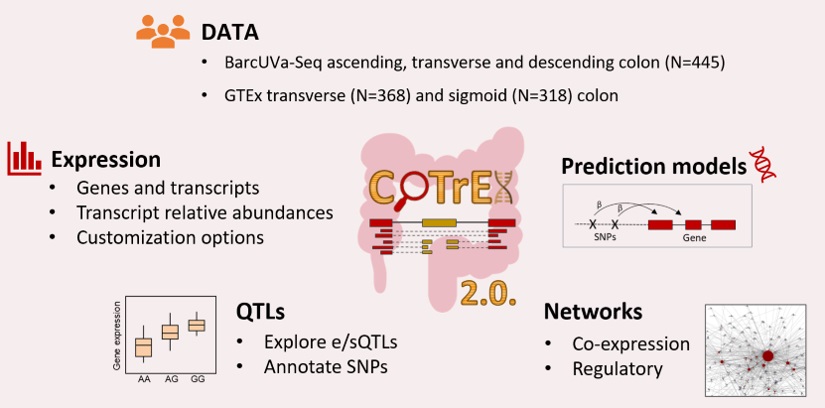
Gene expression data is key for the functional annotation of single nucleotide polymorphisms (SNPs) identified in genome-wide association studies (GWAS). Expression and splicing quantitative trait loci (e/sQTLs) in normal colon tissue, such as those from the University of Barcelona and University of Virginia RNA sequencing project (BarcUVa-Seq) and the Genotype-Tissue Expression project (GTEx), are required to gain biological insight of colon-related diseases risk loci. Moreover, transcriptome-wide association studies (TWAS) rely on reference gene expression imputation panels in the tissue of interest to nominate susceptibility genes. Also, it is of high interest to study the relationships between genes in a network framework. For facilitating these analyses, we have updated and expanded the scope of the Colon Transcriptome Explorer (CoTrEx) to the version 2.0. This web-based resource provides exhaustive visualization and analysis of transcriptome-wide gene expression profiles of normal colon tissue from BarcUVa-Seq and GTEx. In addition to the integration of new datasets, CoTrEx 2.0 provides additional e/sQTLs sets, as well as gene expression prediction models and regulatory and co-expression networks. It is freely available at https://barcuvaseq.org/cotrex/. Overall, it is of high interest for researchers aiming to investigate the genetic susceptibility to colon-related complex traits and diseases.