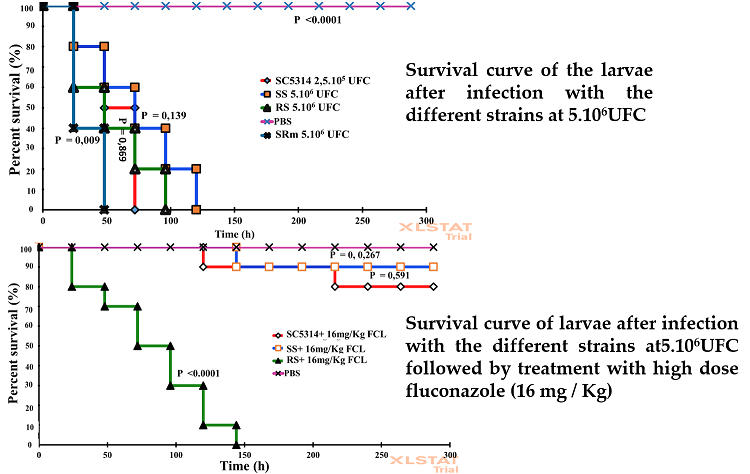
The aim of this study is to have an idea on the molecular mechanisms of C. albicans resistance to fluconazole in Burkina Faso, by studying the polymorphism of the ERG11 gene, and its implication in the C. albicans virulence and resistance in vivo according to the Galleria mellonella model; (2) Methods: Ten (10) clinical strains including, 5 resistant and 5 susceptible and 1 virulent and susceptible reference strain SC5314 are used. For the estimation of virulence, the larvae were inoculated with 10 μL of C. albicans cell suspension at variable concentrations: 2,5.105, 5.105, 1.106, and 5.106 CFU/larva of each strain. For the in vivo efficacy study, fluconazole was administered at 1, 4 and 16 mg/kg respectively to G. mellonella larvae, after infection by inoculum 5.106 CFU / larvae of each strain; (3) Results: Six (6) non-silent mutations in the ERG11 gene (K143R, F145L, G307S, S405F, G448E, V456I on ERG11p) were found in 4 resistant isolates. Larval mortality depended on fungal burden and strain. The inoculum 5.106 CFU caused 100% mortality in 2 days for the 2 CAAL-1 and CAAL-2 strains carrying the F145L mutation, in 3 days for the reference strain SC5314, in 4 days for the ensemble of resistant strains, and in 5 days for the ensemble of susceptible strains. The comparison of the mortality due to the reference strain SC5314 CFU / larva and the average mortality due to the two mutant F145L strains, shows a significant difference (P <0.05).Fluconazole significantly protected (P> 0.05) the larvae from infection by susceptible strains and the reference strain. However, 100% mortality in 6 days after injection of the resistant strains, was observed (4) Conclusions: Certain mutations in the ERG11 gene such as the F145L mutation are thought to be a source of increased virulence in Candida albicans. Fluconazole effectively protected larvae from infection by susceptible strains in vivo, unlike resistant strain