Author summary
Pandemic preparedness requires the development of broad-spectrum antivirals against future and present emerging viruses. In this work, we use publicly available single-cell RNA sequencing data from virally infected tissues to build computational models of how viruses exploit cellular metabolism for their replication and spread. Using these models, we predicted cellular metabolic pathways and enzyme targets essential for viral replication but dispensable for cell survival. To verify our top targets, we selected four specific inhibitors and validated them experimentally as proof-of-concept. Of note, two of our tested compounds, namely phenformin and atpenin A5, showed broad antiviral activity against several viruses with pandemic potential with only negligible cellular toxicity. In vivo, we could confirm the antiviral activity of phenformin against SARS-CoV-2. Hence, our pipeline enormously facilitates and speeds up the identification of drug targets and compounds by a rational-based in silico modeling approach. Furthermore, our study establishes cellular metabolism as druggable for antiviral therapy and provides specific compounds with the potential for further development as broad-spectrum antivirals.
Introduction
From December 2019 on, the outbreak of the novel Severe Acute Respiratory Syndrome coronavirus 2 (SARS-CoV-2) caused a pandemic with dramatic health-related and socioeconomic consequences. This novel virus was highly similar to SARS-CoV, responsible for a global outbreak in 2002 and 2003 (Marty and Jones, 2020). In 2012, another coronavirus spread in the Middle East, causing the Middle East Respiratory Syndrome (MERS), leading to 2,458 reported cases and a high mortality rate of 35% (Azhar et al., 2019). History shows that pandemics have repeatedly plagued humankind. In the last 100 years alone, there have been four major influenza pandemics and multiple epidemics, including the Spanish flu in 1918, with an estimated 17 to 50 million deaths worldwide (Yang et al., 2014), as well as the Asian and Hong Kong flu in 1957/58 and 1968/69 with one to four million deaths worldwide (Honigsbaum, 2020). Pandemics are not only virus-driven: One of the most extensive pandemics was the Black Death from 1331 to 1353, caused by the bacterium Yersinia pestis and is estimated to have killed approximately half of Europe’s population (Arrizabalaga, 2006).
Thus, pandemics and epidemics are recurrent, and more are likely to follow in the future, in particular, due to human impact on the global environment (Marani et al., 2021; Mora et al., 2022). Moreover, the COVID-19 pandemic illustrates its substantial impact on long-term socioeconomic well-being due to the widespread and long-lasting consequences of efforts of pathogen containment (Cutler and Summers, 2020) and the potential occurrence of post-acute sequelae such as long COVID (Shutters, 2021). Therefore, rapidly developing effective treatment strategies and vaccines are vital to mitigate those consequences. However, despite the unprecedented acceleration in the development of treatment and vaccination approaches against SARS-CoV-2, it still took approximately ten months before vaccine approval to treat SARS-CoV-2 and another 6-8 months before production and distribution pipelines became functional for a widespread roll-out. Thus, treatment approaches that contain viral replication of not only a single but a broad array of viruses as first-line therapeutic approaches for novel emerging pathogens are a highly sought-after goal in preparation for future pandemics (Geraghty et al., 2021). Nowadays, pandemic preparedness summarizes efforts to provide such broadly acting antivirals.
Due to the essential dependence of viruses on the metabolic networks of their host for reproduction (Thaker et al., 2019), the utilization of in silico models of virally infected cells provides new avenues to identify druggable targets for antiviral therapy (Zitzmann and Kaderali, 2018). One such approach is represented by constraint-based modeling (Bordbar et al., 2014) with flux balance analysis in particular (FBA, (Orth et al., 2010)) which allows us to predict the metabolic behavior of biological systems. These methods build upon genome-scale metabolic networks that encompass the entire known repertoire of metabolic reactions taking place in an organism (Gu et al., 2019). In the context of viral replication, constraint-based modeling allows the simulation and prediction of the viral capacity to replicate within a host cell by considering nutrient availability, energy resources, and other cellular factors (Aller et al., 2018; Renz et al., 2020). Thus, genome-scale metabolic models of host cells extended to incorporate viral replication (Aller et al., 2018) can be employed to identify host enzymes essential for viral replication but dispensable for cellular viability. The viral biomass reaction is a key element in these networks that represents the sum of all the cellular components and resources consumed or created during viral replication (Aller et al., 2018). Through simulation of the viral biomass reaction, important factors influencing viral replication can be identified, such as essential enzymes or the availability of specific nutrients (Renz et al., 2020). Importantly, these methods also allow for the integration of OMICs data such as transcriptomics, proteomics and metabolomics to reconstruct metabolic models that more accurately reflect the metabolic state of cells in a given condition, so-called context-specific metabolic models (Richelle et al., 2019).
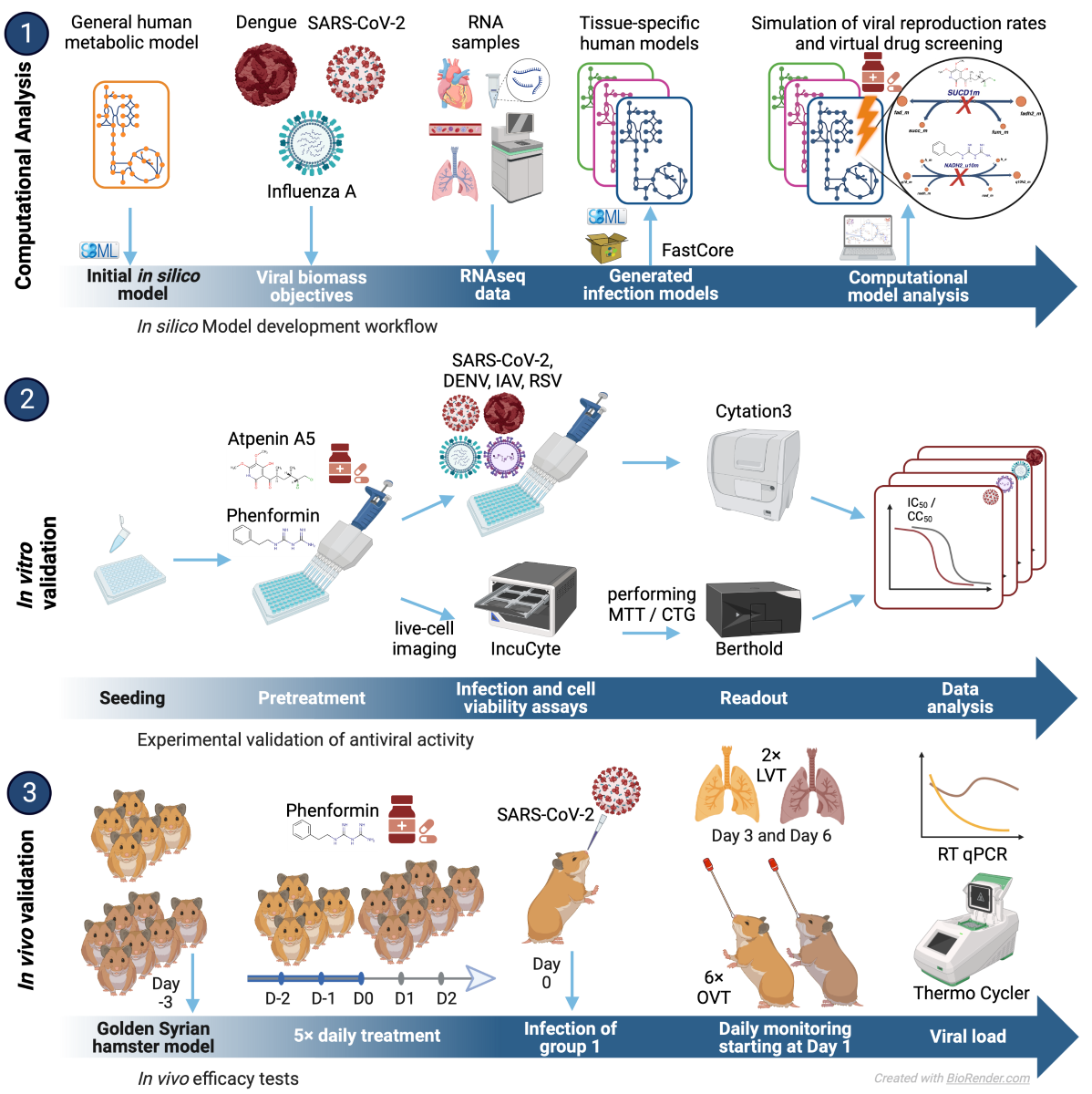
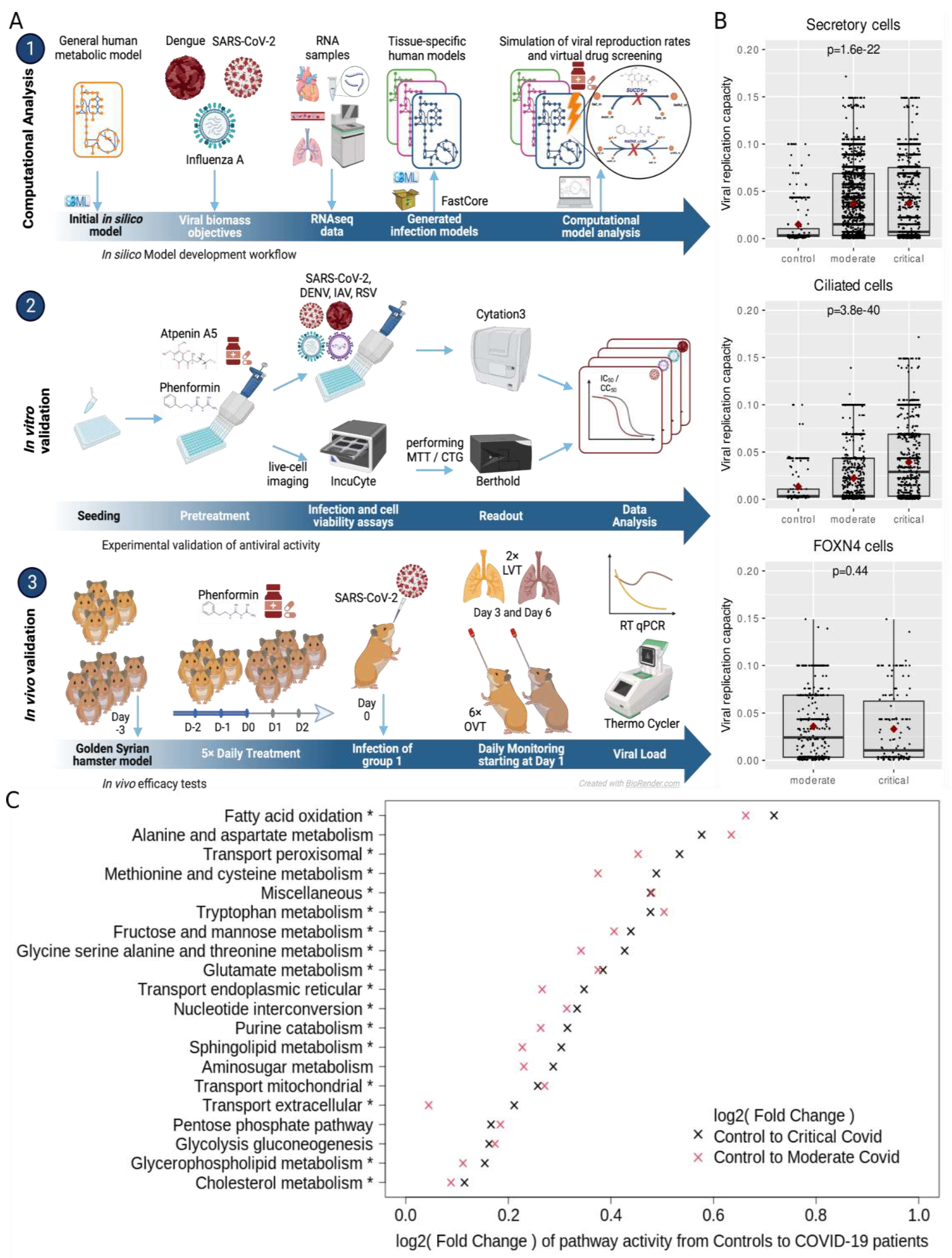
In this work, we introduce a computational pipeline that we developed to integrate a generic metabolic model of a virally infected cell with transcriptomic data to predict metabolic pathways relevant for viral replication (
Figure 1A). We demonstrate the establishment of this pipeline and how it is exploited to rapidly identify druggable targets and human approved compounds with antiviral efficacy
in vitro and
in vivo.
Results
Development of a computational pipeline to predict viral replication capacities and antiviral targets
We developed a computational pipeline to reconstruct the metabolic state of virally infected cells, leveraging both single-cell and bulk sequencing gene expression data (
Figure 1A & Methods). To this end, we expanded the generic human metabolic reconstruction Recon 2.2 (Swainston et al., 2016) by incorporating reactions specific to the substrates essential for the replication of the investigated viruses. Additionally, we considered all metabolites known to exist in the blood as potential inflow to the model (see methods). Subsequently, bulk sequencing or single-cell sequencing data, were preprocessed with StanDep (Joshi et al., 2020) to identify core sets of reactions active in each cell which then were used to identify context-specific metabolic networks that contain those reactions using fastcore (Vlassis et al., 2014). This process facilitated the generation of context-specific models for each cell or gene expression dataset. The resulting computational models of virally infected cells were then employed to predict the cellular capacity to produce virions via flux balance analysis (Orth et al., 2010) and to screen cells for enzymes whose knockout impedes viral replication (
Figure 1A). Moreover, these models enabled us to predict the effect of knockouts on cellular viability and thereby subsetting the target enzymes to those whose inhibition hinders viral replication but not normal cellular metabolism. These predicted targets were further integrated with additional experimental information on the relevance of enzymes for viral replication to subselect candidates for experimental testing.
Identification of drug targets for broad-spectrum antiviral therapy
Following our identification of tier-1 targets that inhibit the individual viruses, we sought to identify potential broad-spectrum antiviral targets. To this end, we collected all predicted tier-1 targets across all cells from the individual datasets and identified 254 enzymes that occurred as shared tier-1 targets across all datasets (
Supplementary Table S3 and Supplementary Data S1). Using the BioGRID database (Oughtred et al., 2019) as a reference, we find that interactions of 158 of these targets with human pathogenic viruses have been reported before, which represents a highly significant enrichment among reported interactions between enzymes and viruses (Fisher’s exact test
p-value = 9.1×10
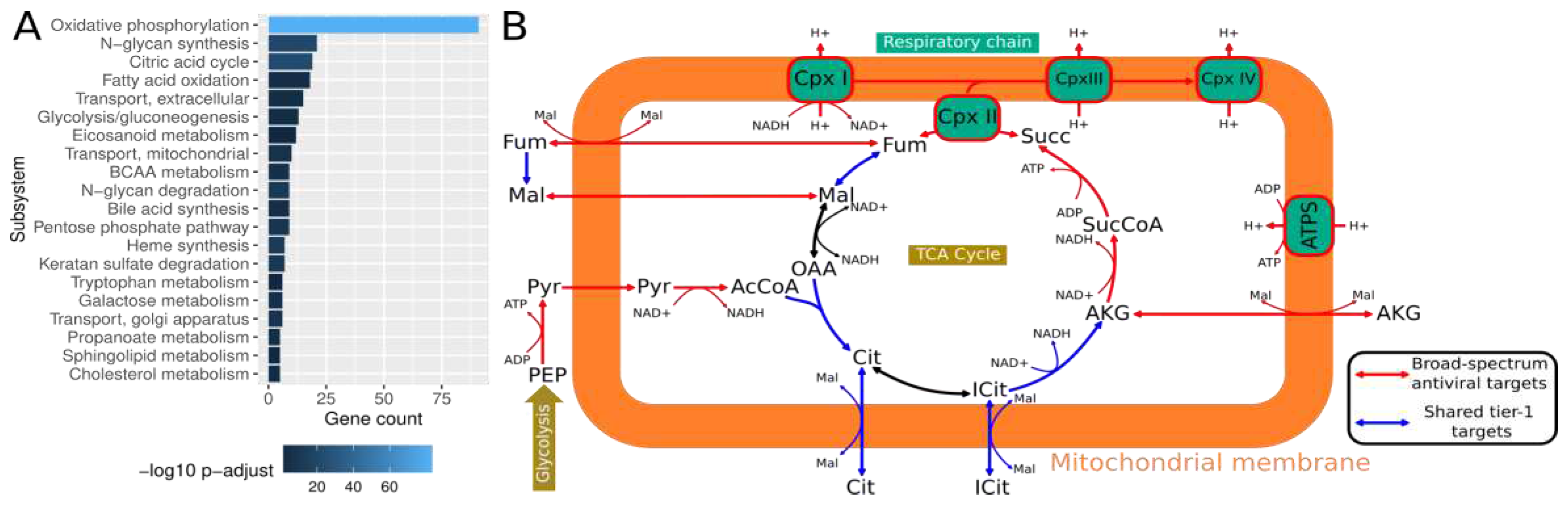
-12, odds ratio 1.9-3.4) and supports their central role in viral replication across diverse viruses. Performing an enrichment analysis, we identified several metabolic subsystems in which shared predicted tier-1 targets were particularly prevalent (
Figure 3A). We observed the most concentrated enrichment among oxidative phosphorylation enzymes, the citric acid cycle, and fatty acid oxidation. This aligns with the high energy need associated with viral replication (Mahmoudabadi et al., 2017) and the mitochondria’s central role in viral replication and antiviral immune responses (Moreno-Altamirano et al., 2019). Thus, viruses often target mitochondrial proteins to deregulate cellular metabolism, providing substrates for viral replication and hampering immune response (Cavallari et al., 2018). Indeed, almost all metabolic reactions in the TCA cycle and the respiratory chain are predicted as targets (
Figure 3B). Along those lines, mitochondrial transport reactions that provide substrates for the TCA cycle are enriched among the predicted targets. Other enriched subsystems are N-glycan synthesis as well as degradation and glycolysis/gluconeogenesis. While protein glycosylation is paramount for enveloped viruses (Y. Li et al., 2021), we did not include glycosylated proteins in the viral biomass reaction due to a lack of information on virus-specific protein glycosylation. Instead, tracing the corresponding metabolic pathways, we found that those pathways were used to recycle sugars contained in glycans such as mannose, fucose, and glucosamine for use as substrates for viral replication (
Supplementary Data S1). Similarly, glycolysis is often found to be induced during viral infection since it also provides crucial building blocks for viral replication (Sanchez and Lagunoff, 2015; Thaker et al., 2019).
To further stratify our hitlist to potential broad-spectrum antiviral targets, we required that each predicted target had an experimentally confirmed interaction with at least two of the three viruses analyzed. Thus, we obtained a list of 39 predicted targets belonging to 22 proteins/protein complexes (
Figure 3B,
Supplementary Table S5). Among those 22 protein complexes, we selected four which encompass 11 of the 39 enzyme targets for further experimental validation based upon additional criteria such as evidence from CRISPR-Cas9 screens (Daniloski et al., 2021; Wang et al., 2021; Wei et al., 2021), relevance in other viral diseases, and the availability of inhibitors as discussed below. These four protein complexes are the NADH:ubiquinone oxidoreductase complex (complex I of the respiratory chain), the succinate dehydrogenase complex (complex II of the respiratory chain), CDP-diacylglycerol—inositol 3-phosphatidyltransferase, and solute carrier family 16 member 1.
Eight targets correspond to various subunits of NADH:ubiquinone oxidoreductase, for which protein-protein interactions with all three viruses have been observed (
Supplementary Material S4). Moreover, interactions of NADH:ubiquinone oxidoreductase subunits with the respiratory syncytial virus (RSV) (Hu et al., 2019), human immunodeficiency virus 1 (HIV-1) (Jäger et al., 2011), human gammaherpesvirus 8 (HHV-8) (Davis et al., 2015), human papillomavirus (HPV) serotype 18 (Rozenblatt-Rosen et al., 2012), hepatitis C virus (HCV) (Tripathi et al., 2010) and Nipah virus (Martinez-Gil et al., 2017) have been reported. An approved drug that inhibits NADH:ubiquinone oxidoreductase is the biguanide metformin (Kelly et al., 2015), the first-line treatment in type II diabetes (Kelly et al., 2015). Intriguingly, previous observational studies have shown reduced mortality of type II diabetic patients taking metformin during COVID-19 (Crouse et al., 2020). The aim to improve the effect of metformin led to the development of phenformin (Sterne, 1963), which showed an improved cellular uptake compared to metformin (Sogame et al., 2009). We hence decided to use phenformin for experimental validation of our approach. The solute carrier family 16 member 1 gene (SLC16A1), a monocarboxylic acid-transporter that transports compounds such as pyruvate, lactate, branched-chain amino acids, and ketone bodies (Garcia et al., 1994), interacts with SARS-CoV-2 and influenza A virus proteins. It was additionally identified as a host cell factor of HCV (Germain et al., 2014) and demonstrated to be a relevant host factor for SARS-CoV-2 infections in two CRISPR-Cas9 knockout screens (Daniloski et al., 2021; Wang et al., 2021).SR13800 was identified as a potential inhibitor of SLC16A1 (Doherty et al., 2014; Morris and Felmlee, 2008). The succinate-ubiquinone oxidoreductase subunit A (SDHA) is part of complex II of the respiratory chain and catalyzes the reaction of succinate to fumarate in the tricarboxylic acid cycle. Besides its interaction with SARS-CoV-2 (Daniloski et al., 2021; Gordon et al., 2020), SDHA also interacts with the Epstein-Barr virus (EBV) (Rozenblatt-Rosen et al., 2012), the influenza A virus (de Chassey et al., 2013), HCV (Zhu et al., 2016), and HHV-8 (Davis et al., 2015). Atpenin A5 is a known SDHB inhibitor (Horsefield et al., 2006) and was previously assessed for its therapeutic effect on cardiac injury in isolated perfused rat hearts (Wojtovich and Brookes, 2009). The CDP-diacylglycerol-inositol 3-phosphatidyltransferase (CDIPT) is involved in the biosynthesis of phosphatidylinositol. It interacts with SARS-CoV-2 as well as influenza A and is reported in three CRISPR-Cas9 studies as a relevant host factor during SARS-CoV-2 infection (Daniloski et al., 2021; Wang et al., 2021; Wei et al., 2021). CDIPT also interacts with proteins of numerous HPV serotypes (Muller et al., 2012; Rozenblatt-Rosen et al., 2012), EBV (Rozenblatt-Rosen et al., 2012), and HIV-1 (Barbosa et al., 2021). Scyllo-inositol is listed as an inhibitor of CDIPT (Chen et al., 2002) and was proposed as a potential drug to prevent Alzheimer’s disease (Michon et al., 2020).
Experimental validation of broad-spectrum antivirals in viral infection assays
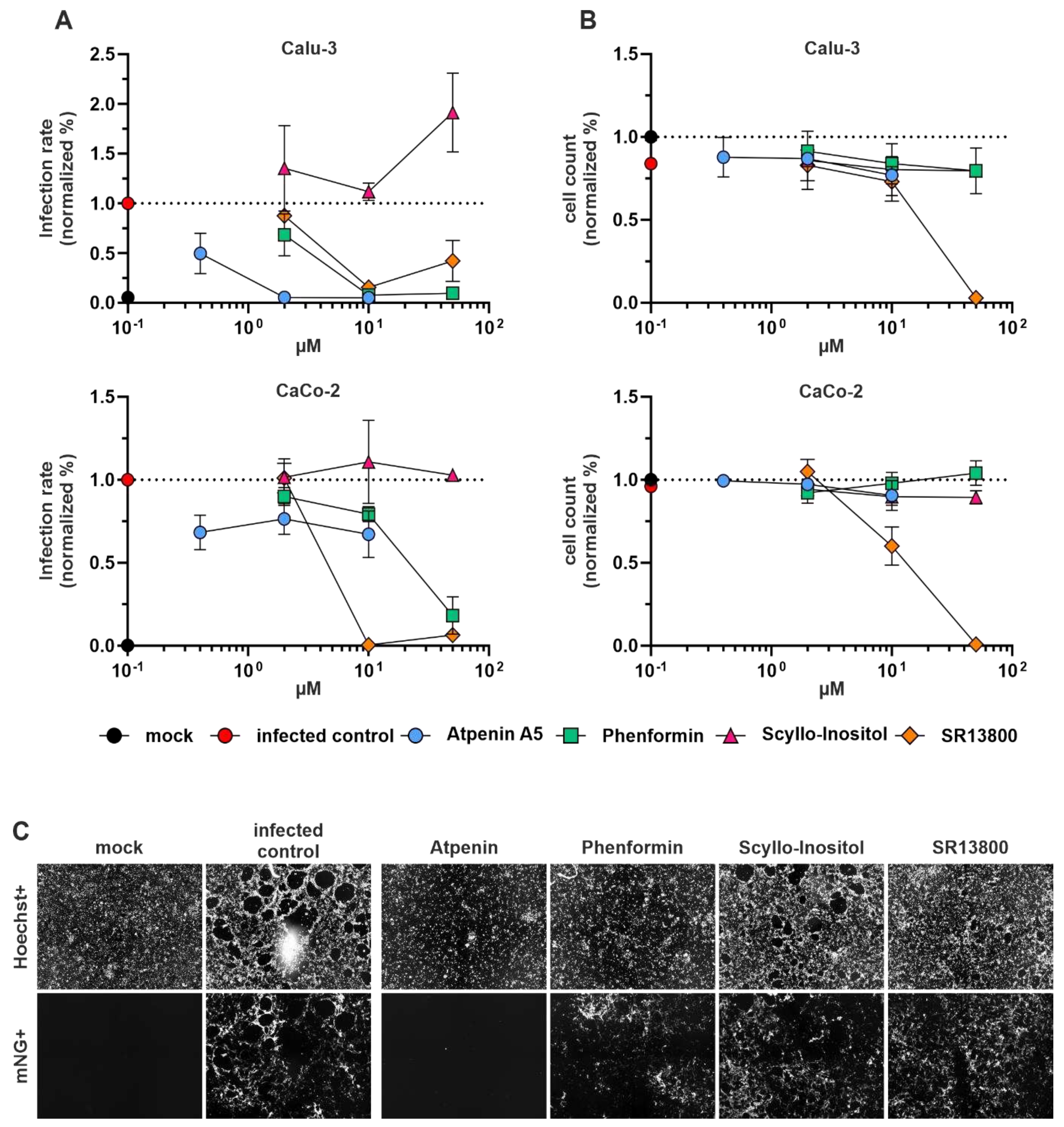
The antiviral activity of the identified compounds phenformin, atpenin A5, SR13800, and scyllo-inositol was first tested in the lung-cancer cell line Calu-3 and the colon-cancer cell line CaCo-2 infected with SARS-CoV-2 (
Figure 4). The infection rate was measured by virus-encoded reporter gene expression (mNeonGreen) 48 hours after infection. The cell viability was assessed by Hoechst staining of the nucleus (see methods) and hence assessment of total cell numbers after 72 hours of treatment. Phenformin and atpenin A5 inhibited SARS-CoV-2 infection of Calu-3 cells at different doses and partly of Caco-2 cells, whereas SR13800 showed toxicity at higher concentrations. Scyllo-inositol showed no activity (
Figure 4A and representative primary images in
Figure 4B).
Phenformin and atpenin A5 showed antiviral activity against mNeonGreen expressing SARS-CoV-2, which is based on an early Wuhan isolate (
Figure 4). Since we identified both compounds
in silico as potential broadly acting antivirals, we assessed their IC
50 in inhibiting SARS-CoV-2 including an Omicron variant, dengue virus (DENV), the respiratory syncytial virus (RSV) and influenza A virus (IAV) (
Figure 5A) and the CC
50 based on total cell numbers (
Figure 5B). Phenformin had an IC
50 of 1.46 µM, and atpenin A5 an IC
50 of 0.45 µM against the original Wuhan strain in Calu-3 cells without cellular toxicity and both compounds were similarly active against the Omicron variant. It was previously reported that type II diabetic patients taking metformin, similar to phenformin, showed a reduced risk of severe COVID-19 compared to non-users (Crouse et al., 2020). We also included metformin in our SARS-CoV-2 infection experiments revealing that it inhibited infection only at very high concentrations at an IC
50 of 459 µM in Calu-3 cells, again without showing any cytotoxicity (
Figure 5 and
Supplementary Figure S1). We therefore decided to do further tests exclusively with phenformin, as metformin was clearly inferior in its antiviral activity.
Phenformin had an IC
50 of 17.15 µM against DENV in Huh7.5 hepatoma cell lines, while atpenin A5 inhibited DENV infection at an IC
50 of 0.36 µM (
Figure 5A and
Supplementary Figure 1). Of note, atpenin A5 also inhibited RSV and IAV infection of the lung cell line A549 at IC
50s of 0.14 µM (IAV) and 0.12 µM (RSV), respectively (
Figure 5A and
Supplementary Figure 1). Phenformin was inactive against RSV and IAV at the concentrations tested. We did not measure any severe cytotoxicity of any of the compounds in the three cell lines tested (
Figure 5B).
Discussion
In this work, we have used constraint-based metabolic modeling to investigate the metabolic state of virally infected cells and identify potential targets for broad-spectrum antiviral treatment. We found that viral infection of a tissue led to pronounced induction of predicted viral replication capacity even in non-infected cells. This indicates that SARS-CoV-2 might induce transcriptional programs in neighboring cells that prime non-infected cells for viral replication along with a pronounced induction of many metabolic pathways, which we previously reported for several types of immune cells in COVID-19 patients (Bernardes et al., 2020). Such pleiotropic effects in the microenvironment of virally infected cells have previously been reported for other viruses such as Epstein Barr Virus and Kaposi’s sarcoma herpesvirus (Magalhaes et al., 2019). In this context, those effects are mediated via extracellular vesicles secreted by virally infected cells that contain viral effectors that modulate the metabolic activity of non-infected cells in the microenvironment. Our observation of a widespread induction of pathways required for viral replication in uninfected cells, in combination with the previous detection of extracellular vesicles containing viral proteins in SARS-CoV-2 infected individuals (Yim et al., 2022) indicate that SARS-CoV-2 infected cells might modulate the permissibility of their microenvironment for viral replication in a similar manner. Other mechanisms that might contribute to the modulation of the microenvironment of virally infected cells could be virus-induced cellular senescence, which entails a senescence-associated secretory phenotype that induces a pro-inflammatory microenvironment upon SARS-CoV-2 infection (Lee et al., 2021). Overall, these results suggest that SARS-CoV-2 strongly relies on a profound induction of metabolic pathways for its replication and supports the notion that the cellular metabolism of virally infected cells is an attractive target for inhibiting viral replication.
Thus, we further exploited our reconstructed metabolic models to identify potential targets for broad-spectrum antiviral therapy. We expanded our analysis to two additional viruses with pandemic potential, dengue virus (DENV), a member of the Flaviviridae and the world’s fastest-spreading insect-borne disease, as well as influenza A virus (IAV), belonging to the Paramyxoviridae. We categorized antiviral targets into tier-1 and tier-2 targets, depending on whether their knockout would impede viral replication but not impact normal metabolism and viability (tier-1 targets) or prevent viral replication with impairment of cell survival (tier-2 targets). We found that the predicted tier-1 targets were highly enriched among experimentally confirmed interaction partners of all three viruses. Of note, this enrichment was much more pronounced for tier-1 targets than tier-2 targets, indicating that all three viruses dysregulate and hijack cellular proteins essential for viral replication but dispensable for cell survival. This indicates a particular host adaptation of viruses to maximize the production of viral progeny without killing the host cell.
Following the identification of tier-1 targets across all datasets, we identified 254 potential enzyme targets, of which 158 have already been reported in association with viral proteins, representing a highly significant enrichment. Those enzyme targets were strongly enriched in pathways known to be highly relevant for viral replication such as the electron transfer chain, the tricarboxylic acid cycle and glycolysis (Moreno-Altamirano et al., 2019; Sanchez and Lagunoff, 2015; Thaker et al., 2019). Further subsetting those targets to those with a reported association with at least two of the three considered viruses, we identified 39 proteins forming parts of 22 protein complexes that could serve as druggable targets for broad-spectrum viral inhibition. Among those candidates, we selected four enzyme complexes comprising eleven identified targets for further experimental validation.
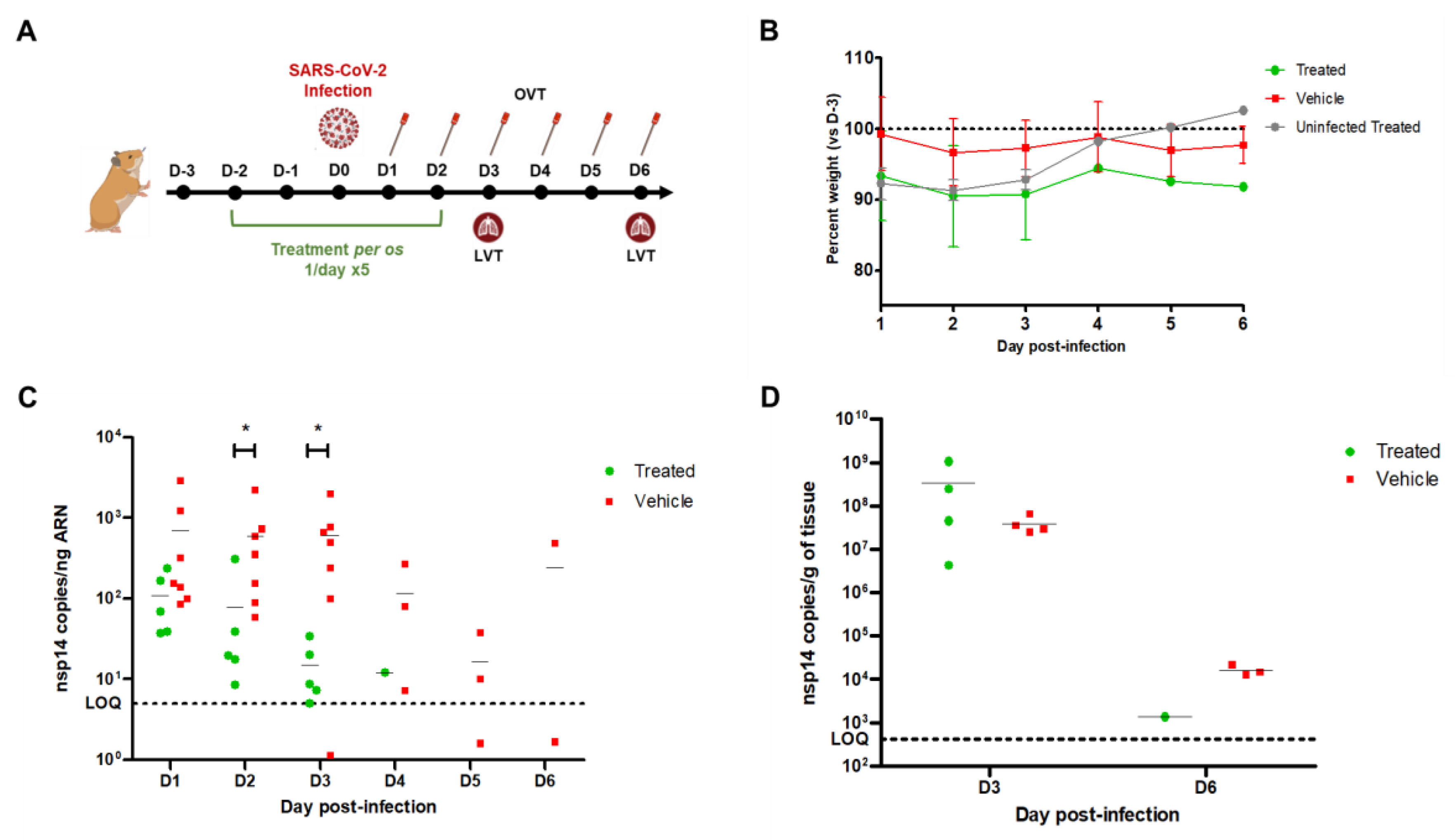
Using inhibitors of those enzymes in cellular infection experiments, we could confirm that targeting the NADH:ubiquinone oxidoreductase complex by phenformin and the succinate dehydrogenase by atpenin A5 blocked viral replication in cell culture experiments with minimal cellular toxicity. Remarkably, while phenformin inhibited the replication of DENV and SARS-CoV-2, atpenin A5 showed excellent antiviral activity against DENV, SARS-Cov-2, RSV, and IAV. All four viruses belong to completely distinct viral taxons, thereby strongly supporting the suitability for broad-spectrum antiviral therapy. Importantly, our target prediction approach did not incorporate data from RSV-infected cells, thus further supporting the notion that the predicted targets also work beyond the three viruses initially included in our analysis.
Phenformin was well tolerated up to high concentrations, which agrees with its previous use as an antidiabetic drug before it was withdrawn from the market in the 1970s due to more frequent fatal cases of lactic acidosis compared to the alternative antidiabetic metformin. The frequency of lactic acidosis was 40 to 64 per 100,000 patients per year and probably related to renal insufficiency similar to metformin therapy (Stang et al., 1999). However, provided that treatment in the context of acute viral infections is short-termed and therefore fundamentally different from the long-term treatment in diabetes, phenformin could represent a viable treatment option. Indeed, biguanides including phenformin showed clinical benefit in diabetic influenza patients and hence the compound was suggested as a COVID treatment option (Lehrer, 2020). Another study identified phenformin as a potential SARS-CoV-2 PL
proinhibitor with potential antiviral activity using molecular dynamics simulations (Kandeel et al., 2021). However, none of the aforementioned studies provided experimental evidence for antiviral activity of phenformin in cell-based or in vivo infection models. Reported plasma levels of phenformin after 50-100 mg of phenformin uptake in patients are ~1.7 µM (Nattrass et al., 1980), while tissue concentrations have not been assessed but are expected to be much higher as reported in animal models (Di Magno et al., 2022). For instance, in rats portal vein concentrations reached 2.5 µM and liver concentrations 147 µM after oral gavage of 50 mg/kg phenformin (Sogame et al., 2011). Thus, tissue concentrations are expected to be much higher than the IC
50 of 1.4 µM that we measured against SARS-CoV-2. Atpenin A5 is, up to now, an experimental compound that was not tested
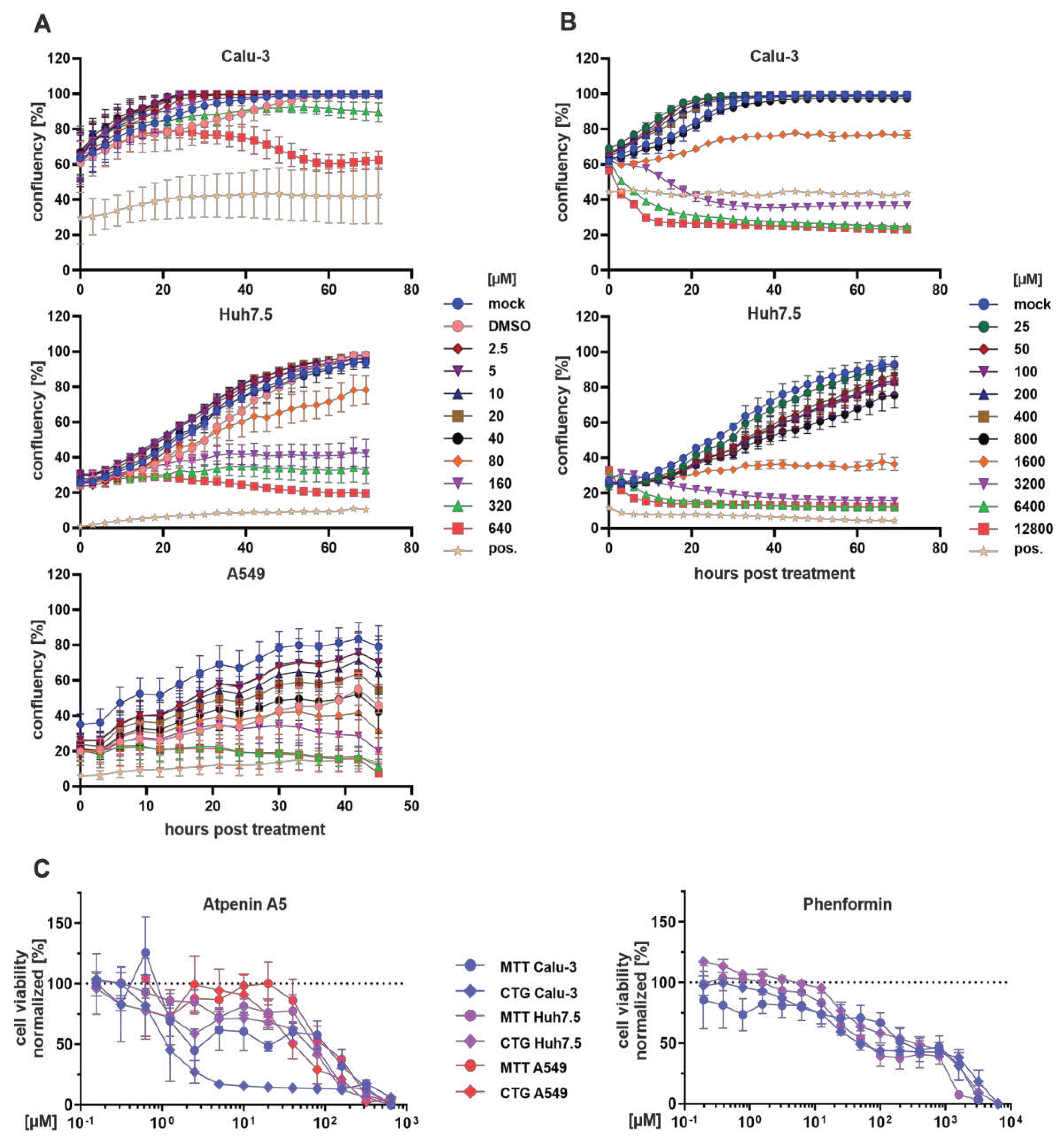
in vivo, possibly due to its expectable multiple adverse effects in various organs. Atpenin A5 is known as a potent complex II inhibitor (Miyadera et al., 2003), thus affecting the redox chain at the mitochondrial membrane and overall energy metabolism. These two features are also reflected by our MTT and CTG assays (
Figure 6 and
Table 1). Nevertheless, complex II inhibition is also discussed as a druggable metabolic pathway for various cancer types (Zhao et al., 2017). Novel atpenin A5-derived leads were developed with a much higher on-target specificity (Wang et al., 2017). These recent developments, combined with our observation that complex II inhibition via the succinate dehydrogenase is a druggable and highly potent target for broad-spectrum antiviral inhibition, raise the possibility for the rapid establishment of novel broadly acting antiviral drugs.
This notion and the reliability of our whole pipeline are supported by our in vivo validation of the antiviral activity of phenformin in the Golden Syrian hamster model. Although further dose-optimization studies are warranted to maximize the antiviral effect in the lower respiratory tract, phenformin showed antiviral activity in the upper respiratory tract, the main anatomic site for viral entry and spread. Hence, early application of phenformin may potentially reduce the time to symptom resolution, prevent or delay spread to the lower respiratory tract and also shorten and limit the transmission window. As already discussed, given the well-elaborated PK/ADME/tox profiles of phenformin in humans, we suggest holding phenformin as a candidate drug for preparedness in case of future outbreaks caused by coronaviruses and potentially other pandemic viruses. Furthermore, recent evidence coming from larger trials analyzing the efficacy of the phenformin-related metformin to ameliorate symptoms of long-covid and also giving benefit to patients during acute infection (Bramante et al., 2023, 2022), supports our findings. Importantly, in our comparative analyses, phenformin strongly outcompeted metformin in terms of its antiviral activity which was ~300 fold higher against SARS-CoV-2 (IC50 of ~1.5µM for phenformin vs ~450µM for metformin). This difference in efficiency is in line with the improved cellular uptake of phenformin over metformin and the much lower typical therapeutic dosage of phenformin compared to metformin in humans (Di Magno et al., 2022).
Nevertheless, there are limitations to our study and open questions remain. The activity of phenformin needs to be characterized in further detail in vitro as well as in vivo prior to upcoming clinical trials. Atpenin A5 is of limited in vivo compatibility, and following its proof-of-concept as broad antiviral agents, more work is necessary to render it biocompatible or find other inhibitors of the same target. Moreover, we have only explored a subset of our predicted targets, and other targets from our list might provide an even higher potency for inhibiting viral replication in a broad set of viruses. Hence, ample follow-up studies to the work presented herein are necessary to further explore the capability of our modeling pipeline to predict antiviral drugs.
Altogether, this study proves that targeting metabolism is a valuable strategy in the context of antiviral therapies (Mayer et al., 2019), with certain advantages. Metabolism-based targets exert a very low variability and are predictably essential for viral replication, resulting in a high resistance barrier and a predictable broad antiviral activity. In this context, our pipeline for identifying antiviral targets, integrating cellular metabolism and data from virally infected cells, as well as the inhibitors we have identified, represents an invaluable resource for pandemic preparedness against future emerging pathogens.
Methods
Single-cell sequencing datasets
Table 2 summarizes the single-cell sequencing datasets used.
Viral replication models based on Recon 2.2
The viral biomass objective function (VBOF) for SARS-CoV-2 was taken from Renz et al. (Renz et al., 2020). The VBOF for dengue was taken from Aller et al. (Aller et al., 2018). No VBOF for influenza A H1N1 was available and, thus, needed to be constructed. The nucleotide and protein sequences with the RefSeq number GCF_001343785.1 were downloaded from NCBI’s Reference Sequence database (W. Li et al., 2021). The genome copy number was assumed to be 1. Influenza A H1N1 has four structural proteins: hemagglutinin (HA), neuraminidase (NA), matrix protein 1 (M1), and matrix protein 2 (M2). The nonstructural proteins include the polymerases PB1, PB2, and PA, the nucleocapsid protein (NP), the nonstructural protein 1 (NS1), the nuclear export protein (NEP), and the PA-X protein (PAX). The detailed list of copy numbers for the structural and nonstructural proteins is given in
Supplementary Table S6. The stoichiometric coefficients for the nucleotides, amino acids, and energy requirements were calculated based on the copy numbers and according to the steps suggested by Aller et al. (Aller et al., 2018). These coefficients are provided in
Supplementary Table S7. The genome-scale metabolic model of humans, Recon 2.2 (Swainston et al., 2016), was expanded with the viral replication reaction for each of the three viruses. The model was conditioned with a medium corresponding to the concentration of metabolites present in human blood (
Supplementary Table S8)(Bernardes et al., 2020). Subsequently, we used fastcore’s fastcc algorithm and the simulated blood serum diet to generate consistent models for each virus. These models are available from the Biomodels database (see data availability).
Cell identity annotation
Cell-type annotation was not available for all datasets analyzed in this study. For deposited data with missing metadata on cell identity, we performed cell type annotation
de novo (see
Supplementary Figure S2A-D). For the datasets BALF1 and dengue, we employed the R-package CHETAH (version 1.8.0) (de Kanter et al., 2019). CHETAH makes use of a reference single-cell RNA dataset which is used to build a classification tree via hierarchical clustering. Input cells are placed and identified as reference cell types or intermediate types in the classification tree.The
BALF1 dataset was annotated using a single-cell lung atlas of idiopathic pulmonary fibrosis, chronic obstructive pulmonary disease, and healthy smoker and non-smoker’s lung cells ((Adams et al., 2020), GSE136831). Only cells annotated as epithelial in the original work were considered.
The
dengue dataset was annotated using an in-house available PBMC single-cell dataset (Zanini et al., 2018). References were prepared according to the CHETAH R-package manual. Briefly, annotation references were created from single-cell expression datasets with known cell type identifications. Single cells of only healthy donors were kept in the reference. Rare cell types with less than 15 cells representing that cell type were dropped from the reference. In the interest of computing times, the remaining cell types were downsampled to a maximum of 300 cells, picked at random. The remaining single cells were normalized sample-wise to an equal sequencing depth of one million, and +1 was added to each gene’s count to allow for log
2 transformation. For an improved classification, ribosomal and housekeeping genes were dropped (
Supplementary Table S9). After creating Seurat objects and performing quality control as described in “scRNA core reaction pre-processing,” cell type annotation was done on the Seurat objects. For this, the classification function CHETAHclassifier was run with the query dataset and the matching reference. The quality of the automated classification was visually controlled with dimension reduction plots.
The influenza H1N1 dataset was manually annotated since CHETAH provided unsatisfactory results. We followed the steps outlined in the accompanying publication’s methods (Medaglia et al., n.d.).
scRNA data processing and reconstruction of context-specific metabolic models
Prior to the reconstruction of single-cell metabolic models, scRNA datasets (Methods: scRNA Datasets) were downloaded from NCBI’s Gene Expression Omnibus (GEO)(Clough and Barrett, 2016) for pre-processing with Seurat (Butler et al., 2018) and StanDep (Joshi et al., 2020). Seurat objects of the respective scRNA datasets were created, and cells were removed if data was of insufficient quality (number of detected genes > 200 and < 6,000; mitochondrial RNA with mapped reads < 10). For datasets with missing cell type information, cell identity annotation was applied to the filtered Seurat objects (see “Cell identity annotation”). Gene level counts were translated into transcripts per million (TPM) values through normalization according to human ENSEMBL gene lengths. ENSEMBL (Howe et al., 2021) gene names were mapped to Recon 2.2 identifiers (Cunningham et al., 2019; Joshi et al., 2020). StanDep was employed with the pre-processed expression data to identify core reactions active in the individual cells. To this end, enzyme expression was log10 transformed into a matrix with rows representing enzymes and columns as bins to identify minimum and maximum enzyme expression. A complete linkage metric with hierarchical clustering and Euclidean distance was used to cluster (number of clusters = 40) genes with respect to their expression. Core reaction matrices were assembled, defined, and used as an input to fastcore (Vlassis et al., 2014) to reconstruct single-cell, context-specific metabolic models. The fastcore algorithm was employed to create cell-type-specific models based on the consistent Recon 2.2 model, including the viral replication reaction and the list of core reactions from StanDep expanded by the biomass objective function and VBOF. The human metabolic models with all constraints for the blood medium and the viral replication reactions have been uploaded to the BioModels Database (Malik-Sheriff et al., 2020) in SBML format (Keating et al., 2020) with hierarchical model composition extension (Smith et al., 2015). Each model entry in BioModels Database contains a base model derived from Recon 2 and tissue-specific models, all wrapped together in an Open Modeling EXchange format (OMEX) (Bergmann et al., 2014) file with annotation (Neal et al., 2019). For links to the respective datasets and models see the data availability statement.
Reactions identified as being active in single-cell metabolic models were counted and summarized into 82 metabolic pathways based on the subsystem annotation of Recon 2.2 to analyze metabolic pathway activity in the BALF2 data. The small number of single-cell models with detected viral RNA were not considered to avoid confounding (87 of 148.420 cells). The resulting counts of active reactions per pathway were checked for too many single cells with zero counts, e.g., metabolic pathways were discarded if they showed zero counts in more than half of the single-cell models across all three patient groups (control, moderate and severe COVID). The remaining 57 metabolic pathways were statistically evaluated individually for their relation to patient groups while controlling for cell type (“Basal,” “Ciliated,” “Ciliated-dif,” “FOXN4,” “Ionocyte,” “IRC,” “outliers_epithelial,” “Secretory,” “Secretory-diff,” “Squamous”). Active reaction counts were modeled as the dependent variable in negative binomial regressions (R-package MASS 7.3-57 function glm.nb (Venables and Ripley, 2012)). Metabolic pathways with inflated zero counts were modeled with negative binomial regression with an additional zero-inflation model. The zero-inflation model took the same independent variables as the primary model, namely patient group and cell type (R-package pscl 1.5.5, function zeroinfl with dist = "negbin" (Zeileis et al., 2008)). The logarithm to the base two was calculated for fold changes between the control group as a baseline and any of the two COVID patient groups.
Prediction of viral replication capacity and antiviral targets
In order to predict viral replication capacity for a cell built from scRNA-Seq data, we used flux balance analysis (Renz et al., 2020) with the applied blood serum diet and maximized the flux through the VBOF. We performed single gene deletions to predict antiviral targets and assessed their effects on host biomass production and viral replication capacity.
Supplemental Data S2 lists the predicted viral replication capacities and predicted tier-1 and tier-2 targets for each cell. Those gene knockouts that were found to decrease viral replication capacity by at least 50% of the initial value while maintaining the host’s biomass minimally at 80% of its initial value were reported as tier-1 targets. Tier-2 targets reduced viral replication capacity by at least 50% but decreased cellular biomass production by more than 20%. While the biomass reaction is typically used to predict maximal growth rates of cells, it consumes all the molecules whose production is continuously required to maintain cellular function and hence can be used as a proxy for metabolic requirements of normal cellular function. Examples of such cellular functions include, for instance, the production of amino acids for protein turnover, nucleotides for DNA repair, and energy equivalents for cellular function. To identify tier-1 and tier-2 targets, we considered only cells with a predicted viral replication rate above 0.01 mmol/gDW (unit of flux measurement in constraint-based models) and detected tier-1 targets. Moreover, we constricted the data to include only virally infected cells where possible (i.e., a sufficient number of virally infected cells). Thus, we only considered infected cells as indicated in the provided metadata for the cell culture data from SARS-CoV-2 infected cells (CALU3, scH1299) and the BALF1 data set. For the BALF2 data set, only a few infected cells were detected. Since we observed priming of non-infected cells for viral replication upon viral infection of the host, we considered all SARS-CoV-2 permissive cell types of SARS-CoV-2 infected individuals for this dataset (i.e., those listed in
Figure 1B). For dengue and influenza A, we considered only cells with >0.1% of reads mapping to the respective viral genomes as indicated in the metadata provided along with the sequencing data.
For the comparison of known interaction partners of the viral proteome with tier-1 and tier-2 targets (
Figure 3), we subsetted tier-1 and tier-2 targets to those occurring in at least 5% of the cells for each data set for which tier-1 targets could be identified (high-confidence tier-1 and tier-2 targets,
Supplementary Table S2). To identify broad-spectrum antiviral targets, we counted for each dataset and enzyme how often it was listed as a tier-1 target. Additionally, we included information on known virus protein interactions (Gordon et al., 2020; Shah et al., 2018; Watanabe et al., 2014). For SARS-CoV-2, we used host proteins that had at least one reported interaction with a SARS-CoV-2 protein at a SAINTexpress Bayesian false-discovery rate (BFDR) ≤ 0.05 (Teo et al., 2014) and an average spectral count ≥ 2. The list of considered interaction partners for each virus is provided in
Supplementary Table S1. To analyze the enrichment of predicted shared tier-1 targets across all datasets among cellular metabolic pathways, we used the human metabolic model Recon2.2 to identify genes associated with each metabolic subsystem annotated in the model. Thus, we mapped each subsystem to the reactions that it contained and identified the associated gene lists via the gene-protein-reaction associations. Subsequently, we used Fisher’s exact test to determine the significance of the enrichment of shared tier-1 targets among the genes associated with each subsystem.
We derived a shortlist of candidate targets by subsetting the list to those enzymes listed as tier-1 targets at least once for each data set and reported as interaction partners of the proteome of at least two of the three viruses (39 candidates). The frequency at which each enzyme was listed as a tier-1 target is provided in
Supplementary Table S5. Further information from the literature, CRISPR-Cas studies (Daniloski et al., 2021; Wang et al., 2021; Wei et al., 2021), and the availability of suitable inhibitors were additionally used to identify candidate targets suitable for experimental testing. To identify the gene’s relevance in other viral diseases, we searched the database of protein, genetic and chemical interactions (BioGRID) (Oughtred et al., 2019) and the molecular INTeraction database (MINT) (Licata et al., 2012). To identify known drugs or inhibitors, we searched the Drug Gene Interaction database (DGIdb) (Cotto et al., 2018), the DrugBank database (Wishart et al., 2018), and the GeneCards suite (Stelzer et al., 2016).
Statistical analysis
Statistical tests were performed with R version 4.1.3 if not indicated otherwise. The individual statistical tests are indicated for each case in which p-values are reported. In the case of multiple tests, false discovery rate control using the Benjamini and Hochberg procedure (Benjamini and Hochberg, 1995) implemented in the p.adjust function of R was used.
Cell culture
A549 cells (human alveolar basal epithelial adenocarcinoma) were maintained at 37 °C with 5% CO2 in RPMI 1640 Medium containing 10% (v/v) inactivated fetal calf serum (FCS) and 100 µg/mL penicillin-streptomycin.
Calu-3 cells (human lung adenocarcinoma) and HEp-2 cells (human epidermoid cancer cells) were maintained at 37 °C with 5% CO2 in Dulbecco’s Modified Eagle Medium (DMEM) containing 10% FCS, GlutaMax, and 100 µg/mL penicillin-streptomycin.
CaCo-2 (human colorectal adenocarcinoma) and Huh7.5 cells (human hepatocellular carcinoma) were maintained at 37 °C with 5% CO2 in DMEM containing 10% FCS, GlutaMax, 1% (v/v) nonessential amino acids and 100 µg/mL penicillin-streptomycin.
Viruses
Two different SARS-CoV-2 strains were used in this study. First is the recombinant SARS-CoV-2 clone expressing mNeonGreen icSARS-CoV-2 mNG (Xie et al., 2020). It was obtained from the World Reference Center of Emerging Viruses and Arboviruses at the University of Texas Medical Branch (UTMB). For virus production, CaCo-2 cells were infected; 48 hours post-infection (hpi), the supernatant was collected, centrifuged, and stored at -80 °C. Second is a clinical SARS-CoV-2 isolate that belongs to the B.1.1.529 (Omicron) BA.1 lineage. It was isolated from a PCR-positive patient by a throat swab at the Institute for Medical Virology and Epidemiology of Viral Diseases, University Hospital Tübingen (UKT). Briefly, 200 µL of patient material was used to inoculate CaCo-2 cells in a six-well plate (150,000/well). At 48 hpi, the supernatant was collected, centrifuged, and stored at -80 °C. After two consecutive passages, samples were prepared for NGS, and the correct SARS-CoV-2 lineage was determined. The MOI was determined for both viruses by titration with serial dilutions. The number of infectious virus particles per mL was calculated as the (MOI × cell number)/(infection volume), where MOI = −ln(1 − infection rate). The recombinant Respiratory Syncytial Virus (RSV rA2-eGFP) was kindly provided by Konstantin Sparrer (Institute of Molecular Virology, University Hospital Ulm, Germany) and Michael N. Teng (University of South Florida Morsani College of Medicine, Tampa, Florida, USA) (Villenave et al., 2015). HEp-2 cells were infected, and the cells and the supernatant were harvested at 72 hpi, sonicated for 10 min at 35 °C, centrifuged, and stored at -80 °C to generate rA2-eGFP stocks. The recombinant influenza Virus A (IAV) expressing GFP (IAV-GFP SC35M) was kindly provided by Konstantin Sparrer (Institute of Molecular Virology, University Hospital Ulm, Germany) and Martin Schwemmle (Institute of Virology, University Hospital Freiburg, Germany) (Reuther et al., 2015).
For the dengue studies, recombinant dengue 2 strains 16681 (Genebank Accession NC_001474) containing a Renilla Luciferase Reporter was used (Fischl and Bartenschlager, 2013). The dengue genome was transcribed as viral RNA in full-length and electroporated into Huh7.5 cells. Seventy-two hours post electroporation, the supernatant was collected, centrifuged, and stored at -80 °C.
Initial screening of four drug candidates against SARS-CoV-2
CaCo-2 and Calu-3 cells were pre-treated with phenformin, SR13800, and scyllo-inositol in concentrations of 50 µM, 10 µM, and 2 µM, and atpenin A5 in concentrations of 10 µM, 2 µM, and 0.4 µM. After 24 hours, cells were infected with icSARS-CoV-2-mNG (Xie et al., 2020) at a multiplicity of infection (MOI) = 0.2 for CaCo-2 cells and at an MOI = 0.5 for Calu-3 cells or mock-infected. After 48 hours post-infection (hpi), cells were fixed with 2% PFA and stained with Hoechst (1 µg / mL final concentration). Images of cell nuclei, mNeonGreen and bright field were taken with Cytation3 (Biotek, Winooski, VT, USA). Hoechst+ and mNeonGreen+ cells were counted by the Gen5 software (Biotek, Winooski, VT, USA), and infection rates were calculated using the ratio of mNeonGreen+/Hoechst+ cells.
IC50 calculation of phenformin, metformin, and atpenin A5
For IC50 calculations with the SARS-CoV-2 strains, Calu-3 cells were pre-treated with phenformin, metformin, or atpenin A5. After 24 hours, cells were infected with icSARS-CoV-2-mNG at an MOI = 0.5 or with SARS-CoV-2 Omicron at an MOI = 1.1. The icSARS-CoV-2-mNG infected cells were fixed, stained, and measured as described before. The SARS-CoV-2 Omicron infected cells were fixed with 80% Acetone and stained by Immunofluorescence with rabbit anti-SARS-CoV-2 nucleocapsid antibody and with goat anti-rabbit AlexaFluor™ 594 antibody. Cell nuclei were stained with DAPI. Images of cell nuclei, AlexaFluor™ 594+ cells, and bright field were taken with the Cytation3 (Biotek, Winooski, VT, USA), and DAPI+ and AlexaFluor™ 594+ cells were automatically counted by the Gen5 software (Biotek, Winooski, VT, USA).
For IC50 calculations with dengue virus, phenformin or atpenin A5 were prediluted in Huh7.5 complete media and added to Huh7.5 cells one hour before adding the virus (MOI = 1). At 24 hours post-infection, a lysis buffer was added to the cells, and the cell lysates were subjected to luciferase assay and measured with Cytation3 (Biotek, Winooski, VT, USA).
For IC50 calculation with IAV or RSV, A549 cells were pre-treated with phenformin or atpenin A5. Cells were infected with IAV or RSV 24 hours post-treatment with an MOI of 0.6 (IAV), MOI of 1.4 (RSV), or mock-infected. After 24 hours (IAV) or 48 hours (RSV), cells were fixed with 2% PFA and stained with Hoechst. The plates were measured with Cytation3 (Biotek, Winooski, VT, USA), and Hoechst+ and GFP+ cells were counted by Gen5 software (Biotek, Winooski, VT, USA).
IC50s of all viruses were calculated as the half-maximal inhibitory dose using the GraphPad Prism 9 Software (GraphPad Software, Inc., San Diego, CA, USA, Version 9).
In vivo infection experiments
Infection experiments in Golden Syrian hamsters were performed in the animal facility of the Université de Lyon, VetAgro Sup, Institut Claude Bourgelat (69280 Marcy l’Etoile, France). The experimental protocol was authorized by the Institutional Ethics Committee of VetAgro Sup (CEEA 18, project number 2066) and the French Ministry (APAFIS#27797-2020100516408472).
Sixteen 9-11 week-old female Golden Syrian hamsters (Janvier Labs, Le Genest-Saint-Isle, France) were randomized according to their weight in two groups of 7 and one group of 2. Animals were housed in micro-isolator cages (maximum 4 animals per cage) in a biosafety 3 controlled environment (22 °C, 30-70% humidity, 12:12 h photoperiods, 10 air cycles/h), with ad libitum access to food and water. On day 0 (D0), the two groups of 7 animals were anesthetized with inhaled isoflurane/oxygen and infected intranasally with 105,5 TCID50 of a SARS-CoV-2 Delta (B.1.617.2) strain in 60 µL of PBS. The group of 2 animals was mock infected with 60 µL of PBS. All animals received one 90 µL gavage per day for 5 days between D-2 and D2 containing 100 mg/kg phenformin or sterile water, as indicated in each case. Animals were weighted and monitored for clinical signs on D-3 (baseline before treatment initiation) and then daily after infection. Oropharyngeal swabs were performed daily between D1 and D6 and immediately stored at -80 °C for further total RNA extraction. A subset of animals were euthanized on D3 and D6 and their lungs were removed and homogenized in cold PBS before total RNA extraction. Total RNA from oropharyngeal swabs and lung homogenates were used for nsp14 gene quantification by RT-qPCR.
Author Contributions
A.R., A.D., and C.K. devised the computational component of the study. M.S. conceived and devised the experimental part of the study. M.R-C., S.M.B., M.P., M.B. and A.P. conceived and devised the animal experiments. A.R., M.H., J.J-S, A.P., A.D., M.S., and C.K. wrote the initial manuscript draft. A.R., J.J-S., L.B., C.K., G.M., N.L., and F.C. analyzed the computational data and contributed to identification of antiviral compounds. M.H., M.B., J.D., R.J., V.D., C.M., M.R-C., A.P., S.M.B., M.P., M.B. and M.S. conducted all virus infection experiments and measurements of cellular metabolic activity and toxicity and analyzed the data. All authors contributed to manuscript editing and approved the final paper.
Data Availability Statement
All data produced in the context of this work is available in the
Supplementary Tables and Data. Scripts are available via GitHub in the repository
https://github.com/draeger-lab/R-DRUGS. The human metabolic models with all constraints for the blood medium and the viral replication reactions have been uploaded to the BioModels Database (Malik-Sheriff et al., 2020). After peer review, all models will become publicly available.
Acknowledgments
This work was supported by the BMBF-funded de.NBI Cloud within the German Network for Bioinformatics Infrastructure (de.NBI) (031A532B, 031A533A, 031A533B, 031A534A, 031A535A, 031A537A, 031A537B, 031A537C, 031A537D, 031A538A). CK acknowledges support by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) within the cluster of excellence “Precision medicine in chronic inflammation” (DFG support code EXC2167), the collaborative research center “Metaorganisms” (DFG support code CRC1182), the research group miTarget (DFG support code FOR5042) and the German Ministry for Education and Research in the frame of iTREAT (BMBF support code 01ZX1902A). The DFG further supported this work under Germany’s Excellence Strategy–EXC 2124–390838134 within the Cluster of Excellence CMFI (Controlling Microbes to Fight Infections). A.D. is funded by the Germany Center for Infection Research (DZIF) within the Deutsche Zentren der Gesundheitsforschung (BMBF-DZG, Germany Centers for Health Research of the Federal Ministry of Education and Research), grant no. 8020708703. M.H. and M.B. were fellows of an MD stipend granted by the German Center for Infection Research (DZIF) under grant no. TI 07.003. The work is also funded by the BMBF and the Baden-Württemberg Ministry of Science as part of the Excellence Strategy of the German Federal and State Governments. We are grateful to Nantia Leonidou for her helpful contributions and to the personnel of the Institut Claude Bourgelat for their expertise and technical support with animal studies. Some figures were created with BioRender.com.
Conflicts of Interest
A.R., M.H., L.B., J.J.S., A.D., M.S., and C.K. have submitted a patent application for using phenformin and atpenin A5 as broad-spectrum antivirals. All other authors declare no competing interests.
References
- Adams TS, Schupp JC, Poli S, Ayaub EA, Neumark N, Ahangari F, Chu SG, Raby BA, DeIuliis G, Januszyk M, Duan Q, Arnett HA, Siddiqui A, Washko GR, Homer R, Yan X, Rosas IO, Kaminski N. 2020. Single-cell RNA-seq reveals ectopic and aberrant lung-resident cell populations in idiopathic pulmonary fibrosis. Sci Adv 6:eaba1983.
- Aller S, Scott A, Sarkar-Tyson M, Soyer OS. 2018. Integrated human-virus metabolic stoichiometric modelling predicts host-based antiviral targets against Chikungunya, Dengue and Zika viruses. J R Soc Interface 15. [CrossRef]
- Arrizabalaga J. 2006. The Black Death, 1346-1353: The Complete History (review). Bulletin of the History of Medicine. [CrossRef]
- Azhar EI, Hui DSC, Memish ZA, Drosten C, Zumla A. 2019. The Middle East Respiratory Syndrome (MERS). Infect Dis Clin North Am 33:891–905.
- Barbosa JAF, Sparapani S, Boulais J, Lodge R, Cohen ÉA. 2021. Human Immunodeficiency Virus Type 1 Vpr Mediates Degradation of APC1, a Scaffolding Component of the Anaphase-Promoting Complex/Cyclosome. J Virol 95:e0097120.
- Benjamini Y, Hochberg Y. 1995. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society: Series B (Methodological). [CrossRef]
- Bergmann FT, Adams R, Moodie S, Cooper J, Glont M, Golebiewski M, Hucka M, Laibe C, Miller AK, Nickerson DP, Olivier BG, Rodriguez N, Sauro HM, Scharm M, Soiland-Reyes S, Waltemath D, Yvon F, Le Novère N. 2014. COMBINE archive and OMEX format: one file to share all information to reproduce a modeling project. BMC Bioinformatics 15:369.
- Bernardes JP, Mishra N, Tran F, Bahmer T, Best L, Blase JI, Bordoni D, Franzenburg J, Geisen U, Josephs-Spaulding J, Köhler P, Künstner A, Rosati E, Aschenbrenner AC, Bacher P, Baran N, Boysen T, Brandt B, Bruse N, Dörr J, Dräger A, Elke G, Ellinghaus D, Fischer J, Forster M, Franke A, Franzenburg S, Frey N, Friedrichs A, Fuß J, Glück A, Hamm J, Hinrichsen F, Hoeppner MP, Imm S, Junker R, Kaiser S, Kan YH, Knoll R, Lange C, Laue G, Lier C, Lindner M, Marinos G, Markewitz R, Nattermann J, Noth R, Pickkers P, Rabe KF, Renz A, Röcken C, Rupp J, Schaffarzyk A, Scheffold A, Schulte-Schrepping J, Schunk D, Skowasch D, Ulas T, Wandinger K-P, Wittig M, Zimmermann J, Busch H, Hoyer BF, Kaleta C, Heyckendorf J, Kox M, Rybniker J, Schreiber S, Schultze JL, Rosenstiel P, HCA Lung Biological Network, Deutsche COVID-19 Omics Initiative (DeCOI). 2020. Longitudinal Multi-omics Analyses Identify Responses of Megakaryocytes, Erythroid Cells, and Plasmablasts as Hallmarks of Severe COVID-19. Immunity 53:1296–1314.e9.
- Bordbar A, Monk JM, King ZA, Palsson BO. 2014. Constraint-based models predict metabolic and associated cellular functions. Nat Rev Genet 15:107–120.
- Bramante CT, Buse JB, Liebovitz DM, Nicklas JM, Puskarich MA, Cohen K, Belani HK, Anderson BJ, Huling JD, Tignanelli CJ, Thompson JL, Pullen M, Wirtz EL, Siegel LK, Proper JL, Odde DJ, Klatt NR, Sherwood NE, Lindberg SM, Karger AB, Beckman KB, Erickson SM, Fenno SL, Hartman KM, Rose MR, Mehta T, Patel B, Griffiths G, Bhat NS, Murray TA, Boulware DR. 2023. Outpatient treatment of COVID-19 and incidence of post-COVID-19 condition over 10 months (COVID-OUT): a multicentre, randomised, quadruple-blind, parallel-group, phase 3 trial. Lancet Infect Dis. [CrossRef]
- Bramante CT, Huling JD, Tignanelli CJ, Buse JB, Liebovitz DM, Nicklas JM, Cohen K, Puskarich MA, Belani HK, Proper JL, Siegel LK, Klatt NR, Odde DJ, Luke DG, Anderson B, Karger AB, Ingraham NE, Hartman KM, Rao V, Hagen AA, Patel B, Fenno SL, Avula N, Reddy NV, Erickson SM, Lindberg S, Fricton R, Lee S, Zaman A, Saveraid HG, Tordsen WJ, Pullen MF, Biros M, Sherwood NE, Thompson JL, Boulware DR, Murray TA, COVID-OUT Trial Team. 2022. Randomized Trial of Metformin, Ivermectin, and Fluvoxamine for Covid-19. N Engl J Med 387:599–610.
- Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. 2018. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36:411–420.
- Cavallari I, Scattolin G, Silic-Benussi M, Raimondi V, D’Agostino DM, Ciminale V. 2018. Mitochondrial Proteins Coded by Human Tumor Viruses. Front Microbiol 9:81.
- Chan JF-W, Zhang AJ, Yuan S, Poon VK-M, Chan CC-S, Lee AC-Y, Chan W-M, Fan Z, Tsoi H-W, Wen L, Liang R, Cao J, Chen Y, Tang K, Luo C, Cai J-P, Kok K-H, Chu H, Chan K-H, Sridhar S, Chen Z, Chen H, To KK-W, Yuen K-Y. 2020. Simulation of the Clinical and Pathological Manifestations of Coronavirus Disease 2019 (COVID-19) in a Golden Syrian Hamster Model: Implications for Disease Pathogenesis and Transmissibility. Clin Infect Dis 71:2428–2446.
- Chen X, Ji ZL, Chen YZ. 2002. TTD: Therapeutic Target Database. Nucleic Acids Res 30:412–415.
- Chua RL, Lukassen S, Trump S, Hennig BP, Wendisch D, Pott F, Debnath O, Thürmann L, Kurth F, Völker MT, Kazmierski J, Timmermann B, Twardziok S, Schneider S, Machleidt F, Müller-Redetzky H, Maier M, Krannich A, Schmidt S, Balzer F, Liebig J, Loske J, Suttorp N, Eils J, Ishaque N, Liebert UG, von Kalle C, Hocke A, Witzenrath M, Goffinet C, Drosten C, Laudi S, Lehmann I, Conrad C, Sander L-E, Eils R. 2020. COVID-19 severity correlates with airway epithelium-immune cell interactions identified by single-cell analysis. Nat Biotechnol 38:970–979.
- Clough E, Barrett T. 2016. The Gene Expression Omnibus Database. Methods in Molecular Biology. [CrossRef]
- Cotto KC, Wagner AH, Feng Y-Y, Kiwala S, Coffman AC, Spies G, Wollam A, Spies NC, Griffith OL, Griffith M. 2018. DGIdb 3.0: a redesign and expansion of the drug-gene interaction database. Nucleic Acids Res 46:D1068–D1073.
- Crouse AB, Grimes T, Li P, Might M, Ovalle F, Shalev A. 2020. Metformin Use Is Associated With Reduced Mortality in a Diverse Population With COVID-19 and Diabetes. Front Endocrinol 11:600439.
- Cunningham F, Achuthan P, Akanni W, Allen J, Amode MR, Armean IM, Bennett R, Bhai J, Billis K, Boddu S, Cummins C, Davidson C, Dodiya KJ, Gall A, Girón CG, Gil L, Grego T, Haggerty L, Haskell E, Hourlier T, Izuogu OG, Janacek SH, Juettemann T, Kay M, Laird MR, Lavidas I, Liu Z, Loveland JE, Marugán JC, Maurel T, McMahon AC, Moore B, Morales J, Mudge JM, Nuhn M, Ogeh D, Parker A, Parton A, Patricio M, Abdul Salam AI, Schmitt BM, Schuilenburg H, Sheppard D, Sparrow H, Stapleton E, Szuba M, Taylor K, Threadgold G, Thormann A, Vullo A, Walts B, Winterbottom A, Zadissa A, Chakiachvili M, Frankish A, Hunt SE, Kostadima M, Langridge N, Martin FJ, Muffato M, Perry E, Ruffier M, Staines DM, Trevanion SJ, Aken BL, Yates AD, Zerbino DR, Flicek P. 2019. Ensembl 2019. Nucleic Acids Res 47:D745–D751.
- Cutler DM, Summers LH. 2020. The COVID-19 Pandemic and the $16 Trillion Virus. JAMA 324:1495–1496.
- Daniloski Z, Jordan TX, Wessels H-H, Hoagland DA, Kasela S, Legut M, Maniatis S, Mimitou EP, Lu L, Geller E, Danziger O, Rosenberg BR, Phatnani H, Smibert P, Lappalainen T, tenOever BR, Sanjana NE. 2021. Identification of Required Host Factors for SARS-CoV-2 Infection in Human Cells. Cell 184:92–105.e16.
- Davis ZH, Verschueren E, Jang GM, Kleffman K, Johnson JR, Park J, Von Dollen J, Maher MC, Johnson T, Newton W, Jäger S, Shales M, Horner J, Hernandez RD, Krogan NJ, Glaunsinger BA. 2015. Global mapping of herpesvirus-host protein complexes reveals a transcription strategy for late genes. Mol Cell 57:349–360.
- de Chassey B, Aublin-Gex A, Ruggieri A, Meyniel-Schicklin L, Pradezynski F, Davoust N, Chantier T, Tafforeau L, Mangeot P-E, Ciancia C, Perrin-Cocon L, Bartenschlager R, André P, Lotteau V. 2013. The interactomes of influenza virus NS1 and NS2 proteins identify new host factors and provide insights for ADAR1 playing a supportive role in virus replication. PLoS Pathog 9:e1003440.
- de Kanter JK, Lijnzaad P, Candelli T, Margaritis T, Holstege FCP. 2019. CHETAH: a selective, hierarchical cell type identification method for single-cell RNA sequencing. Nucleic Acids Res 47:e95.
- Deprez M, Zaragosi L-E, Truchi M, Becavin C, Ruiz García S, Arguel M-J, Plaisant M, Magnone V, Lebrigand K, Abelanet S, Brau F, Paquet A, Pe’er D, Marquette C-H, Leroy S, Barbry P. 2020. A Single-Cell Atlas of the Human Healthy Airways. Am J Respir Crit Care Med 202:1636–1645.
- Di Magno L, Di Pastena F, Bordone R, Coni S, Canettieri G. 2022. The Mechanism of Action of Biguanides: New Answers to a Complex Question. Cancers 14. [CrossRef]
- Doherty JR, Yang C, Scott KEN, Cameron MD, Fallahi M, Li W, Hall MA, Amelio AL, Mishra JK, Li F, Tortosa M, Genau HM, Rounbehler RJ, Lu Y, Dang CV, Kumar KG, Butler AA, Bannister TD, Hooper AT, Unsal-Kacmaz K, Roush WR, Cleveland JL. 2014. Blocking lactate export by inhibiting the Myc target MCT1 Disables glycolysis and glutathione synthesis. Cancer Res 74:908–920.
- Fischl W, Bartenschlager R. 2013. High-throughput screening using dengue virus reporter genomes. Methods Mol Biol 1030:205–219.
- Garcia CK, Goldstein JL, Pathak RK, Anderson RG, Brown MS. 1994. Molecular characterization of a membrane transporter for lactate, pyruvate, and other monocarboxylates: implications for the Cori cycle. Cell 76:865–873.
- Gawron P, Ostaszewski M, Satagopam V, Gebel S, Mazein A, Kuzma M, Zorzan S, McGee F, Otjacques B, Balling R, Schneider R. 2016. MINERVA-a platform for visualization and curation of molecular interaction networks. NPJ Syst Biol Appl 2:16020.
- Geraghty RJ, Aliota MT, Bonnac LF. 2021. Broad-Spectrum Antiviral Strategies and Nucleoside Analogues. Viruses 13. [CrossRef]
- Germain M-A, Chatel-Chaix L, Gagné B, Bonneil É, Thibault P, Pradezynski F, de Chassey B, Meyniel-Schicklin L, Lotteau V, Baril M, Lamarre D. 2014. Elucidating novel hepatitis C virus-host interactions using combined mass spectrometry and functional genomics approaches. Mol Cell Proteomics 13:184–203.
- Gordon DE, Jang GM, Bouhaddou M, Xu J, Obernier K, White KM, O’Meara MJ, Rezelj VV, Guo JZ, Swaney DL, Tummino TA, Hüttenhain R, Kaake RM, Richards AL, Tutuncuoglu B, Foussard H, Batra J, Haas K, Modak M, Kim M, Haas P, Polacco BJ, Braberg H, Fabius JM, Eckhardt M, Soucheray M, Bennett MJ, Cakir M, McGregor MJ, Li Q, Meyer B, Roesch F, Vallet T, Mac Kain A, Miorin L, Moreno E, Naing ZZC, Zhou Y, Peng S, Shi Y, Zhang Z, Shen W, Kirby IT, Melnyk JE, Chorba JS, Lou K, Dai SA, Barrio-Hernandez I, Memon D, Hernandez-Armenta C, Lyu J, Mathy CJP, Perica T, Pilla KB, Ganesan SJ, Saltzberg DJ, Rakesh R, Liu X, Rosenthal SB, Calviello L, Venkataramanan S, Liboy-Lugo J, Lin Y, Huang X-P, Liu Y, Wankowicz SA, Bohn M, Safari M, Ugur FS, Koh C, Savar NS, Tran QD, Shengjuler D, Fletcher SJ, O’Neal MC, Cai Y, Chang JCJ, Broadhurst DJ, Klippsten S, Sharp PP, Wenzell NA, Kuzuoglu-Ozturk D, Wang H-Y, Trenker R, Young JM, Cavero DA, Hiatt J, Roth TL, Rathore U, Subramanian A, Noack J, Hubert M, Stroud RM, Frankel AD, Rosenberg OS, Verba KA, Agard DA, Ott M, Emerman M, Jura N, von Zastrow M, Verdin E, Ashworth A, Schwartz O, d’Enfert C, Mukherjee S, Jacobson M, Malik HS, Fujimori DG, Ideker T, Craik CS, Floor SN, Fraser JS, Gross JD, Sali A, Roth BL, Ruggero D, Taunton J, Kortemme T, Beltrao P, Vignuzzi M, García-Sastre A, Shokat KM, Shoichet BK, Krogan NJ. 2020. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 583:459–468.
- Gu C, Kim GB, Kim WJ, Kim HU, Lee SY. 2019. Current status and applications of genome-scale metabolic models. Genome Biol 20:121.
- He J, Cai S, Feng H, Cai B, Lin L, Mai Y, Fan Y, Zhu A, Huang H, Shi J, Li D, Wei Y, Li Y, Zhao Y, Pan Y, Liu H, Mo X, He X, Cao S, Hu F, Zhao J, Wang J, Zhong N, Chen X, Deng X, Chen J. 2020. Single-cell analysis reveals bronchoalveolar epithelial dysfunction in COVID-19 patients. Protein Cell.
- Honigsbaum M. 2020. Revisiting the 1957 and 1968 influenza pandemics. Lancet 395:1824–1826.
- Horsefield R, Yankovskaya V, Sexton G, Whittingham W, Shiomi K, Omura S, Byrne B, Cecchini G, Iwata S. 2006. Structural and computational analysis of the quinone-binding site of complex II (succinate-ubiquinone oxidoreductase): a mechanism of electron transfer and proton conduction during ubiquinone reduction. J Biol Chem 281:7309–7316.
- Howe KL, Achuthan P, Allen J, Allen J, Alvarez-Jarreta J, Amode MR, Armean IM, Azov AG, Bennett R, Bhai J, Billis K, Boddu S, Charkhchi M, Cummins C, Da Rin Fioretto L, Davidson C, Dodiya K, El Houdaigui B, Fatima R, Gall A, Garcia Giron C, Grego T, Guijarro-Clarke C, Haggerty L, Hemrom A, Hourlier T, Izuogu OG, Juettemann T, Kaikala V, Kay M, Lavidas I, Le T, Lemos D, Gonzalez Martinez J, Marugán JC, Maurel T, McMahon AC, Mohanan S, Moore B, Muffato M, Oheh DN, Paraschas D, Parker A, Parton A, Prosovetskaia I, Sakthivel MP, Salam AIA, Schmitt BM, Schuilenburg H, Sheppard D, Steed E, Szpak M, Szuba M, Taylor K, Thormann A, Threadgold G, Walts B, Winterbottom A, Chakiachvili M, Chaubal A, De Silva N, Flint B, Frankish A, Hunt SE, IIsley GR, Langridge N, Loveland JE, Martin FJ, Mudge JM, Morales J, Perry E, Ruffier M, Tate J, Thybert D, Trevanion SJ, Cunningham F, Yates AD, Zerbino DR, Flicek P. 2021. Ensembl 2021. Nucleic Acids Res 49:D884–D891.
- Hu M, Bogoyevitch MA, Jans DA. 2019. Subversion of Host Cell Mitochondria by RSV to Favor Virus Production is Dependent on Inhibition of Mitochondrial Complex I and ROS Generation. Cells 8. [CrossRef]
- Jäger S, Cimermancic P, Gulbahce N, Johnson JR, McGovern KE, Clarke SC, Shales M, Mercenne G, Pache L, Li K, Hernandez H, Jang GM, Roth SL, Akiva E, Marlett J, Stephens M, D’Orso I, Fernandes J, Fahey M, Mahon C, O’Donoghue AJ, Todorovic A, Morris JH, Maltby DA, Alber T, Cagney G, Bushman FD, Young JA, Chanda SK, Sundquist WI, Kortemme T, Hernandez RD, Craik CS, Burlingame A, Sali A, Frankel AD, Krogan NJ. 2011. Global landscape of HIV-human protein complexes. Nature 481:365–370.
- Joshi CJ, Schinn S-M, Richelle A, Shamie I, O’Rourke EJ, Lewis NE. 2020. StanDep: Capturing transcriptomic variability improves context-specific metabolic models. PLoS Comput Biol 16:e1007764.
- Kandeel M, Abdelrahman AHM, Oh-Hashi K, Ibrahim A, Venugopala KN, Morsy MA, Ibrahim MAA. 2021. Repurposing of FDA-approved antivirals, antibiotics, anthelmintics, antioxidants, and cell protectives against SARS-CoV-2 papain-like protease. J Biomol Struct Dyn 39:5129–5136.
- Keating SM, Waltemath D, König M, Zhang F, Dräger A, Chaouiya C, Bergmann FT, Finney A, Gillespie CS, Helikar T, Hoops S, Malik-Sheriff RS, Moodie SL, Moraru II, Myers CJ, Naldi A, Olivier BG, Sahle S, Schaff JC, Smith LP, Swat MJ, Thieffry D, Watanabe L, Wilkinson DJ, Blinov ML, Begley K, Faeder JR, Gómez HF, Hamm TM, Inagaki Y, Liebermeister W, Lister AL, Lucio D, Mjolsness E, Proctor CJ, Raman K, Rodriguez N, Shaffer CA, Shapiro BE, Stelling J, Swainston N, Tanimura N, Wagner J, Meier-Schellersheim M, Sauro HM, Palsson B, Bolouri H, Kitano H, Funahashi A, Hermjakob H, Doyle JC, Hucka M, SBML Level 3 Community members. 2020. SBML Level 3: an extensible format for the exchange and reuse of biological models. Mol Syst Biol 16:e9110.
- Kelly B, Tannahill GM, Murphy MP, O’Neill LAJ. 2015. Metformin Inhibits the Production of Reactive Oxygen Species from NADH:Ubiquinone Oxidoreductase to Limit Induction of Interleukin-1β (IL-1β) and Boosts Interleukin-10 (IL-10) in Lipopolysaccharide (LPS)-activated Macrophages. J Biol Chem 290:20348–20359.
- Lee S, Yu Y, Trimpert J, Benthani F, Mairhofer M, Richter-Pechanska P, Wyler E, Belenki D, Kaltenbrunner S, Pammer M, Kausche L, Firsching TC, Dietert K, Schotsaert M, Martínez-Romero C, Singh G, Kunz S, Niemeyer D, Ghanem R, Salzer HJF, Paar C, Mülleder M, Uccellini M, Michaelis EG, Khan A, Lau A, Schönlein M, Habringer A, Tomasits J, Adler JM, Kimeswenger S, Gruber AD, Hoetzenecker W, Steinkellner H, Purfürst B, Motz R, Di Pierro F, Lamprecht B, Osterrieder N, Landthaler M, Drosten C, García-Sastre A, Langer R, Ralser M, Eils R, Reimann M, Fan DNY, Schmitt CA. 2021. Virus-induced senescence is a driver and therapeutic target in COVID-19. Nature 599:283–289.
- Lehrer S. 2020. Inhaled biguanides and mTOR inhibition for influenza and coronavirus (Review). World Acad Sci J 2. [CrossRef]
- Liao M, Liu Y, Yuan J, Wen Y, Xu G, Zhao J, Cheng L, Li J, Wang X, Wang F, Liu L, Amit I, Zhang S, Zhang Z. 2020. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat Med 26:842–844.
- Licata L, Briganti L, Peluso D, Perfetto L, Iannuccelli M, Galeota E, Sacco F, Palma A, Nardozza AP, Santonico E, Castagnoli L, Cesareni G. 2012. MINT, the molecular interaction database: 2012 update. Nucleic Acids Res 40:D857–61.
- Li W, O’Neill KR, Haft DH, DiCuccio M, Chetvernin V, Badretdin A, Coulouris G, Chitsaz F, Derbyshire MK, Durkin AS, Gonzales NR, Gwadz M, Lanczycki CJ, Song JS, Thanki N, Wang J, Yamashita RA, Yang M, Zheng C, Marchler-Bauer A, Thibaud-Nissen F. 2021. RefSeq: expanding the Prokaryotic Genome Annotation Pipeline reach with protein family model curation. Nucleic Acids Res 49:D1020–D1028.
- Li Y, Liu D, Wang Y, Su W, Liu G, Dong W. 2021. The Importance of Glycans of Viral and Host Proteins in Enveloped Virus Infection. Front Immunol 12:638573.
- Magalhaes I, Yogev O, Mattsson J, Schurich A. 2019. The Metabolic Profile of Tumor and Virally Infected Cells Shapes Their Microenvironment Counteracting T Cell Immunity. Front Immunol 10:2309.
- Mahmoudabadi G, Milo R, Phillips R. 2017. Energetic cost of building a virus. Proc Natl Acad Sci U S A 114:E4324–E4333.
- Malik-Sheriff RS, Glont M, Nguyen TVN, Tiwari K, Roberts MG, Xavier A, Vu MT, Men J, Maire M, Kananathan S, Fairbanks EL, Meyer JP, Arankalle C, Varusai TM, Knight-Schrijver V, Li L, Dueñas-Roca C, Dass G, Keating SM, Park YM, Buso N, Rodriguez N, Hucka M, Hermjakob H. 2020. BioModels-15 years of sharing computational models in life science. Nucleic Acids Res 48:D407–D415.
- Marani M, Katul GG, Pan WK, Parolari AJ. 2021. Intensity and frequency of extreme novel epidemics. Proc Natl Acad Sci U S A 118. https://doi.org/10.1073/pnas.2105482118. [CrossRef]
- Martinez-Gil L, Vera-Velasco NM, Mingarro I. 2017. Exploring the Human-Nipah Virus Protein-Protein Interactome. J Virol 91. [CrossRef]
- Marty AM, Jones MK. 2020. The novel Coronavirus (SARS-CoV-2) is a one health issue. One Health. [CrossRef]
- Mayer KA, Stöckl J, Zlabinger GJ, Gualdoni GA. 2019. Hijacking the Supplies: Metabolism as a Novel Facet of Virus-Host Interaction. Front Immunol 10:1533.
- Medaglia C, Kolpakov I, Zhu Y, Constant S, Huang S, Zwygart AC-A, Cagno V, Dermitzakis ET, Stellacci F, Xenarios I, Tapparel C. n.d. A novel anti-influenza combined therapy assessed by single cell RNA-sequencing. [CrossRef]
- Michon C, Kang C-M, Karpenko S, Tanaka K, Ishikawa S, Yoshida K-I. 2020. A bacterial cell factory converting glucose into scyllo-inositol, a therapeutic agent for Alzheimer’s disease. Commun Biol 3:93.
- Miyadera H, Shiomi K, Ui H, Yamaguchi Y, Masuma R, Tomoda H, Miyoshi H, Osanai A, Kita K, Omura S. 2003. Atpenins, potent and specific inhibitors of mitochondrial complex II (succinate-ubiquinone oxidoreductase). Proc Natl Acad Sci U S A 100:473–477.
- Mora C, McKenzie T, Gaw IM, Dean JM, von Hammerstein H, Knudson TA, Setter RO, Smith CZ, Webster KM, Patz JA, Franklin EC. 2022. Over half of known human pathogenic diseases can be aggravated by climate change. Nat Clim Chang 1–7.
- Moreno-Altamirano MMB, Kolstoe SE, Sánchez-García FJ. 2019. Virus Control of Cell Metabolism for Replication and Evasion of Host Immune Responses. Front Cell Infect Microbiol 9:95.
- Morris ME, Felmlee MA. 2008. Overview of the proton-coupled MCT (SLC16A) family of transporters: characterization, function and role in the transport of the drug of abuse gamma-hydroxybutyric acid. AAPS J 10:311–321.
- Muller M, Jacob Y, Jones L, Weiss A, Brino L, Chantier T, Lotteau V, Favre M, Demeret C. 2012. Large scale genotype comparison of human papillomavirus E2-host interaction networks provides new insights for e2 molecular functions. PLoS Pathog 8:e1002761.
- Nattrass M, Sizer K, Alberti KG. 1980. Correlation of plasma phenformin concentration with metabolic effects in normal subjects. Clin Sci 58:153–155.
- Neal ML, König M, Nickerson D, Mısırlı G, Kalbasi R, Dräger A, Atalag K, Chelliah V, Cooling MT, Cook DL, Crook S, de Alba M, Friedman SH, Garny A, Gennari JH, Gleeson P, Golebiewski M, Hucka M, Juty N, Myers C, Olivier BG, Sauro HM, Scharm M, Snoep JL, Touré V, Wipat A, Wolkenhauer O, Waltemath D. 2019. Harmonizing semantic annotations for computational models in biology. Brief Bioinform 20:540–550.
- Noronha A, Daníelsdóttir AD, Gawron P, Jóhannsson F, Jónsdóttir S, Jarlsson S, Gunnarsson JP, Brynjólfsson S, Schneider R, Thiele I, Fleming RMT. 2017. ReconMap: an interactive visualization of human metabolism. Bioinformatics 33:605–607.
- Orth JD, Thiele I, Palsson BØ. 2010. What is flux balance analysis? Nat Biotechnol 28:245–248.
- Oughtred R, Stark C, Breitkreutz B-J, Rust J, Boucher L, Chang C, Kolas N, O’Donnell L, Leung G, McAdam R, Zhang F, Dolma S, Willems A, Coulombe-Huntington J, Chatr-Aryamontri A, Dolinski K, Tyers M. 2019. The BioGRID interaction database: 2019 update. Nucleic Acids Res 47:D529–D541.
- Ravindra NG, Alfajaro MM, Gasque V, Huston NC, Wan H, Szigeti-Buck K, Yasumoto Y, Greaney AM, Habet V, Chow RD, Chen JS, Wei J, Filler RB, Wang B, Wang G, Niklason LE, Montgomery RR, Eisenbarth SC, Chen S, Williams A, Iwasaki A, Horvath TL, Foxman EF, Pierce RW, Pyle AM, van Dijk D, Wilen CB. 2021. Single-cell longitudinal analysis of SARS-CoV-2 infection in human airway epithelium identifies target cells, alterations in gene expression, and cell state changes. PLoS Biol 19:e3001143.
- Renz A, Widerspick L, Dräger A. 2020. FBA reveals guanylate kinase as a potential target for antiviral therapies against SARS-CoV-2. Bioinformatics 36:i813–i821.
- Reuther P, Göpfert K, Dudek AH, Heiner M, Herold S, Schwemmle M. 2015. Generation of a variety of stable Influenza A reporter viruses by genetic engineering of the NS gene segment. Sci Rep 5:11346.
- Richelle A, Chiang AWT, Kuo C-C, Lewis NE. 2019. Increasing consensus of context-specific metabolic models by integrating data-inferred cell functions. PLoS Comput Biol 15:e1006867.
- Rozenblatt-Rosen O, Deo RC, Padi M, Adelmant G, Calderwood MA, Rolland T, Grace M, Dricot A, Askenazi M, Tavares M, Pevzner SJ, Abderazzaq F, Byrdsong D, Carvunis A-R, Chen AA, Cheng J, Correll M, Duarte M, Fan C, Feltkamp MC, Ficarro SB, Franchi R, Garg BK, Gulbahce N, Hao T, Holthaus AM, James R, Korkhin A, Litovchick L, Mar JC, Pak TR, Rabello S, Rubio R, Shen Y, Singh S, Spangle JM, Tasan M, Wanamaker S, Webber JT, Roecklein-Canfield J, Johannsen E, Barabási A-L, Beroukhim R, Kieff E, Cusick ME, Hill DE, Münger K, Marto JA, Quackenbush J, Roth FP, DeCaprio JA, Vidal M. 2012. Interpreting cancer genomes using systematic host network perturbations by tumour virus proteins. Nature 487:491–495.
- Sanchez EL, Lagunoff M. 2015. Viral activation of cellular metabolism. Virology 479-480:609–618.
- Shah PS, Link N, Jang GM, Sharp PP, Zhu T, Swaney DL, Johnson JR, Von Dollen J, Ramage HR, Satkamp L, Newton B, Hüttenhain R, Petit MJ, Baum T, Everitt A, Laufman O, Tassetto M, Shales M, Stevenson E, Iglesias GN, Shokat L, Tripathi S, Balasubramaniam V, Webb LG, Aguirre S, Willsey AJ, Garcia-Sastre A, Pollard KS, Cherry S, Gamarnik AV, Marazzi I, Taunton J, Fernandez-Sesma A, Bellen HJ, Andino R, Krogan NJ. 2018. Comparative Flavivirus-Host Protein Interaction Mapping Reveals Mechanisms of Dengue and Zika Virus Pathogenesis. Cell 175:1931–1945.e18.
- Shutters ST. 2021. Modelling long-term COVID-19 impacts on the U.S. workforce of 2029. PLoS One 16:e0260797.
- Sia SF, Yan L-M, Chin AWH, Fung K, Choy K-T, Wong AYL, Kaewpreedee P, Perera RAPM, Poon LLM, Nicholls JM, Peiris M, Yen H-L. 2020. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature 583:834–838.
- Smith LP, Hucka M, Hoops S, Finney A, Ginkel M, Myers CJ, Moraru I, Liebermeister W. 2015. SBML Level 3 package: Hierarchical Model Composition, Version 1 Release 3. J Integr Bioinform 12:268.
- Sogame Y, Kitamura A, Yabuki M, Komuro S. 2011. Liver uptake of biguanides in rats. Biomed Pharmacother 65:451–455.
- Sogame Y, Kitamura A, Yabuki M, Komuro S. 2009. A comparison of uptake of metformin and phenformin mediated by hOCT1 in human hepatocytes. Biopharm Drug Dispos 30:476–484.
- Stang M, Wysowski DK, Butler-Jones D. 1999. Incidence of lactic acidosis in metformin users. Diabetes Care 22:925–927.
- Stelzer G, Rosen N, Plaschkes I, Zimmerman S, Twik M, Fishilevich S, Stein TI, Nudel R, Lieder I, Mazor Y, Kaplan S, Dahary D, Warshawsky D, Guan-Golan Y, Kohn A, Rappaport N, Safran M, Lancet D. 2016. The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Curr Protoc Bioinformatics 54:1.30.1–1.30.33.
- Sterne J. 1963. [Report on 5-years’ experience with dimethylbiguanide (metformin, glucophage) in diabetic therapy]. Wien Med Wochenschr 113:599–602.
- Sumbria D, Berber E, Mathayan M, Rouse BT. 2020. Virus Infections and Host Metabolism-Can We Manage the Interactions? Front Immunol 11:594963.
- Swainston N, Smallbone K, Hefzi H, Dobson PD, Brewer J, Hanscho M, Zielinski DC, Ang KS, Gardiner NJ, Gutierrez JM, Kyriakopoulos S, Lakshmanan M, Li S, Liu JK, Martínez VS, Orellana CA, Quek L-E, Thomas A, Zanghellini J, Borth N, Lee D-Y, Nielsen LK, Kell DB, Lewis NE, Mendes P. 2016. Recon 2.2: from reconstruction to model of human metabolism. Metabolomics 12:109.
- Teo G, Liu G, Zhang J, Nesvizhskii AI, Gingras A-C, Choi H. 2014. SAINTexpress: improvements and additional features in Significance Analysis of INTeractome software. J Proteomics 100:37–43.
- Thaker SK, Ch’ng J, Christofk HR. 2019. Viral hijacking of cellular metabolism. BMC Biol 17:59.
- Touré V, Dräger A, Luna A, Dogrusoz U, Rougny A. 2021. The Systems Biology Graphical Notation: Current Status and Applications in Systems Medicine. Systems Medicine. [CrossRef]
- Tripathi LP, Kataoka C, Taguwa S, Moriishi K, Mori Y, Matsuura Y, Mizuguchi K. 2010. Network based analysis of hepatitis C virus core and NS4B protein interactions. Mol Biosyst 6:2539–2553.
- Venables WN, Ripley BD. 2012. Modern Applied Statistics with S. Springer.
- Villenave R, Broadbent L, Douglas I, Lyons JD, Coyle PV, Teng MN, Tripp RA, Heaney LG, Shields MD, Power UF. 2015. Induction and Antagonism of Antiviral Responses in Respiratory Syncytial Virus-Infected Pediatric Airway Epithelium. J Virol 89:12309–12318.
- Vlassis N, Pacheco MP, Sauter T. 2014. Fast reconstruction of compact context-specific metabolic network models. PLoS Comput Biol 10:e1003424.
- Wang H, Huwaimel B, Verma K, Miller J, Germain TM, Kinarivala N, Pappas D, Brookes PS, Trippier PC. 2017. Synthesis and Antineoplastic Evaluation of Mitochondrial Complex II (Succinate Dehydrogenase) Inhibitors Derived from Atpenin A5. ChemMedChem 12:1033–1044.
- Wang R, Simoneau CR, Kulsuptrakul J, Bouhaddou M, Travisano KA, Hayashi JM, Carlson-Stevermer J, Zengel JR, Richards CM, Fozouni P, Oki J, Rodriguez L, Joehnk B, Walcott K, Holden K, Sil A, Carette JE, Krogan NJ, Ott M, Puschnik AS. 2021. Genetic Screens Identify Host Factors for SARS-CoV-2 and Common Cold Coronaviruses. Cell 184:106–119.e14.
- Watanabe T, Kawakami E, Shoemaker JE, Lopes TJS, Matsuoka Y, Tomita Y, Kozuka-Hata H, Gorai T, Kuwahara T, Takeda E, Nagata A, Takano R, Kiso M, Yamashita M, Sakai-Tagawa Y, Katsura H, Nonaka N, Fujii H, Fujii K, Sugita Y, Noda T, Goto H, Fukuyama S, Watanabe S, Neumann G, Oyama M, Kitano H, Kawaoka Y. 2014. Influenza virus-host interactome screen as a platform for antiviral drug development. Cell Host Microbe 16:795–805.
- Wei J, Alfajaro MM, DeWeirdt PC, Hanna RE, Lu-Culligan WJ, Cai WL, Strine MS, Zhang S-M, Graziano VR, Schmitz CO, Chen JS, Mankowski MC, Filler RB, Ravindra NG, Gasque V, de Miguel FJ, Patil A, Chen H, Oguntuyo KY, Abriola L, Surovtseva YV, Orchard RC, Lee B, Lindenbach BD, Politi K, van Dijk D, Kadoch C, Simon MD, Yan Q, Doench JG, Wilen CB. 2021. Genome-wide CRISPR Screens Reveal Host Factors Critical for SARS-CoV-2 Infection. Cell 184:76–91.e13.
- Wishart DS, Feunang YD, Guo AC, Lo EJ, Marcu A, Grant JR, Sajed T, Johnson D, Li C, Sayeeda Z, Assempour N, Iynkkaran I, Liu Y, Maciejewski A, Gale N, Wilson A, Chin L, Cummings R, Le D, Pon A, Knox C, Wilson M. 2018. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res 46:D1074–D1082.
- Wojtovich AP, Brookes PS. 2009. The complex II inhibitor atpenin A5 protects against cardiac ischemia-reperfusion injury via activation of mitochondrial KATP channels. Basic Res Cardiol 104:121–129.
- Wyler E, Mösbauer K, Franke V, Diag A, Gottula LT, Arsiè R, Klironomos F, Koppstein D, Hönzke K, Ayoub S, Buccitelli C, Hoffmann K, Richter A, Legnini I, Ivanov A, Mari T, Del Giudice S, Papies J, Praktiknjo S, Meyer TF, Müller MA, Niemeyer D, Hocke A, Selbach M, Akalin A, Rajewsky N, Drosten C, Landthaler M. 2021. Transcriptomic profiling of SARS-CoV-2 infected human cell lines identifies HSP90 as target for COVID-19 therapy. iScience 24:102151.
- Xie X, Muruato A, Lokugamage KG, Narayanan K, Zhang X, Zou J, Liu J, Schindewolf C, Bopp NE, Aguilar PV, Plante KS, Weaver SC, Makino S, LeDuc JW, Menachery VD, Shi P-Y. 2020. An Infectious cDNA Clone of SARS-CoV-2. Cell Host Microbe 27:841–848.e3.
- Yang W, Petkova E, Shaman J. 2014. The 1918 influenza pandemic in New York City: age-specific timing, mortality, and transmission dynamics. Influenza Other Respi Viruses 8:177–188.
- Yim KHW, Borgoni S, Chahwan R. 2022. Serum extracellular vesicles profiling is associated with COVID-19 progression and immune responses. J Extracell Biol 1:e37.
- Zanini F, Robinson ML, Croote D, Sahoo MK, Sanz AM, Ortiz-Lasso E, Albornoz LL, Rosso F, Montoya JG, Goo L, Pinsky BA, Quake SR, Einav S. 2018. Virus-inclusive single-cell RNA sequencing reveals the molecular signature of progression to severe dengue. Proc Natl Acad Sci U S A 115:E12363–E12369.
- Zeileis A, Kleiber C, Jackman S. 2008. Regression Models for Count Data inR. Journal of Statistical Software. [CrossRef]
- Zhao T, Mu X, You Q. 2017. Succinate: An initiator in tumorigenesis and progression. Oncotarget 8:53819–53828.
- Zhu S-L, Wang L, Cao Z-Y, Wang J, Jing M-Z, Xia Z-C, Ao F, Ye L-B, Liu S, Zhu Y. 2016. Inducible CYP4F12 enhances Hepatitis C virus infection via association with viral nonstructural protein 5B. Biochem Biophys Res Commun 471:95–102.
- Zitzmann C, Kaderali L. 2018. Mathematical Analysis of Viral Replication Dynamics and Antiviral Treatment Strategies: From Basic Models to Age-Based Multi-Scale Modeling. Front Microbiol 9:1546.
Figure 2.
| Enrichment of viral interaction partners among enzymes relevant for viral replication in SARS-CoV-2, dengue virus, and influenza A virus. For each dataset, the fraction of genes that are experimentally-determined interaction partners of the viral proteomes among tier-1 and tier-2 targets occurring in at least 5% of all cells of a dataset with tier-1 targets was determined.
P-values were corrected for multiple testing using false discovery rate control. The number of interacting proteins relative to the number of all proteins in each category is shown below each bar. For the host-virus-interaction data set and the list of tier-1 and tier-2 targets for each data set, see
Supplementary Tables S2 and S3. ScRNA-Seq datasets: SARS-CoV-2, BALF1 (Liao et al., 2020); SARS-CoV-2, BALF2 (Chua et al., 2020); SARS-CoV-2, CALU-3 cell culture (Wyler et al., 2021); SARS-CoV-2, scH1299 cell culture (Wyler et al., 2021); influenza H1N1 (Medaglia et al., n.d.), Human airway epithelia (MucilAirTM) (Deprez et al., 2020); dengue, PBMC, (Zanini et al., 2018). Protein-protein-interaction datasets: SARS-CoV-2 (Gordon et al., 2020); dengue virus (Shah et al., 2018); influenza A virus (Watanabe et al., 2014).
Figure 2.
| Enrichment of viral interaction partners among enzymes relevant for viral replication in SARS-CoV-2, dengue virus, and influenza A virus. For each dataset, the fraction of genes that are experimentally-determined interaction partners of the viral proteomes among tier-1 and tier-2 targets occurring in at least 5% of all cells of a dataset with tier-1 targets was determined.
P-values were corrected for multiple testing using false discovery rate control. The number of interacting proteins relative to the number of all proteins in each category is shown below each bar. For the host-virus-interaction data set and the list of tier-1 and tier-2 targets for each data set, see
Supplementary Tables S2 and S3. ScRNA-Seq datasets: SARS-CoV-2, BALF1 (Liao et al., 2020); SARS-CoV-2, BALF2 (Chua et al., 2020); SARS-CoV-2, CALU-3 cell culture (Wyler et al., 2021); SARS-CoV-2, scH1299 cell culture (Wyler et al., 2021); influenza H1N1 (Medaglia et al., n.d.), Human airway epithelia (MucilAirTM) (Deprez et al., 2020); dengue, PBMC, (Zanini et al., 2018). Protein-protein-interaction datasets: SARS-CoV-2 (Gordon et al., 2020); dengue virus (Shah et al., 2018); influenza A virus (Watanabe et al., 2014).
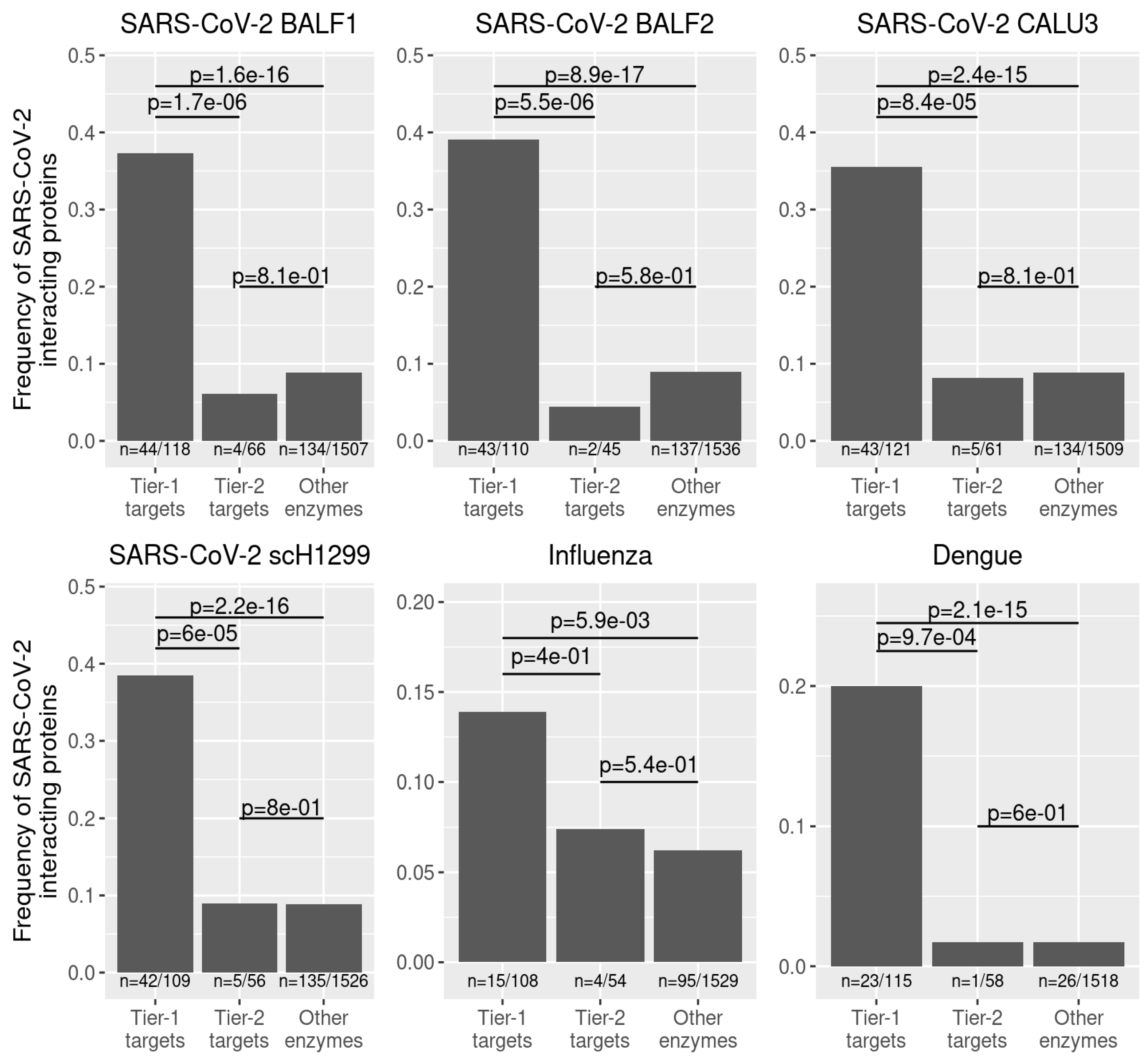
Figure 3.
| Shared tier-1 and broad-spectrum antiviral targets.
A Enrichment of metabolic subsystems among shared tier-1 targets. The 20 most significantly enriched metabolic subsystems are shown. Abbreviations: BCAA metabolism, branched-chain amino acid metabolism.
B Shared tier-1 targets and broad-spectrum antiviral targets in central metabolic pathways. Broad-spectrum antiviral targets are also shared tier-1 targets. Please note that transport reactions involving malate are anti-porters. See
Supplemental Data S1 for a complete map of human metabolism. Abbreviations: AcCoA, acetyl-CoA; AKG, alpha-ketoglutarate; ATPS, ATP synthase; Cit, citrate; Cpx I-IV, complex I - IV; Fum, fumarate; ICit, isocitrate; Mal, malate; PEP, phosphoenolpyruvate; Pyr, pyruvate; Succ, succinate; SuccCoA, succinate-CoA.
Figure 3.
| Shared tier-1 and broad-spectrum antiviral targets.
A Enrichment of metabolic subsystems among shared tier-1 targets. The 20 most significantly enriched metabolic subsystems are shown. Abbreviations: BCAA metabolism, branched-chain amino acid metabolism.
B Shared tier-1 targets and broad-spectrum antiviral targets in central metabolic pathways. Broad-spectrum antiviral targets are also shared tier-1 targets. Please note that transport reactions involving malate are anti-porters. See
Supplemental Data S1 for a complete map of human metabolism. Abbreviations: AcCoA, acetyl-CoA; AKG, alpha-ketoglutarate; ATPS, ATP synthase; Cit, citrate; Cpx I-IV, complex I - IV; Fum, fumarate; ICit, isocitrate; Mal, malate; PEP, phosphoenolpyruvate; Pyr, pyruvate; Succ, succinate; SuccCoA, succinate-CoA.
Figure 4.
| Atpenin A5 and phenformin show antiviral activity against SARS-CoV-2. Calu-3 and CaCo-2 cells were pretreated for 24 hours with atpenin A5, phenformin, scyllo-inositol, and SR13800 in indicated concentrations and infected with icSARS-CoV-2-mNG (MOI 0.2 for CaCo-2 and MOI 0.5 for Calu-3). Fourty-eight hours post-infection, cells were fixed, stained with Hoechst, and measured with a Cytation3 multiplate imager. n = 3. Data represent means ± S.E.M. A Infection rate (mNG+/Hoechst+ cells) normalized to control/DMSO-treated infected cells. B Overall cell count (Hoechst+ cells) normalized to mock. C Representative fluorescence microscopy images were taken at 4-fold magnification to detect cell nuclei count (Hoechst+) and infected cells count (mNeonGreen+). Shown are Calu-3 cells treated with 2 µM of the respective compounds.
Figure 4.
| Atpenin A5 and phenformin show antiviral activity against SARS-CoV-2. Calu-3 and CaCo-2 cells were pretreated for 24 hours with atpenin A5, phenformin, scyllo-inositol, and SR13800 in indicated concentrations and infected with icSARS-CoV-2-mNG (MOI 0.2 for CaCo-2 and MOI 0.5 for Calu-3). Fourty-eight hours post-infection, cells were fixed, stained with Hoechst, and measured with a Cytation3 multiplate imager. n = 3. Data represent means ± S.E.M. A Infection rate (mNG+/Hoechst+ cells) normalized to control/DMSO-treated infected cells. B Overall cell count (Hoechst+ cells) normalized to mock. C Representative fluorescence microscopy images were taken at 4-fold magnification to detect cell nuclei count (Hoechst+) and infected cells count (mNeonGreen+). Shown are Calu-3 cells treated with 2 µM of the respective compounds.
Figure 5.
| IC50 determination against different RNA viruses in various cell lines of atpenin A5 and phenformin. Atpenin A5 and phenformin were tested for antiviral activity against two SARS-CoV-2 strains, icSARS-CoV-2-mNG (MOI 0,5), and the variant of concern Omicron (MOI 1.1), a respiratory syncytial reporter virus (RSV, MOI 0.6) and an influenza A reporter virus (IAV, MOI 1.4) expressing GFP as well as a luciferase-expressing dengue reporter virus. Cells were pretreated with the compounds for 24 hours, respectively for 1 hour (DENV, MOI 1). Fourty-eight hours post-infection, cells were fixed, stained with Hoechst, and measured with a Cytation3 multiplate reader. DENV-infected cells were lysed to measure luciferase activity. A Graphs show the infection rate normalized to untreated infected cells. µM values in the graphs refer to the specific IC50 values. B Cell count (Hoechst+ cells) normalized to mock. n =3; n = 2 (IAV, atpenin A5); n = 1 (IAV, phenformin). Data represents means ± S.E.M.
Figure 5.
| IC50 determination against different RNA viruses in various cell lines of atpenin A5 and phenformin. Atpenin A5 and phenformin were tested for antiviral activity against two SARS-CoV-2 strains, icSARS-CoV-2-mNG (MOI 0,5), and the variant of concern Omicron (MOI 1.1), a respiratory syncytial reporter virus (RSV, MOI 0.6) and an influenza A reporter virus (IAV, MOI 1.4) expressing GFP as well as a luciferase-expressing dengue reporter virus. Cells were pretreated with the compounds for 24 hours, respectively for 1 hour (DENV, MOI 1). Fourty-eight hours post-infection, cells were fixed, stained with Hoechst, and measured with a Cytation3 multiplate reader. DENV-infected cells were lysed to measure luciferase activity. A Graphs show the infection rate normalized to untreated infected cells. µM values in the graphs refer to the specific IC50 values. B Cell count (Hoechst+ cells) normalized to mock. n =3; n = 2 (IAV, atpenin A5); n = 1 (IAV, phenformin). Data represents means ± S.E.M.
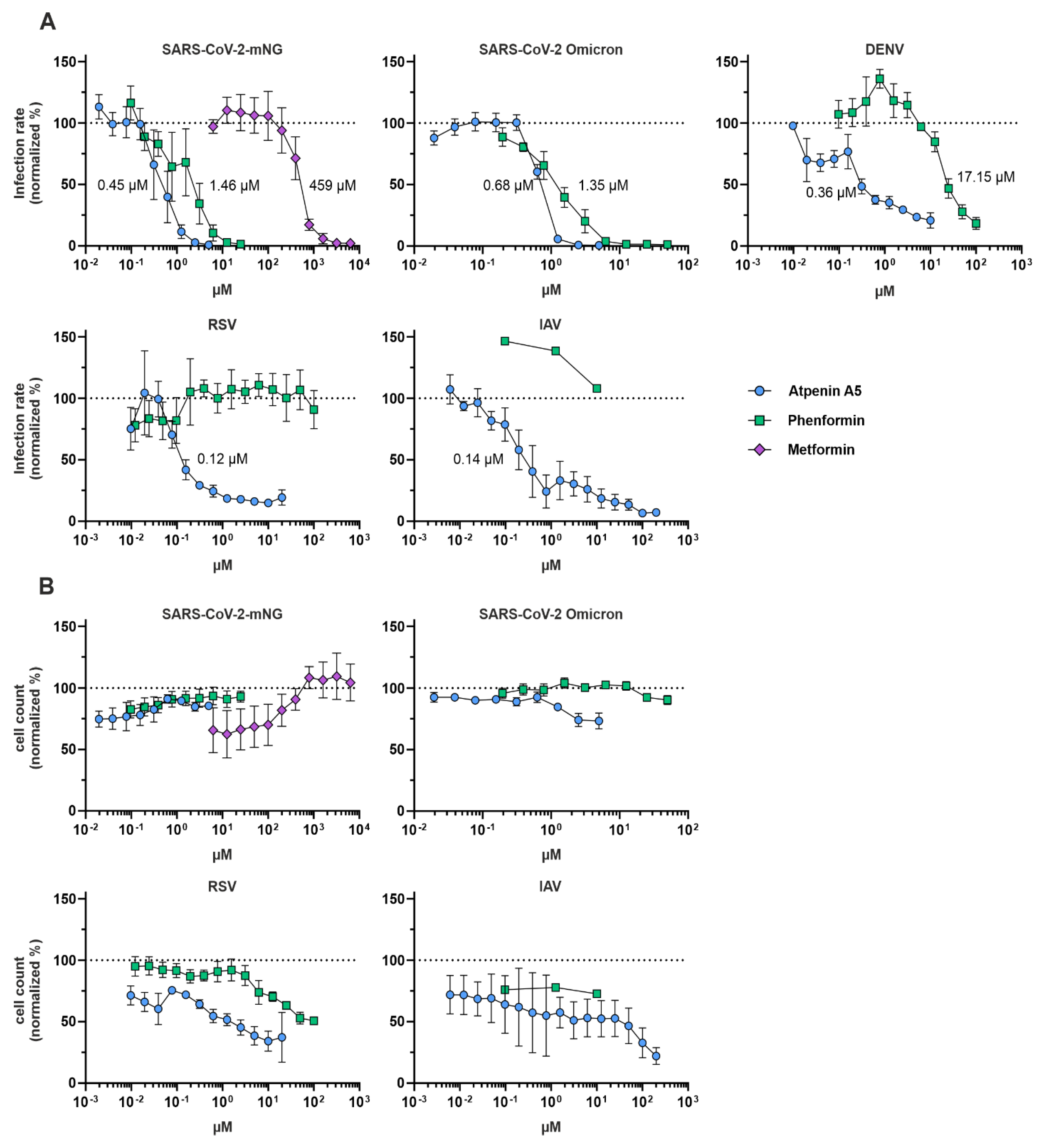
Figure 6.
| Effects of atpenin A5 and phenformin on the growth and viability of various cell lines. Calu-3, Huh7.5, and A549 cells were treated with the indicated concentrations of A atpenin A5 and B phenformin for 48 hours (A549) or 72 hours (Calu-3, Huh7.5). DMSO (1.3%) is a solvent control. The positive control is 50% DMSO to inhibit cell growth. Plates were imaged in the IncuCyte® at 37 °C, 5% CO2, every three hours to assess cell confluency. For analysis, IncuCyte® S3 Software was used. Number of replicates: Calu-3, n = 2 / 3; Huh7.5, n = 3; A549, n = 5. Data represent means ± interplate S.E.M. C Cellular metabolic activity was measured using CellTiter-Glo® or MTT assay as described in the material and methods section. Graphs show normalized MTT signal to non-treated cells or normalized luminescence signal to non-treated cells. n = 3; Data represent means ± S.E.M.
Figure 6.
| Effects of atpenin A5 and phenformin on the growth and viability of various cell lines. Calu-3, Huh7.5, and A549 cells were treated with the indicated concentrations of A atpenin A5 and B phenformin for 48 hours (A549) or 72 hours (Calu-3, Huh7.5). DMSO (1.3%) is a solvent control. The positive control is 50% DMSO to inhibit cell growth. Plates were imaged in the IncuCyte® at 37 °C, 5% CO2, every three hours to assess cell confluency. For analysis, IncuCyte® S3 Software was used. Number of replicates: Calu-3, n = 2 / 3; Huh7.5, n = 3; A549, n = 5. Data represent means ± interplate S.E.M. C Cellular metabolic activity was measured using CellTiter-Glo® or MTT assay as described in the material and methods section. Graphs show normalized MTT signal to non-treated cells or normalized luminescence signal to non-treated cells. n = 3; Data represent means ± S.E.M.
Figure 7.
| Antiviral activity of phenformin in vivo. A Golden Syrian hamsters were infected intranasally on day 0 (D0) with 105,5 TCID50 of SARS-CoV-2 Delta (B.1.617.2) virus. “Treated” (n = 5) and “Uninfected Treated” animals received 100 mg/kg phenformin by gavage once daily for five days, starting on D-2. “Vehicle” animals (n = 7) received PBS. B Weight loss was monitored on D-3 (baseline before treatment) and daily from D0 to D6. Data are shown as means ± sd. C Oropharyngeal swabs were collected daily from D1 to D6 and viral titers were assessed by RT-qPCR. D On D3 and D6 a subset of animals were euthanized and lungs were harvested to assess viral loads by RT-qPCR. Data are shown as means ± sd. *, p < 0.05 as compared by Mann Whitney test. OVT: oropharyngeal viral titers; LVT: lung viral titers; LOQ: limit of quantification.
Figure 7.
| Antiviral activity of phenformin in vivo. A Golden Syrian hamsters were infected intranasally on day 0 (D0) with 105,5 TCID50 of SARS-CoV-2 Delta (B.1.617.2) virus. “Treated” (n = 5) and “Uninfected Treated” animals received 100 mg/kg phenformin by gavage once daily for five days, starting on D-2. “Vehicle” animals (n = 7) received PBS. B Weight loss was monitored on D-3 (baseline before treatment) and daily from D0 to D6. Data are shown as means ± sd. C Oropharyngeal swabs were collected daily from D1 to D6 and viral titers were assessed by RT-qPCR. D On D3 and D6 a subset of animals were euthanized and lungs were harvested to assess viral loads by RT-qPCR. Data are shown as means ± sd. *, p < 0.05 as compared by Mann Whitney test. OVT: oropharyngeal viral titers; LVT: lung viral titers; LOQ: limit of quantification.
Table 1.
| Selectivity indices of atpenin A5 and phenformin against various pandemic RNA viruses in different cell lines.
Table 1.
| Selectivity indices of atpenin A5 and phenformin against various pandemic RNA viruses in different cell lines.
| Atpenin A5 |
IC50
|
CC50
|
selectivity index:
CC50/IC50
|
ic-SARS-CoV-2-mNG
(in Calu-3 cells)
|
0.45 µM |
MTT: 3.6 µM
CTG: 1.0 µM
IncuCyte: > 160 µM |
MTT: 8
CTG: 2.2
IncuCyte: > 355 |
SARS-CoV-2 Omicron
(in Calu-3 cells)
|
0.68 µM |
MTT: 3.6 µM
CTG: 1.0 µM
IncuCyte: > 160 µM |
MTT: 5.3
CTG: 1.5
IncuCyte: > 235 |
Dengue Virus
(in Huh7.5 cells)
|
0.36 µM |
MTT: 83.3 µM
CTG: 85.4 µM
IncuCyte: 115 µM |
MTT: 231.4
CTG: 258.8
IncuCyte: 319.4 |
Influenza A Virus
(in A549 cells)
|
0.14 µM (n = 2)
|
MTT: 114.7 µM
CTG: 49.5 µM
Incucyte: 67 µM
|
MTT: 819.3
CTG: 353.6
Incucyte: 478.6
|
RSV
(in A549 cells)
|
0.12 µM |
MTT: 114.7 µM
CTG: 49.5 µM
IncuCyte: 67 µM |
MTT: 955.8
CTG: 412.5
Incucyte: 558.3 |
| Phenformin |
IC50
|
CC50
|
selectivity index:
CC50/IC50
|
ic-SARS-CoV-2-mNG
(in Calu-3 cells)
|
1.46 µM |
MTT: 456 µM
CTG: 26.3 µM
IncuCyte: 2060 µM |
MTT: 312.3
CTG: 18.0
IncuCyte: 1411 |
SARS-CoV-2 Omicron
(in Calu-3 cells)
|
1.35 µM |
MTT: 456 µM
CTG: 26.3 µM
IncuCyte: 2060 µM |
MTT: 337.8
CTG: 19.5
IncuCyte: 1525.9 |
Dengue Virus
(in Huh7.5 cells)
|
17.15 µM |
MTT: 64.2 µM
CTG: 80.2 µM
IncuCyte: 1313 µM |
MTT: 3.7
CTG: 4.7
IncuCyte: 76.6 |
Influenza A Virus
(in A549 cells)
|
not active |
|
|
RSV
(in A459 cells)
|
not active |
|
|
Table 2.
| Single-cell sequencing datasets.
Table 2.
| Single-cell sequencing datasets.
| Dataset |
Reference |
Accession Number |
Celltype annotation source |
| BALF1 |
(Liao et al., 2020) |
GSE145926 |
Annotation via CHETAH version 1.8.0 with single cell lung atlas (GSE136831,(Adams et al., 2020)) |
| BALF2 |
(Chua et al., 2020) |
EGAS00001004481 |
Original authors (https://doi.org/10.6084/m9.figshare.12436517) |
| CALU-3 |
(Wyler et al., 2021) |
GSE148729 |
Single cell experiment |
| scH1299 |
(Wyler et al., 2021) |
GSE148729 |
Single cell experiment |
| Influenza H1N1 |
(Medaglia et al., n.d.) |
GSE191176 |
Manual annotation following methods of original publication |
| Dengue |
(Zanini et al., 2018) |
GSE116672 |
Annotation via CHETAH version 1.8.0 with PBMC single cell reference 62
|
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).