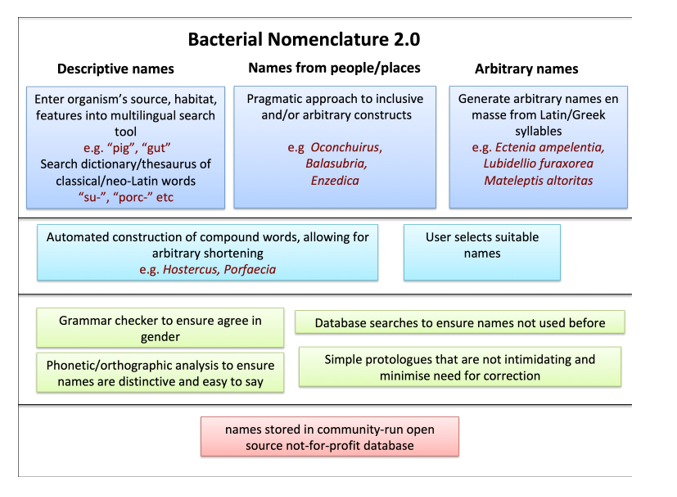
The remarkable success of taxonomic discovery, powered by culturomics, genomics and metagenomics, creates a pressing need for new bacterial names, while holding a mirror up to the slow pace of change in bacterial nomenclature. Here, I take a fresh look at bacterial nomenclature, exploring how we might create a system fit for the age of genomics, playing to the strengths of current practice, while minimising difficulties. Adoption of linguistic pragmatism, obeying the rules while treating recommendations as merely optional will make it easier to create names derived from descriptions, from people or places or even arbitrarily. Simpler protologues and a relaxed approach to recommendations will also remove much of the need for expert linguistic quality control. Automated computer-based approaches will allow names to be created en masse before they are needed, while also relieving microbiologists of the need for competence in Latin. The result will be a system that is accessible, inclusive and digital, while also fully capable of naming the unnamed millions of bacteria.