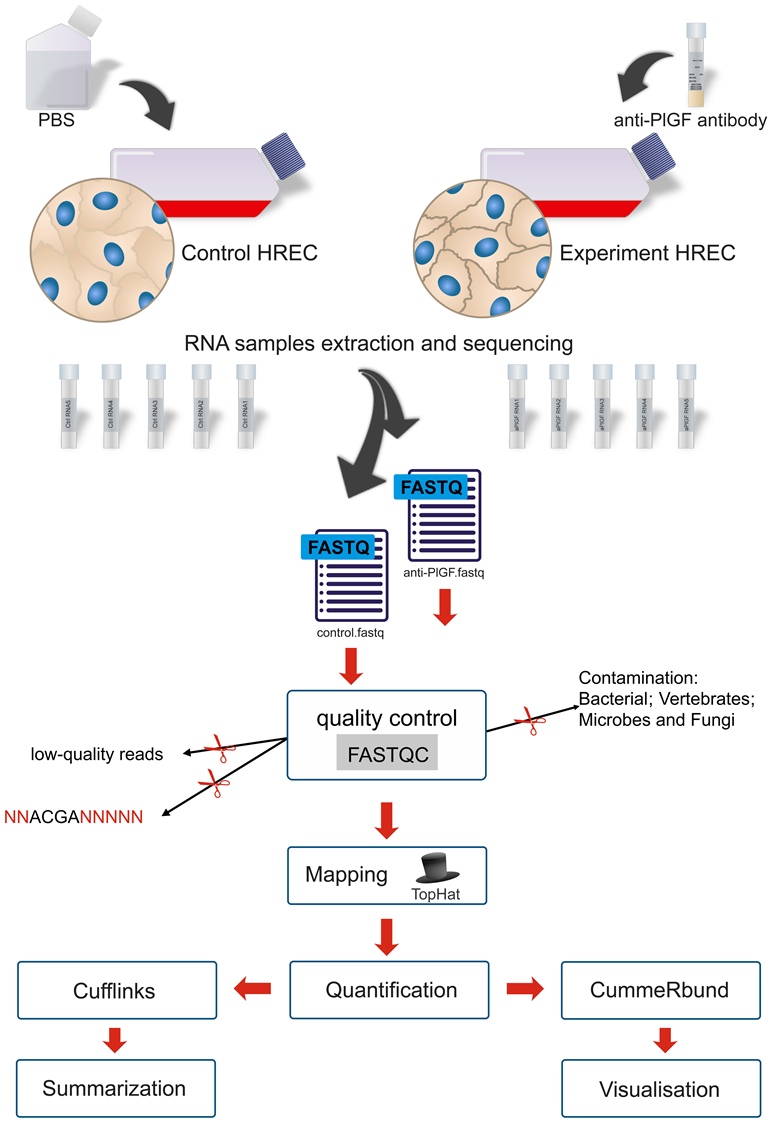
Placental growth factor (PlGF or PGF) is a member of the VEGF family, which is known to play a critical role in pathological angiogenesis, inflammation, and endothelial cell barrier function. However, the molecular mechanisms by which PlGF mediates its effects in non-proliferative diabetic retinopathy (DR) remain elusive. In this study, we performed transcriptome-wide profiling of differential gene expression for human retinal endothelial cells (HRECs) treated with PlGF antibody. The effect of antibody treatment on the samples was validated using trans-endothelial electric resistance (TEER), and western blot. A total of 3760 genes (1750 upregulated and 2010 downregulated) were found to be differentially expressed between the control and PlGF antibody treatment group. These differentially expressed genes (DEGs) were used for gene ontology and enrichment analysis to identify gene function, signal pathway, and interaction networks. The gene ontology results revealed that catalytic activity (GO:0003824) of molecular function, cell (GO:0005623) of the cellular component, and cellular process (GO:0009987) were among the most enriched biological processes. Pathways such as TGF-β, VEGF-VEGFR2, p53, apoptosis, pentose phosphate pathway, and ubiquitin-proteasome pathway, were among the most enriched, and TGF-β1 was identified as a primary upstream regulator. These data provide new insights into the underlying molecular mechanisms of PlGF in mediating biological functions, in relation to DR.