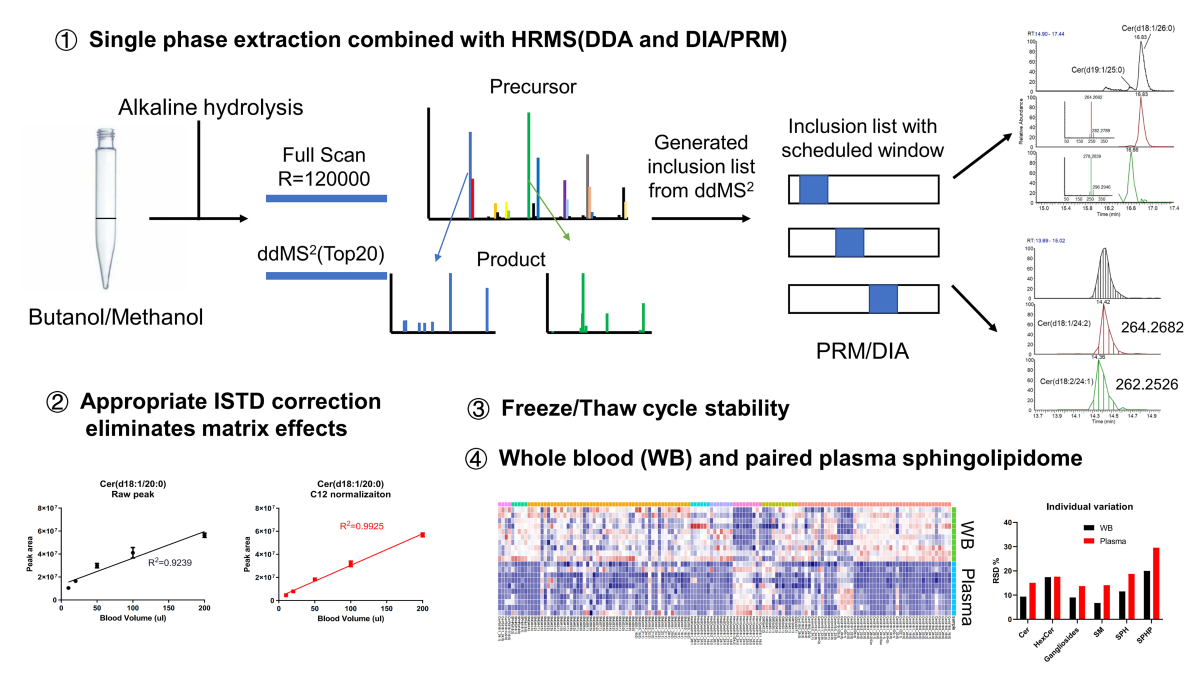
Plasma and serum are the most widely used blood-derived biofluids for metabolomics and lipidomics assays, but the isolation of these products from blood may introduce additional bias as indicated by the fact that many analytes that are present at high concentrations in blood cells cannot be measured and evaluated in those samples. Of particular concern, variable hemolysis during the pre-processing of blood products could compromise accurate and reproducible quantification. Compared with plasma or serum, whole blood may be a better alternative due to simplicity of processing. In this study, we provide a comprehensive method for quantification of the whole blood sphingolipidome and the concentrations were compared with those from plasma. Combining a single-phase extraction method with liquid-chromatography high resolution mass spectrometry (R=120, 000), assisted by alkaline hydrolysis, we were able to identify and simultaneously quantify more than 150 sphingolipids. Furthermore, most of sphingolipids remained stable after a freeze/thaw cycle. Whole blood contained a higher concentration of most sphingolipids than corresponding plasma. Moreover, individual variations in the levels of sphingolipids were lower for whole blood than plasma. These findings demonstrate that whole blood could be a better alternative to plasma, and potentially guide the evaluation of sphinglipidome for biomarker discovery.