Submitted:
30 December 2022
Posted:
03 January 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
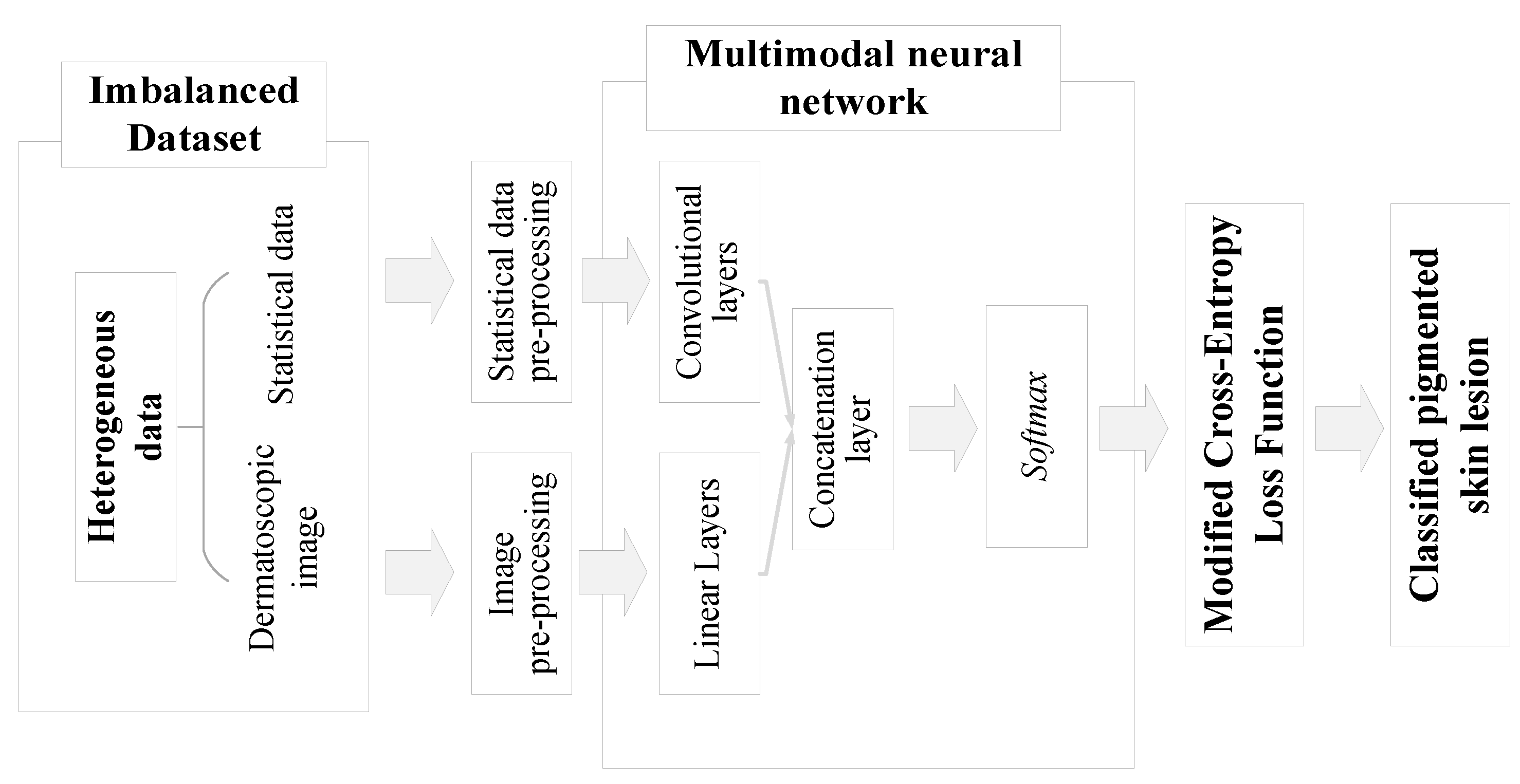
2. Materials and Methods
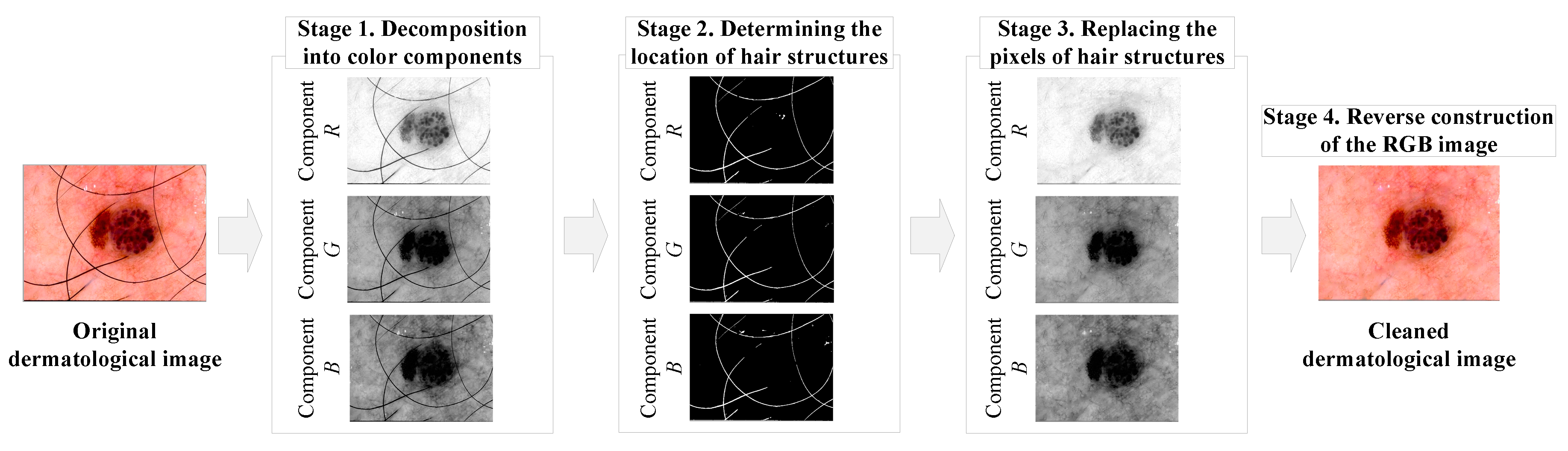
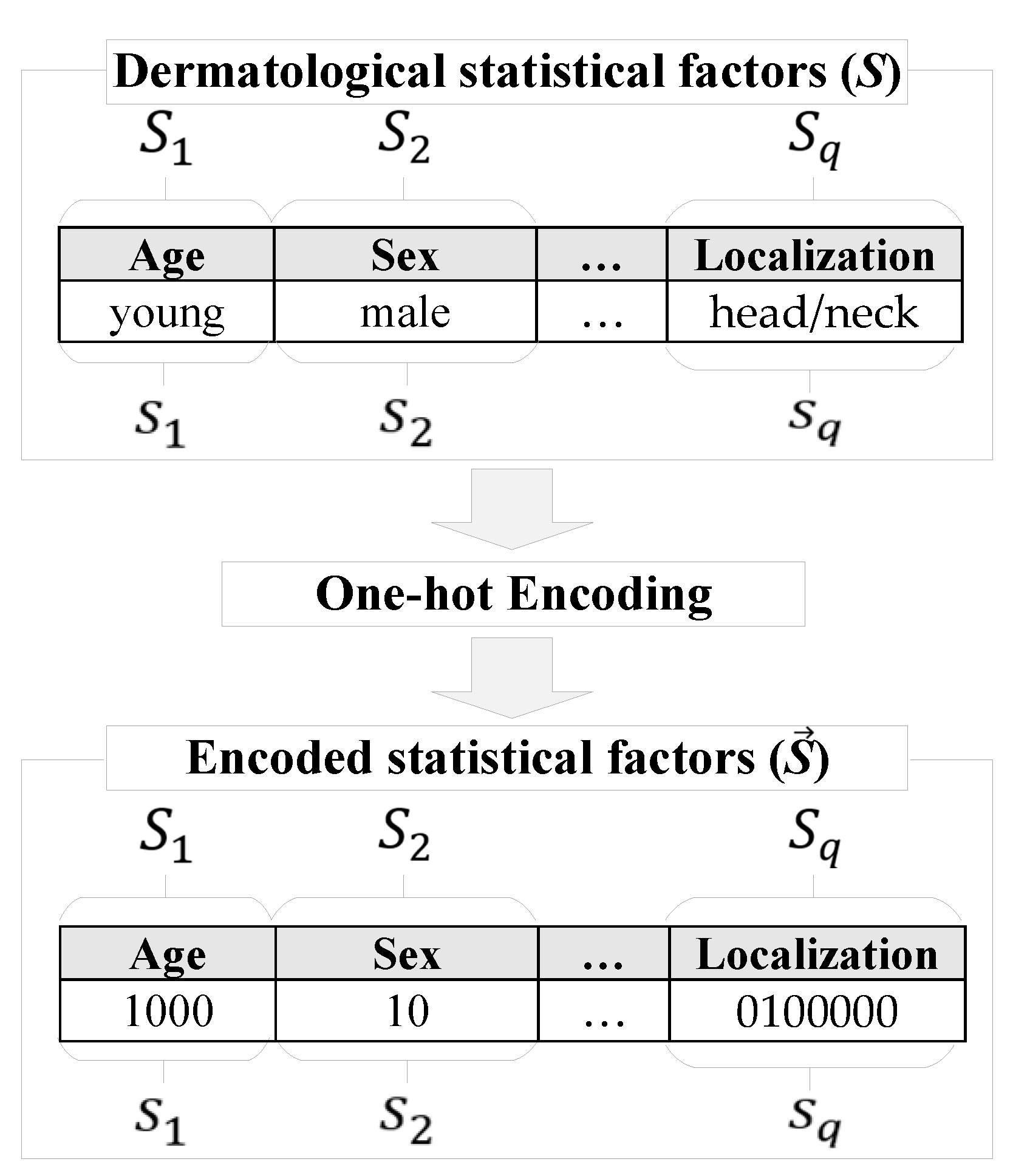
2.1. Pre-processing of heterogeneous dermatological data
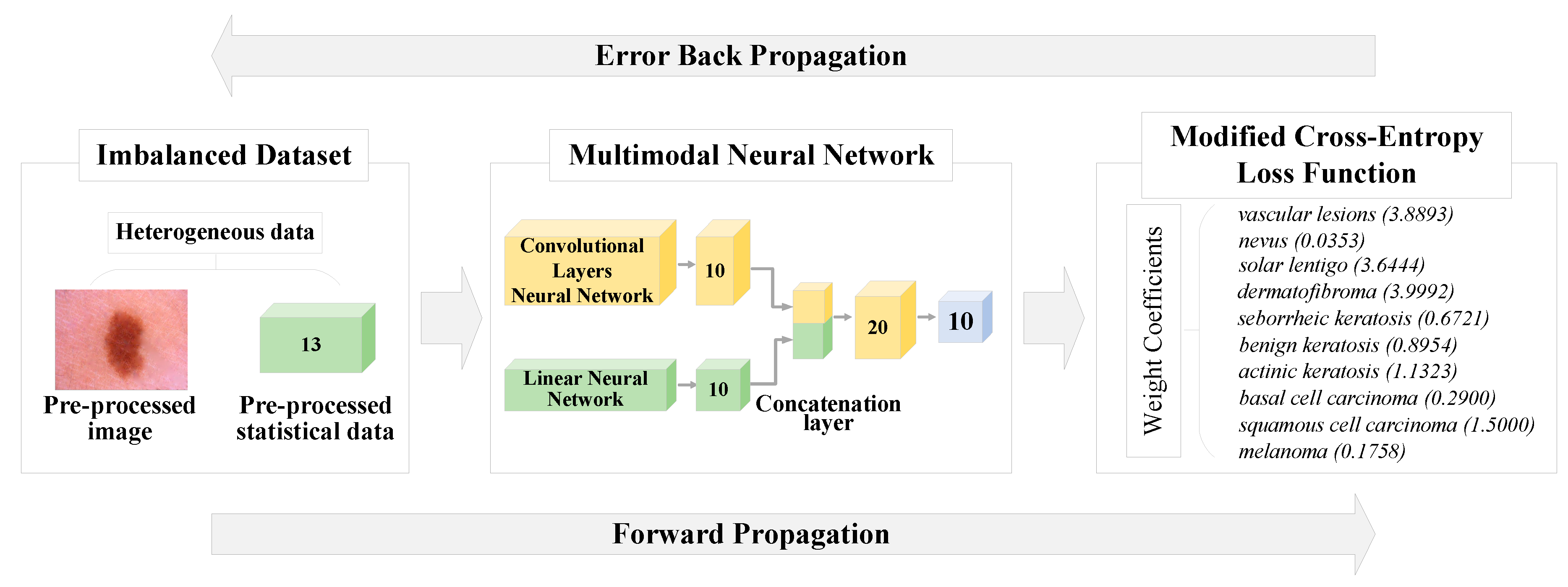
2.2. Modification of the cross-entropy loss function using weight coefficients
2.3. Multimodal neural network system for the analysis of unbalanced dermatological data
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Apalla, Z.; Lallas, A.; Sotiriou, E.; Lazaridou, E.; Ioannides, D. Epidemiological Trends in Skin Cancer. Dermatol Pract Concept 2017, 7, 1. [Google Scholar] [CrossRef]
- Hu, W.; Fang, L.; Ni, R.; Zhang, H.; Pan, G. Changing Trends in the Disease Burden of Non-Melanoma Skin Cancer Globally from 1990 to 2019 and Its Predicted Level in 25 Years. BMC Cancer 2022, 22, 1–11. [Google Scholar] [CrossRef]
- Garbe, C.; Leiter, U. Epidemiology of Melanoma and Nonmelanoma Skin Cancer-the Role of Sunlight. Adv Exp Med Biol 2008, 624, 89–103. [Google Scholar] [CrossRef]
- Fitzpatrick, T.B. Pathophysiology of Hypermelanoses. Clinical Drug Investigation 1995 10:2 2012, 10, 17–26. [Google Scholar] [CrossRef]
- Pathak, M.A.; Jimbow, K.; Szabo, G.; Fitzpatrick, T.B. Sunlight and Melanin Pigmentation. Photochemical and Photobiological Reviews 1976, 211–239. [Google Scholar] [CrossRef]
- Ciążyńska, M.; Kamińska-Winciorek, G.; Lange, D.; Lewandowski, B.; Reich, A.; Sławińska, M.; Pabianek, M.; Szczepaniak, K.; Hankiewicz, A.; Ułańska, M.; et al. The Incidence and Clinical Analysis of Non-Melanoma Skin Cancer. Scientific Reports 2021 11:1 2021, 11, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Melanoma Awareness Month 2022 – IARC. Available online: https://www.iarc.who.int/news-events/melanoma-awareness-month-2022/ (accessed on 14 December 2022).
- Arnold, M.; Singh, D.; Laversanne, M.; Vignat, J.; Vaccarella, S.; Meheus, F.; Cust, A.E.; de Vries, E.; Whiteman, D.C.; Bray, F. Global Burden of Cutaneous Melanoma in 2020 and Projections to 2040. JAMA Dermatol 2022, 158, 495–503. [Google Scholar] [CrossRef] [PubMed]
- Allais, B.S.; Beatson, M.; Wang, H.; Shahbazi, S.; Bijelic, L.; Jang, S.; Venna, S. Five-Year Survival in Patients with Nodular and Superficial Spreading Melanomas in the US Population. J Am Acad Dermatol 2021, 84, 1015–1022. [Google Scholar] [CrossRef]
- Balch, C.M.; Soong, S.J.; Murad, T.M.; Ingalls, A.L.; Maddox, W.A. A Multifactorial Analysis of Melanoma: III. Prognostic Factors in Melanoma Patients with Lymph Node Metastases (Stage II). Ann Surg 1981, 193, 377. [Google Scholar] [CrossRef]
- Lideikaitė, A.; Mozūraitienė, J.; Letautienė, S. Analysis of Prognostic Factors for Melanoma Patients. Acta Med Litu 2017, 24, 25–34. [Google Scholar] [CrossRef]
- Amaral, T.; Garbe, C. Non-Melanoma Skin Cancer: New and Future Synthetic Drug Treatments. Expert Opin Pharmacother 2017, 18, 689–699. [Google Scholar] [CrossRef] [PubMed]
- Leigh, I.M. Progress in Skin Cancer: The U.K. Experience. British Journal of Dermatology 2014, 171, 443–445. [Google Scholar] [CrossRef] [PubMed]
- Eide, M.J.; Krajenta, R.; Johnson, D.; Long, J.J.; Jacobsen, G.; Asgari, M.M.; Lim, H.W.; Johnson, C.C. Identification of Patients With Nonmelanoma Skin Cancer Using Health Maintenance Organization Claims Data. Am J Epidemiol 2010, 171, 123–128. [Google Scholar] [CrossRef] [PubMed]
- Lucena, S.R.; Salazar, N.; Gracia-Cazaña, T.; Zamarrón, A.; González, S.; Juarranz, Á.; Gilaberte, Y. Combined Treatments with Photodynamic Therapy for Non-Melanoma Skin Cancer. International Journal of Molecular Sciences 2015, Vol. 16, Pages 25912-25933 2015, 16, 25912–25933. [Google Scholar] [CrossRef] [PubMed]
- Chua, B.; Jackson, J.E.; Lin, C.; Veness, M.J. Radiotherapy for Early Non-Melanoma Skin Cancer. Oral Oncol 2019, 98, 96–101. [Google Scholar] [CrossRef] [PubMed]
- Organization, W.H. ICD-11 for Mortality and Morbidity Statistics (2018). 2018.
- Fung, K.W.; Xu, J.; Bodenreider, O. The New International Classification of Diseases 11th Edition: A Comparative Analysis with ICD-10 and ICD-10-CM. Journal of the American Medical Informatics Association 2020, 27, 738–746. [Google Scholar] [CrossRef] [PubMed]
- Ciążyńska, M.; Kamińska-Winciorek, G.; Lange, D.; Lewandowski, B.; Reich, A.; Sławińska, M.; Pabianek, M.; Szczepaniak, K.; Hankiewicz, A.; Ułańska, M.; et al. The Incidence and Clinical Analysis of Non-Melanoma Skin Cancer. Scientific Reports 2021 11:1 2021, 11, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Linares, M.A.; Zakaria, A.; Nizran, P. Skin Cancer. Primary Care - Clinics in Office Practice 2015, 42, 645–659. [Google Scholar] [CrossRef] [PubMed]
- Apalla, Z.; Lallas, A.; Sotiriou, E.; Lazaridou, E.; Ioannides, D. Epidemiological Trends in Skin Cancer. Dermatol Pract Concept 2017, 7, 1. [Google Scholar] [CrossRef]
- Juszczak, A.M.; Wöelfle, U.; Končić, M.Z.; Tomczyk, M. Skin Cancer, Including Related Pathways and Therapy and the Role of Luteolin Derivatives as Potential Therapeutics. Med Res Rev 2022, 42, 1423–1462. [Google Scholar] [CrossRef]
- Collier, V.; Musicante, M; Patel, T; Liu-Smith, F. Sex Disparity in Skin Carcinogenesis and Potential Influence of Sex Hormones. Skin Health and Disease 2021, 1, e27. [Google Scholar] [CrossRef] [PubMed]
- Apalla, Z.; Calzavara-Pinton, P.; Lallas, A.; Argenziano, G.; Kyrgidis, A.; Crotti, S.; Facchetti, F.; Monari, P.; Gualdi, G. Histopathological Study of Perilesional Skin in Patients Diagnosed with Nonmelanoma Skin Cancer. Clin Exp Dermatol 2016, 41, 21–25. [Google Scholar] [CrossRef]
- Ring, C.; Cox, N.; Lee, J.B. Dermatoscopy. Clin Dermatol 2021, 39, 635–642. [Google Scholar] [CrossRef] [PubMed]
- Nami, N.; Giannini, E.; Burroni, M.; Fimiani, M.; Rubegni, P. Teledermatology: State-of-the-Art and Future Perspectives. Expert Review of Dermatology 2014, 7, 1–3. [Google Scholar] [CrossRef]
- Sinz, C.; Tschandl, P.; Rosendahl, C.; Akay, B.N.; Argenziano, G.; Blum, A.; Braun, R.P.; Cabo, H.; Gourhant, J.Y.; Kreusch, J.; et al. Accuracy of Dermatoscopy for the Diagnosis of Nonpigmented Cancers of the Skin. J Am Acad Dermatol 2017, 77, 1100–1109. [Google Scholar] [CrossRef] [PubMed]
- Vestergaard, M.E.; Macaskill, P.; Holt, P.E.; Menzies, S.W. Dermoscopy Compared with Naked Eye Examination for the Diagnosis of Primary Melanoma: A Meta-Analysis of Studies Performed in a Clinical Setting. British Journal of Dermatology 2008, 159, 669–676. [Google Scholar] [CrossRef] [PubMed]
- Haenssle, H.A.; Fink, C.; Schneiderbauer, R.; Toberer, F.; Buhl, T.; Blum, A.; Kalloo, A.; ben Hadj Hassen, A.; Thomas, L.; Enk, A.; et al. Man against Machine: Diagnostic Performance of a Deep Learning Convolutional Neural Network for Dermoscopic Melanoma Recognition in Comparison to 58 Dermatologists. Annals of Oncology 2018, 29, 1836–1842. [Google Scholar] [CrossRef] [PubMed]
- Brochez, L.; Verhaeghe, E.; Grosshans, E.; Haneke, E.; Piérard, G.; Ruiter, D.; Naeyaert, J.M. Inter-Observer Variation in the Histopathological Diagnosis of Clinically Suspicious Pigmented Skin Lesions. J Pathol 2002, 196, 459–466. [Google Scholar] [CrossRef] [PubMed]
- Lodha, S.; Saggar, S.; Celebi, J.T.; Silvers, D.N. Discordance in the Histopathologic Diagnosis of Difficult Melanocytic Neoplasms in the Clinical Setting. J Cutan Pathol 2008, 35, 349–352. [Google Scholar] [CrossRef] [PubMed]
- Haggenmüller, S.; Maron, R.C.; Hekler, A.; Utikal, J.S.; Barata, C.; Barnhill, R.L.; Beltraminelli, H.; Berking, C.; Betz-Stablein, B.; Blum, A.; et al. Skin Cancer Classification via Convolutional Neural Networks: Systematic Review of Studies Involving Human Experts. Eur J Cancer 2021, 156, 202–216. [Google Scholar] [CrossRef]
- Yan, Y.; Hong, S.; Zhang, W.; Li, H. Artificial Intelligence in Skin Diseases: Fulfilling Its Potentials to Meet the Real Needs in Dermatology Practice. Health Data Science 2022, 2022, 1–2. [Google Scholar] [CrossRef]
- Wiens, J.; Saria, S.; Sendak, M.; Ghassemi, M.; Liu, V.X.; Doshi-Velez, F.; Jung, K.; Heller, K.; Kale, D.; Saeed, M.; et al. Author Correction: Do No Harm: A Roadmap for Responsible Machine Learning for Health Care (Nature Medicine, (2019), 25, 9, (1337-1340), 10.1038/S41591-019-0548-6). Nat Med 2019, 25, 1627. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; Chen, L.; Shang, T. Stream of Unbalanced Medical Big Data Using Convolutional Neural Network. IEEE Access 2020, 8, 81310–81319. [Google Scholar] [CrossRef]
- Yang, H.; Li, X.; Cao, H.; Cui, Y.; Luo, Y.; Liu, J.; Zhang, Y. Using Machine Learning Methods to Predict Hepatic Encephalopathy in Cirrhotic Patients with Unbalanced Data. Comput Methods Programs Biomed 2021, 211, 106420. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Barea, F.J.; Jerez, J.M.; Franco, L. Improving Classification Accuracy Using Data Augmentation on Small Data Sets. Expert Syst Appl 2020, 161, 113696. [Google Scholar] [CrossRef]
- Buda, M.; Maki, A.; Mazurowski, M.A. A Systematic Study of the Class Imbalance Problem in Convolutional Neural Networks. Neural Networks 2018, 106, 249–259. [Google Scholar] [CrossRef] [PubMed]
- Czarnowski, I. Weighted Ensemble with One-Class Classification and Over-Sampling and Instance Selection (WECOI): An Approach for Learning from Imbalanced Data Streams. J Comput Sci 2022, 61, 101614. [Google Scholar] [CrossRef]
- Bhowan, U.; Johnston, M.; Zhang, M.; Yao, X. Evolving Diverse Ensembles Using Genetic Programming for Classification with Unbalanced Data. IEEE Transactions on Evolutionary Computation 2013, 17, 368–386. [Google Scholar] [CrossRef]
- Mikołajczyk, A.; Grochowski, M. Data Augmentation for Improving Deep Learning in Image Classification Problem. 2018 International Interdisciplinary PhD Workshop, IIPhDW 2018 2018, 117–122. [CrossRef]
- Chen, N.; Xu, Z.; Liu, Z.; Chen, Y.; Miao, Y.; Li, Q.; Hou, Y.; Wang, L. Data Augmentation and Intelligent Recognition in Pavement Texture Using a Deep Learning. IEEE Transactions on Intelligent Transportation Systems 2022. [Google Scholar] [CrossRef]
- Sadhukhan, P.; Palit, S. Reverse-Nearest Neighborhood Based Oversampling for Imbalanced, Multi-Label Datasets. Pattern Recognit Lett 2019, 125, 813–820. [Google Scholar] [CrossRef]
- Koziarski, M. Radial-Based Undersampling for Imbalanced Data Classification. Pattern Recognit 2020, 102, 107262. [Google Scholar] [CrossRef]
- Zhu, Y.; Jia, C.; Li, F.; Song, J. Inspector: A Lysine Succinylation Predictor Based on Edited Nearest-Neighbor Undersampling and Adaptive Synthetic Oversampling. Anal Biochem 2020, 593, 113592. [Google Scholar] [CrossRef] [PubMed]
- Kubus, M. Evaluation of Resampling Methods in the Class Unbalance Problem. Econometrics. Ekonometria. Advances in Applied Data Analytics 2020, 24, 39–50. [Google Scholar] [CrossRef]
- Wang, C.; Deng, C.; Wang, S. Imbalance-XGBoost: Leveraging Weighted and Focal Losses for Binary Label-Imbalanced Classification with XGBoost. Pattern Recognit Lett 2020, 136, 190–197. [Google Scholar] [CrossRef]
- Ryou, S.; Jeong, S.-G.; Perona, P. Anchor Loss: Modulating Loss Scale Based on Prediction Difficulty 2019, 5992–6001.
- Yu, H.; Sun, C.; Yang, X.; Zheng, S.; Wang, Q.; Xi, X. LW-ELM: A Fast and Flexible Cost-Sensitive Learning Framework for Classifying Imbalanced Data. IEEE Access 2018, 6, 28488–28500. [Google Scholar] [CrossRef]
- Turkay, C.; Lundervold, A.; Lundervold, A.J.; Hauser, H. Hypothesis Generation by Interactive Visual Exploration of Heterogeneous Medical Data. Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics) 2013, 7947 LNCS, 1–12. [Google Scholar] [CrossRef]
- Yue, L.; Tian, D.; Chen, W.; Han, X.; Yin, M. Deep Learning for Heterogeneous Medical Data Analysis. World Wide Web 2020, 23, 2715–2737. [Google Scholar] [CrossRef]
- Cios, K.J.; William Moore, G. Uniqueness of Medical Data Mining. Artif Intell Med 2002, 26, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Lama, N.; Kasmi, R.; Hagerty, J.R.; Stanley, R.J.; Young, R.; Miinch, J.; Nepal, J.; Nambisan, A.; Stoecker, W. v. ChimeraNet: U-Net for Hair Detection in Dermoscopic Skin Lesion Images. Journal of Digital Imaging 2022, 2022, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Abbas, Q.; Celebi, M.E.; García, I.F. Hair Removal Methods: A Comparative Study for Dermoscopy Images. Biomed Signal Process Control 2011, 6, 395–404. [Google Scholar] [CrossRef]
- Lyakhov, P.A.; Lyakhova, U.A.; Nagornov, N.N. System for the Recognizing of Pigmented Skin Lesions with Fusion and Analysis of Heterogeneous Data Based on a Multimodal Neural Network. Cancers 2022, Vol. 14, Page 1819 2022, 14, 1819. [Google Scholar] [CrossRef]
- Cerda, P.; Varoquaux, G.; Kégl, B. Similarity Encoding for Learning with Dirty Categorical Variables. Mach Learn 2018, 107, 1477–1494. [Google Scholar] [CrossRef]
- Al-Shehari, T.; Alsowail, R.A. An Insider Data Leakage Detection Using One-Hot Encoding, Synthetic Minority Oversampling and Machine Learning Techniques. Entropy 2021, 23, 1258. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, P.; Bautista, M.A.; Gonzàlez, J.; Escalera, S. Beyond One-Hot Encoding: Lower Dimensional Target Embedding. Image Vis Comput 2018, 75, 21–31. [Google Scholar] [CrossRef]
- Potdar, K.; Pardawala, T.S.; Pai, C.D. A Comparative Study of Categorical Variable Encoding Techniques for Neural Network Classifiers. Int J Comput Appl 2017, 175, 7–9. [Google Scholar] [CrossRef]
- Wang, Q.; Ma, Y.; Zhao, K.; Tian, Y. A Comprehensive Survey of Loss Functions in Machine Learning. Annals of Data Science 2022, 9, 187–212. [Google Scholar] [CrossRef]
- Kim, Y.; Lee, Y.; Jeon, M. Imbalanced Image Classification with Complement Cross Entropy. Pattern Recognit Lett 2021, 151, 33–40. [Google Scholar] [CrossRef]
- Ho, Y.; Wookey, S. The Real-World-Weight Cross-Entropy Loss Function: Modeling the Costs of Mislabeling. IEEE Access 2020, 8, 4806–4813. [Google Scholar] [CrossRef]
- Jodelet, Q.; Liu, X.; Murata, T. Balanced Softmax Cross-Entropy for Incremental Learning. Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics) 2021, 12892 LNCS, 385–396. [Google Scholar] [CrossRef]
- Tasci, E.; Zhuge, Y.; Camphausen, K.; Krauze, A. v. Bias and Class Imbalance in Oncologic Data—Towards Inclusive and Transferrable AI in Large Scale Oncology Data Sets. Cancers 2022, Vol. 14, Page 2897 2022, 14, 2897. [Google Scholar] [CrossRef]
- Huynh, T.; Nibali, A.; He, Z. Semi-Supervised Learning for Medical Image Classification Using Imbalanced Training Data. Comput Methods Programs Biomed 2022, 216, 106628. [Google Scholar] [CrossRef] [PubMed]
- Foahom Gouabou, A.C.; Iguernaissi, R.; Damoiseaux, J.L.; Moudafi, A.; Merad, D. End-to-End Decoupled Training: A Robust Deep Learning Method for Long-Tailed Classification of Dermoscopic Images for Skin Lesion Classification. Electronics 2022, Vol. 11, Page 3275 2022, 11, 3275. [Google Scholar] [CrossRef]
- Vo, N.H.; Won, Y. Classification of Unbalanced Medical Data with Weighted Regularized Least Squares. Proceedings of the Frontiers in the Convergence of Bioscience and Information Technologies, FBIT 2007 2007, 347–352. [CrossRef]
- Aurelio, Y.S.; de Almeida, G.M.; de Castro, C.L.; Braga, A.P. Learning from Imbalanced Data Sets with Weighted Cross-Entropy Function. Neural Process Lett 2019, 50, 1937–1949. [Google Scholar] [CrossRef]
- Dong, Y.; Shen, X.; Jiang, Z.; Wang, H. Recognition of Imbalanced Underwater Acoustic Datasets with Exponentially Weighted Cross-Entropy Loss. Applied Acoustics 2021, 174, 107740. [Google Scholar] [CrossRef]
- Wang, S.; Yin, Y.; Wang, D.; Wang, Y.; Jin, Y. Interpretability-Based Multimodal Convolutional Neural Networks for Skin Lesion Diagnosis. IEEE Trans Cybern 2021. [Google Scholar] [CrossRef] [PubMed]
- Goh, G.; †, N.C.; †, C.V.; Carter, S.; Petrov, M.; Schubert, L.; Radford, A.; Olah, C. Multimodal Neurons in Artificial Neural Networks. Distill 2021, 6, e30. [Google Scholar] [CrossRef]
- Liu, K.; Li, Y.; Xu, N.; Natarajan, P. Learn to Combine Modalities in Multimodal Deep Learning. 2018.
- O’Shea, K.; Nash, R. An Introduction to Convolutional Neural Networks. 2015. [CrossRef]
- Lyu, J.; Shi, H.; Zhang, J.; Norvilitis, J. Prediction Model for Suicide Based on Back Propagation Neural Network and Multilayer Perceptron. Front Neuroinform 2022, 16, 79. [Google Scholar] [CrossRef]
- Siegel, J.A.; Korgavkar, K.; Weinstock, M.A. Current Perspective on Actinic Keratosis: A Review. British Journal of Dermatology 2017, 177, 350–358. [Google Scholar] [CrossRef]
- Huang, G.; Liu, Z.; van der Maaten, L.; Weinberger, K.Q. Densely Connected Convolutional Networks 2017, 4700–4708.
- Szegedy, C.; Ioffe, S.; Vanhoucke, V.; Alemi, A.A. Inception-v4, Inception-ResNet and the Impact of Residual Connections on Learning. 31st AAAI Conference on Artificial Intelligence, AAAI 2017 2016, 4278–4284. [CrossRef]
- Xie, S.; Girshick, R.; Dollár, P.; Tu, Z.; He, K. Aggregated Residual Transformations for Deep Neural Networks. Proceedings - 30th IEEE Conference on Computer Vision and Pattern Recognition, CVPR 2017 2016, 2017-January, 5987–5995. [CrossRef]
- Alzubaidi, L.; Zhang, J.; Humaidi, A.J.; Al-Dujaili, A.; Duan, Y.; Al-Shamma, O.; Santamaría, J.; Fadhel, M.A.; Al-Amidie, M.; Farhan, L. Review of Deep Learning: Concepts, CNN Architectures, Challenges, Applications, Future Directions. Journal of Big Data 2021 8:1 2021, 8, 1–74. [Google Scholar] [CrossRef] [PubMed]
- Chicco, D.; Jurman, G. The Advantages of the Matthews Correlation Coefficient (MCC) over F1 Score and Accuracy in Binary Classification Evaluation. BMC Genomics 2020, 21, 1–13. [Google Scholar] [CrossRef]
- Yap, J.; Yolland, W.; Tschandl, P. Multimodal Skin Lesion Classification Using Deep Learning. Exp Dermatol 2018, 27, 1261–1267. [Google Scholar] [CrossRef]
- He, X.; Wang, Y.; Zhao, S.; Chen, X. Co-Attention Fusion Network for Multimodal Skin Cancer Diagnosis. Pattern Recognit 2023, 133, 108990. [Google Scholar] [CrossRef]
- Chen, Q.; Li, M.; Chen, C.; Zhou, P.; Lv, X.; Chen, C. MDFNet: Application of Multimodal Fusion Method Based on Skin Image and Clinical Data to Skin Cancer Classification. J Cancer Res Clin Oncol 2022, 1–13. [Google Scholar] [CrossRef]
- Brinker, T.J.; Hekler, A.; Enk, A.H.; Klode, J.; Hauschild, A.; Berking, C.; Schilling, B.; Haferkamp, S.; Schadendorf, D.; Holland-Letz, T.; et al. Deep Learning Outperformed 136 of 157 Dermatologists in a Head-to-Head Dermoscopic Melanoma Image Classification Task. Eur J Cancer 2019, 113, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Esteva, A.; Kuprel, B.; Novoa, R.A.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-Level Classification of Skin Cancer with Deep Neural Networks. Nature 2017, 542, 115–118. [Google Scholar] [CrossRef] [PubMed]
- Brinker, T.J.; Hekler, A.; Enk, A.H.; Berking, C.; Haferkamp, S.; Hauschild, A.; Weichenthal, M.; Klode, J.; Schadendorf, D.; Holland-Letz, T.; et al. Deep Neural Networks Are Superior to Dermatologists in Melanoma Image Classification. Eur J Cancer 2019, 119, 11–17. [Google Scholar] [CrossRef] [PubMed]
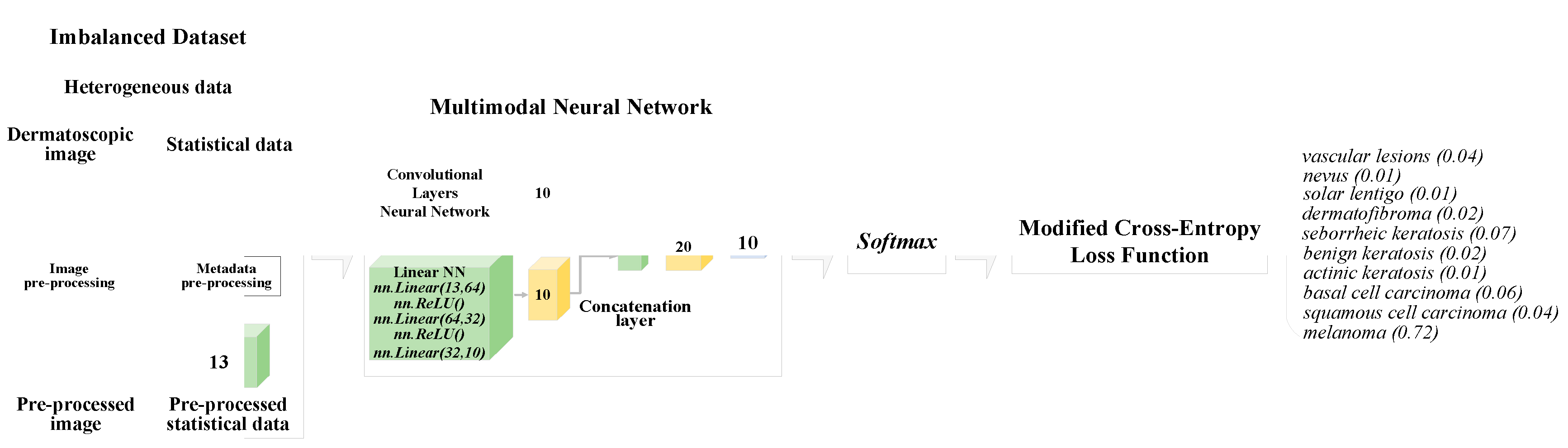
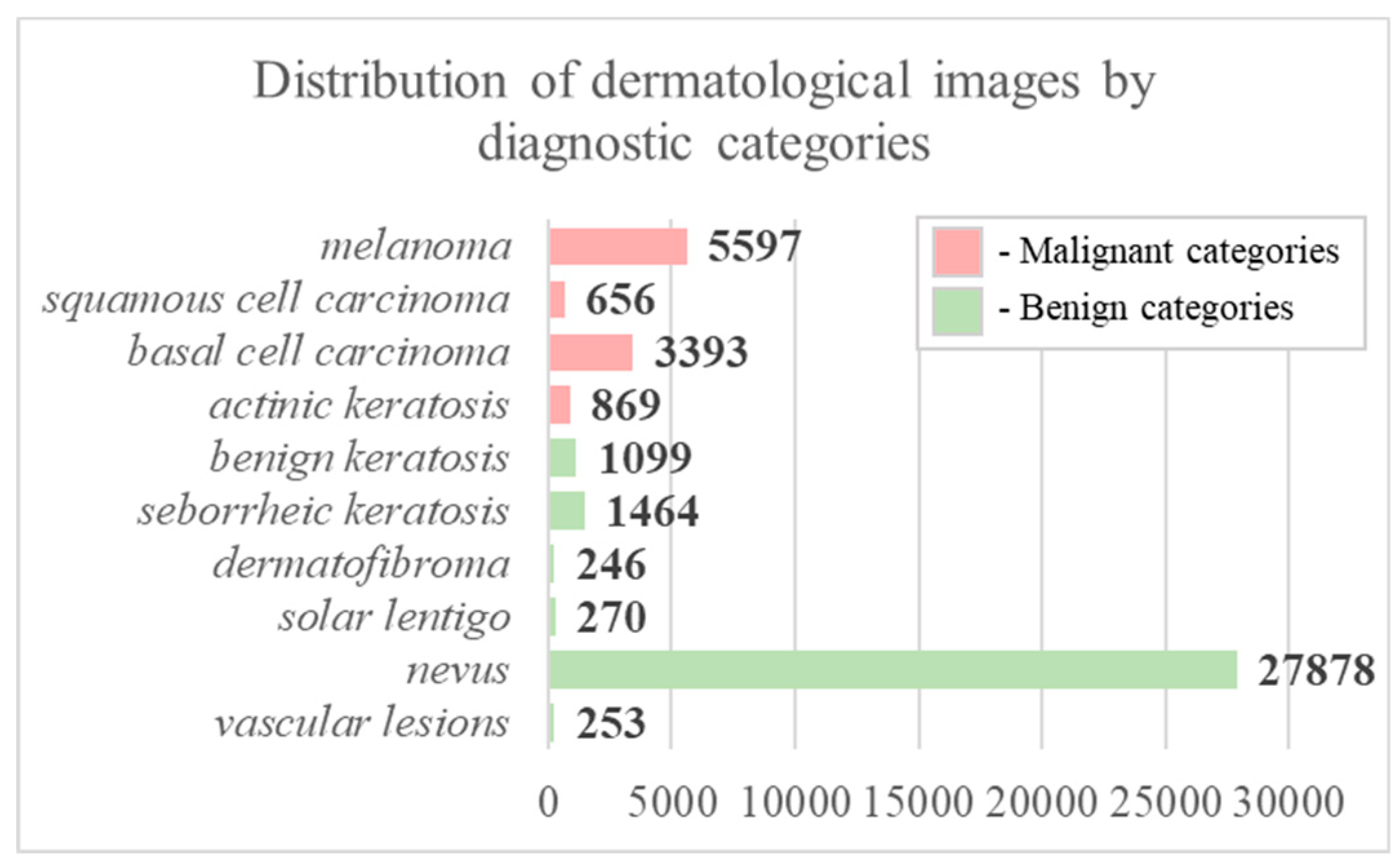
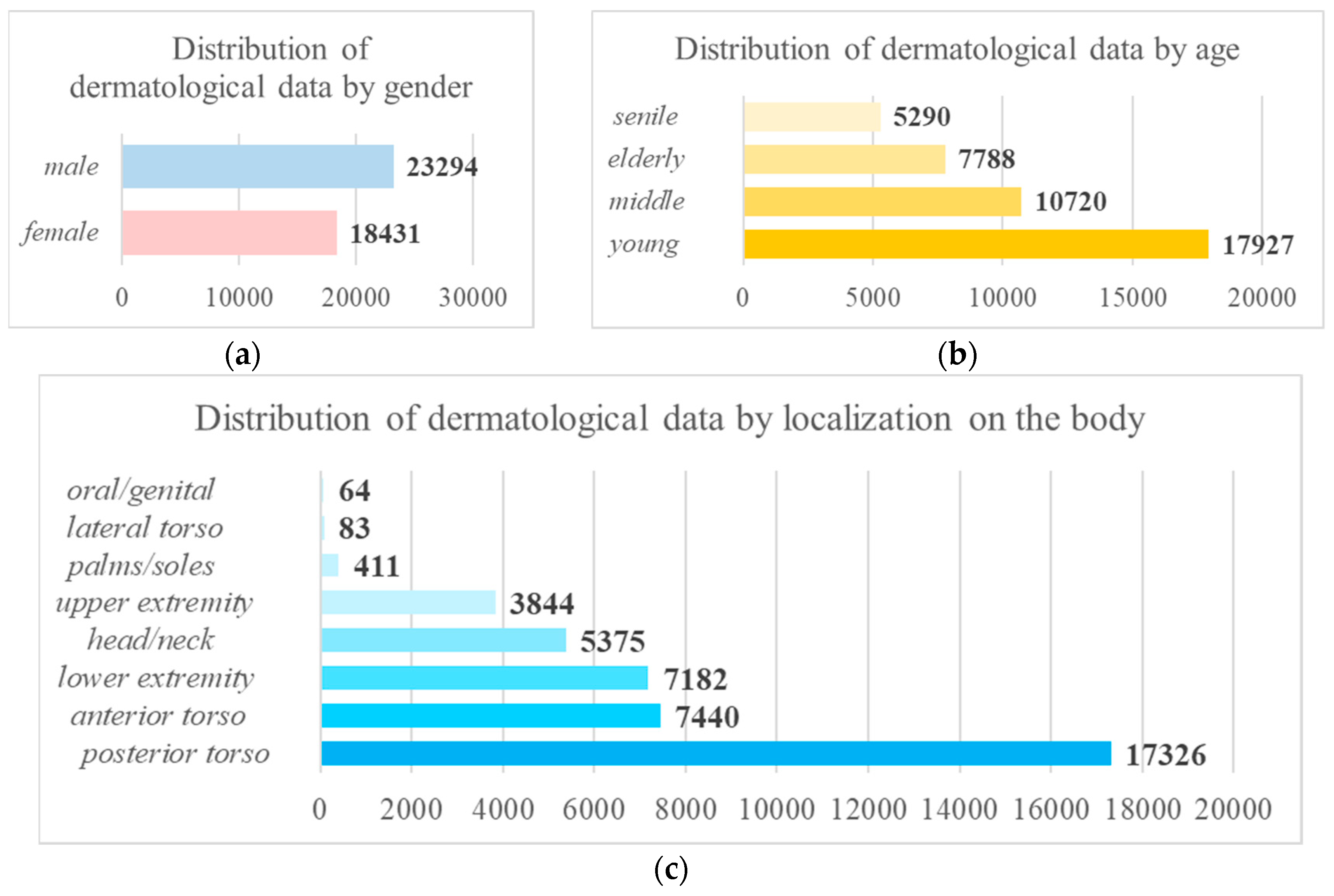
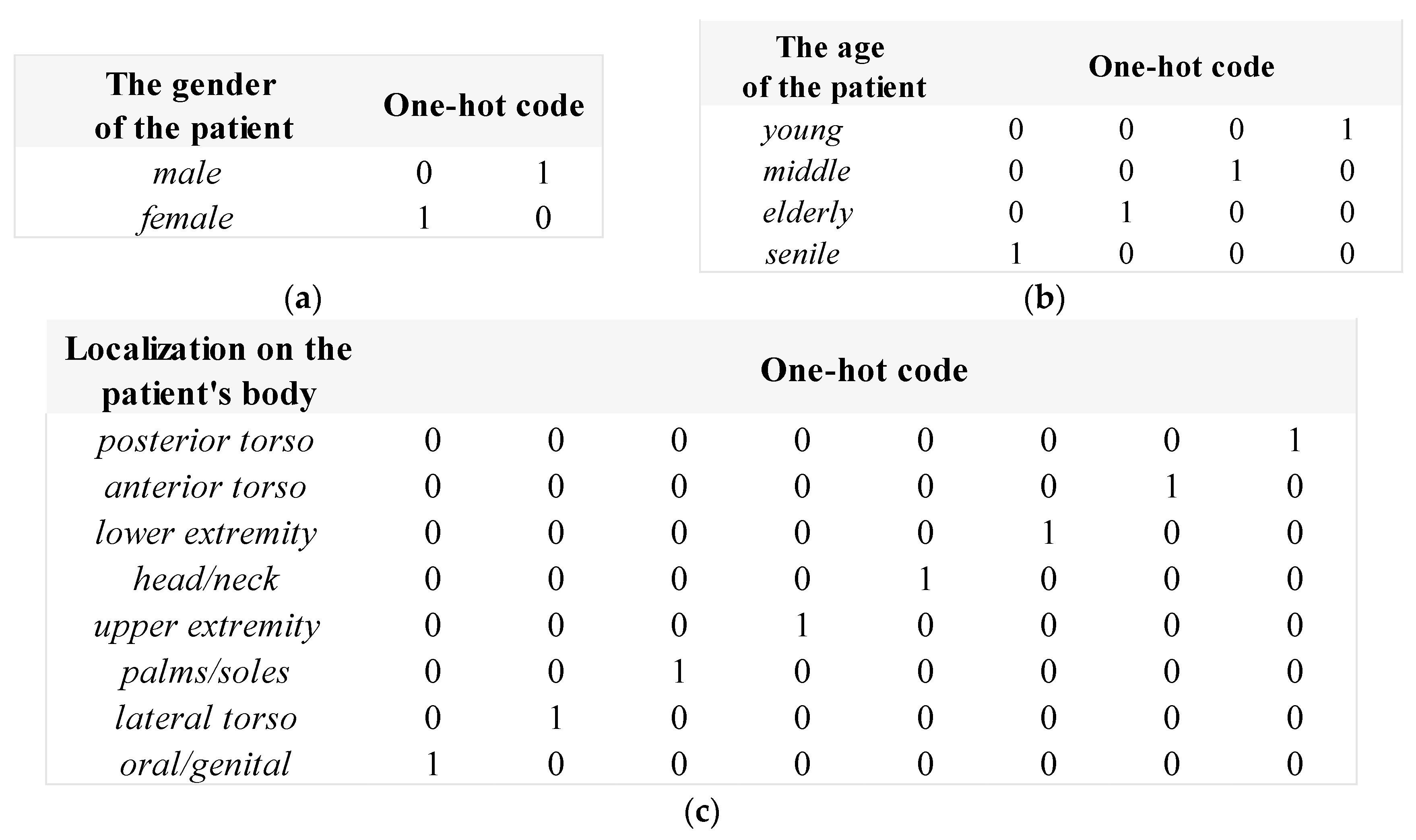
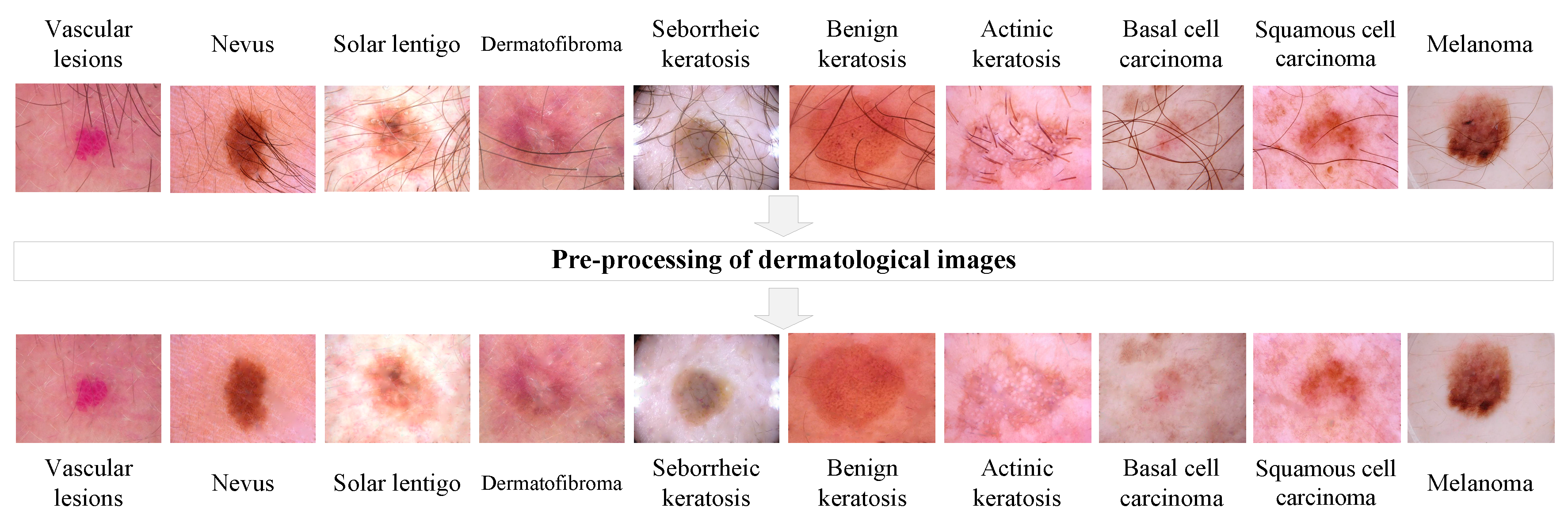
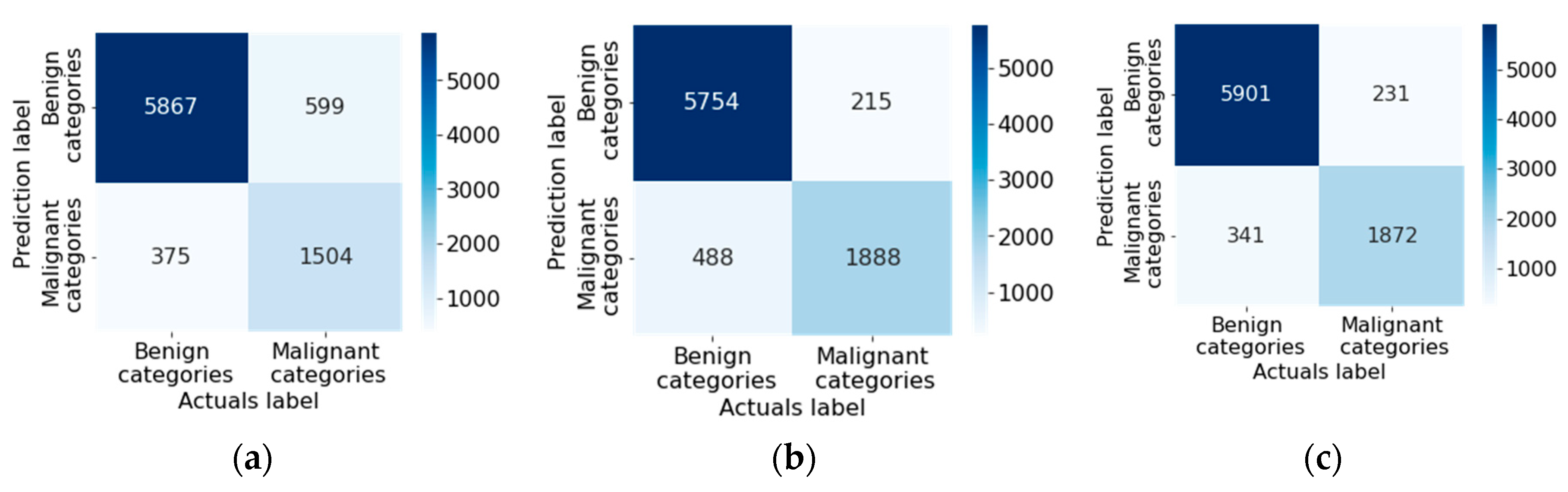
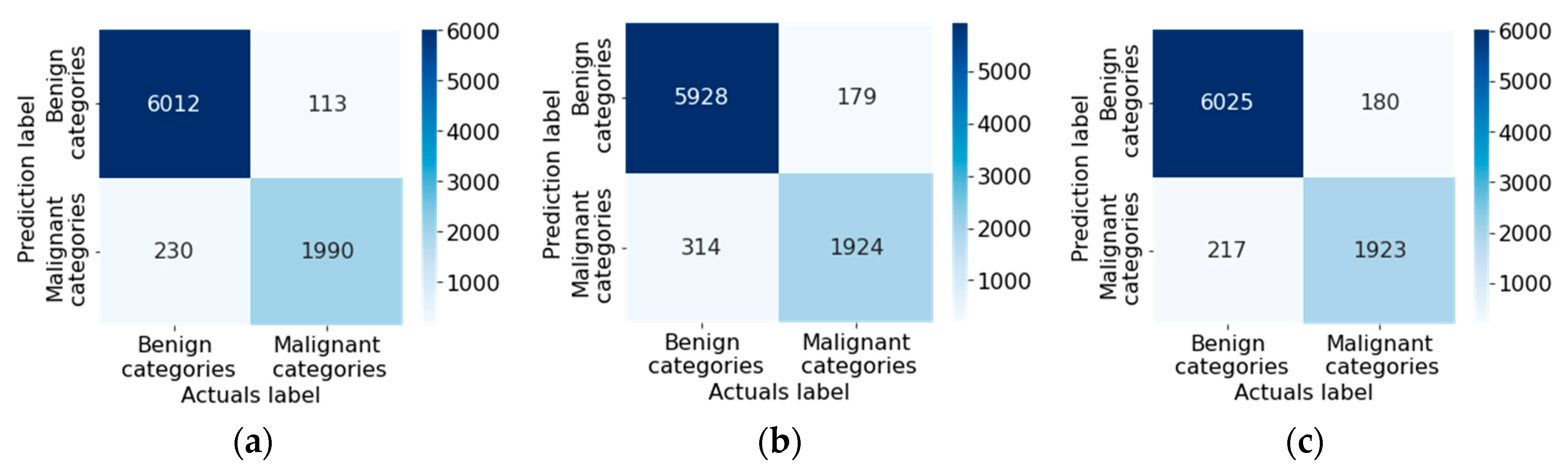
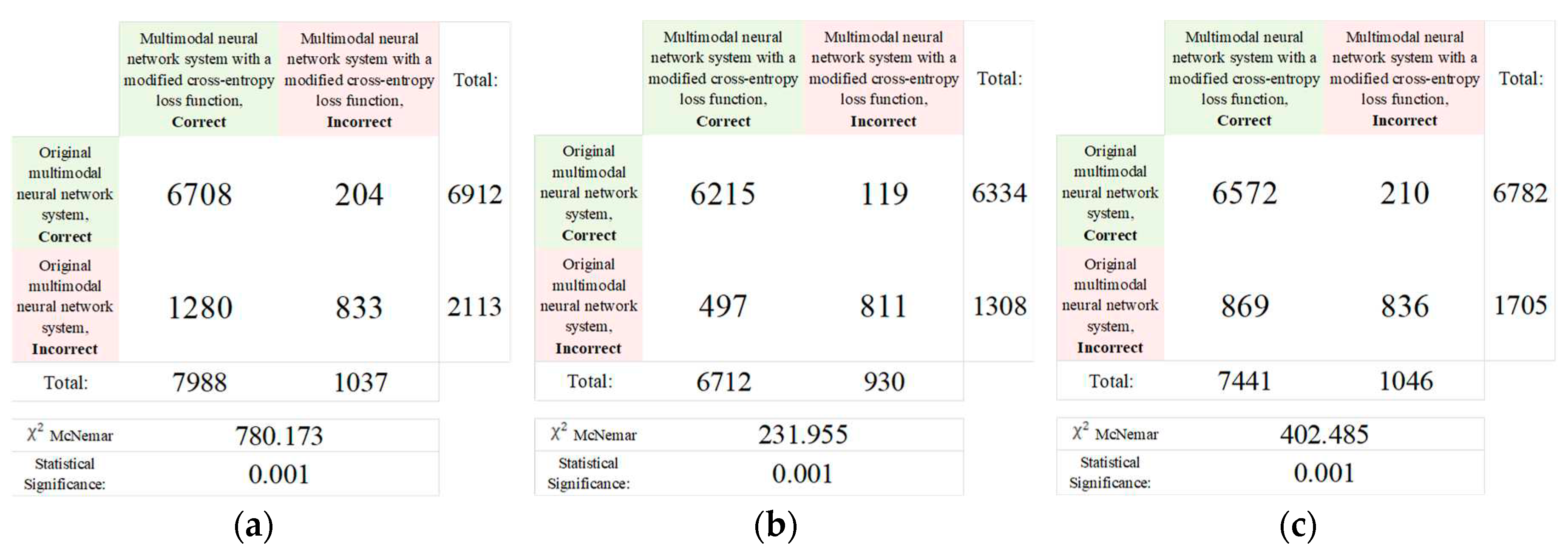
| № | Statistical factor | Cardinality |
|---|---|---|
| 1 | Gender | 2 |
| 2 | Age | 18 |
| 3 | Localization on the body | 8 |
| TOTAL | 28 | |
| № | Statistical factor | Cardinality |
|---|---|---|
| 1 | Gender | 2 |
| 2 | Age | 4 |
| 3 | Localization on the body | 8 |
| TOTAL | 14 | |
| № | Diagnostic category | Weight coefficient |
|---|---|---|
| 1 | Vascular lesions | 3.8893 |
| 2 | Nevus | 0.0353 |
| 3 | Solar lentigo | 3.6444 |
| 4 | Dermatofibroma | 3.9992 |
| 5 | Seborrheic keratosis | 0.6721 |
| 6 | Benign keratosis | 0.8954 |
| 7 | Actinic keratosis | 1.1323 |
| 8 | Basal cell carcinoma | 0.2900 |
| 9 | Squamous cell carcinoma | 1.5000 |
| 10 | Melanoma | 0.1758 |
|
CNN architecture |
Results of test | ||
| Original multimodal neural network system, % | Multimodal neural network system with a modified cross-entropy loss function, % | Difference in recognition accuracy between original and proposed multimodal neural network systems, % | |
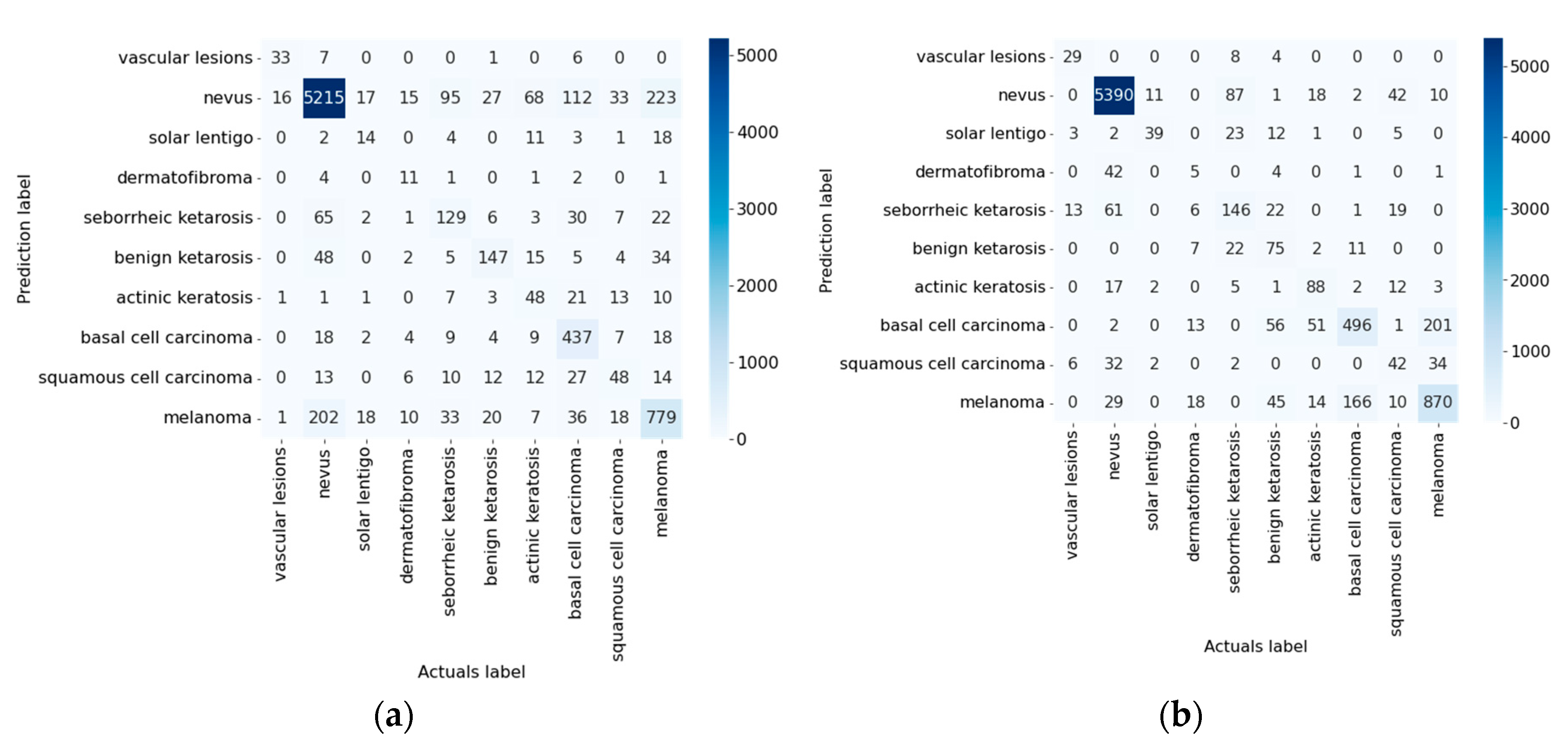
| DenseNet_161 [76] | 81.15 | 85.19 | 4.04 |
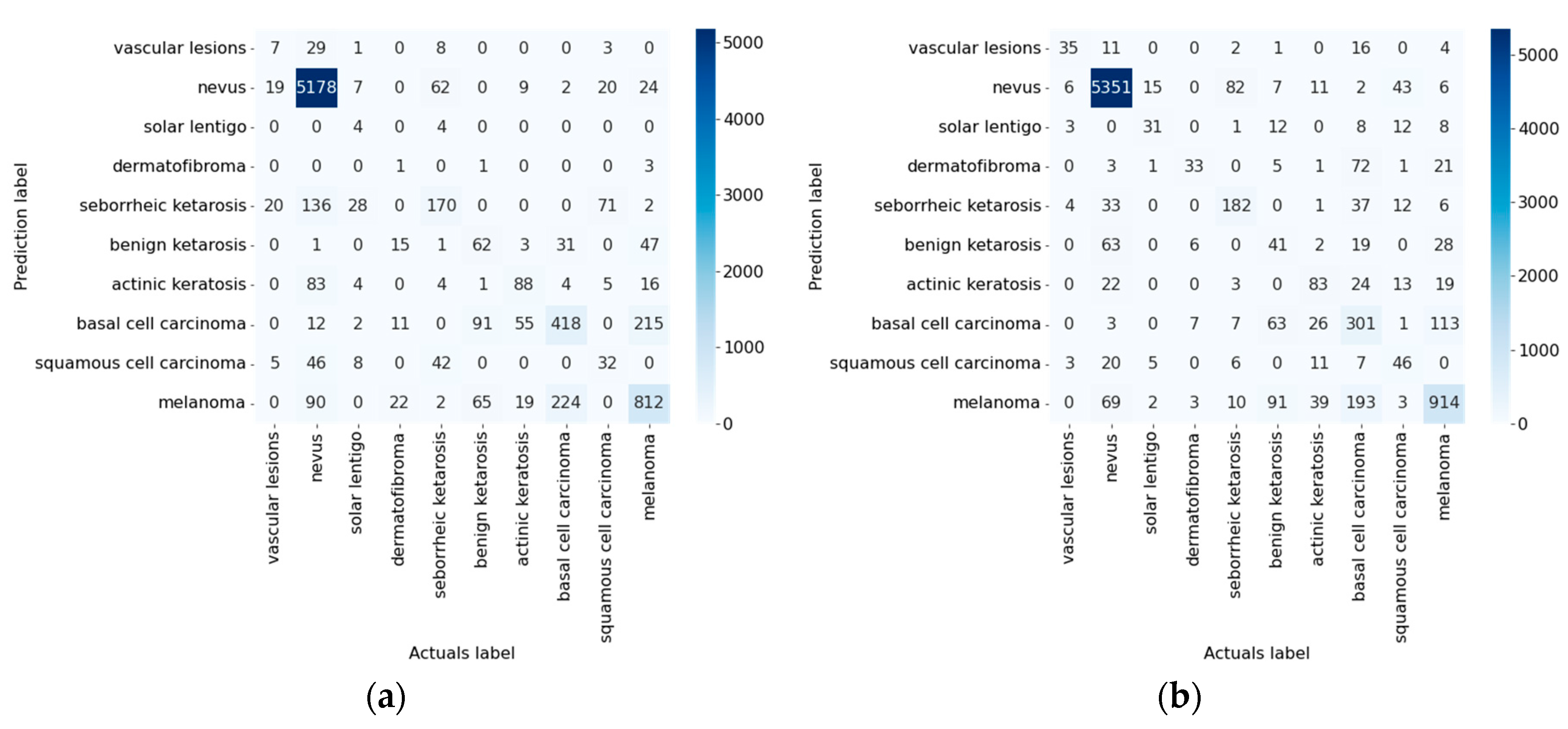
| Inception_v4 [77] | 82.42 | 83.86 | 1.44 |
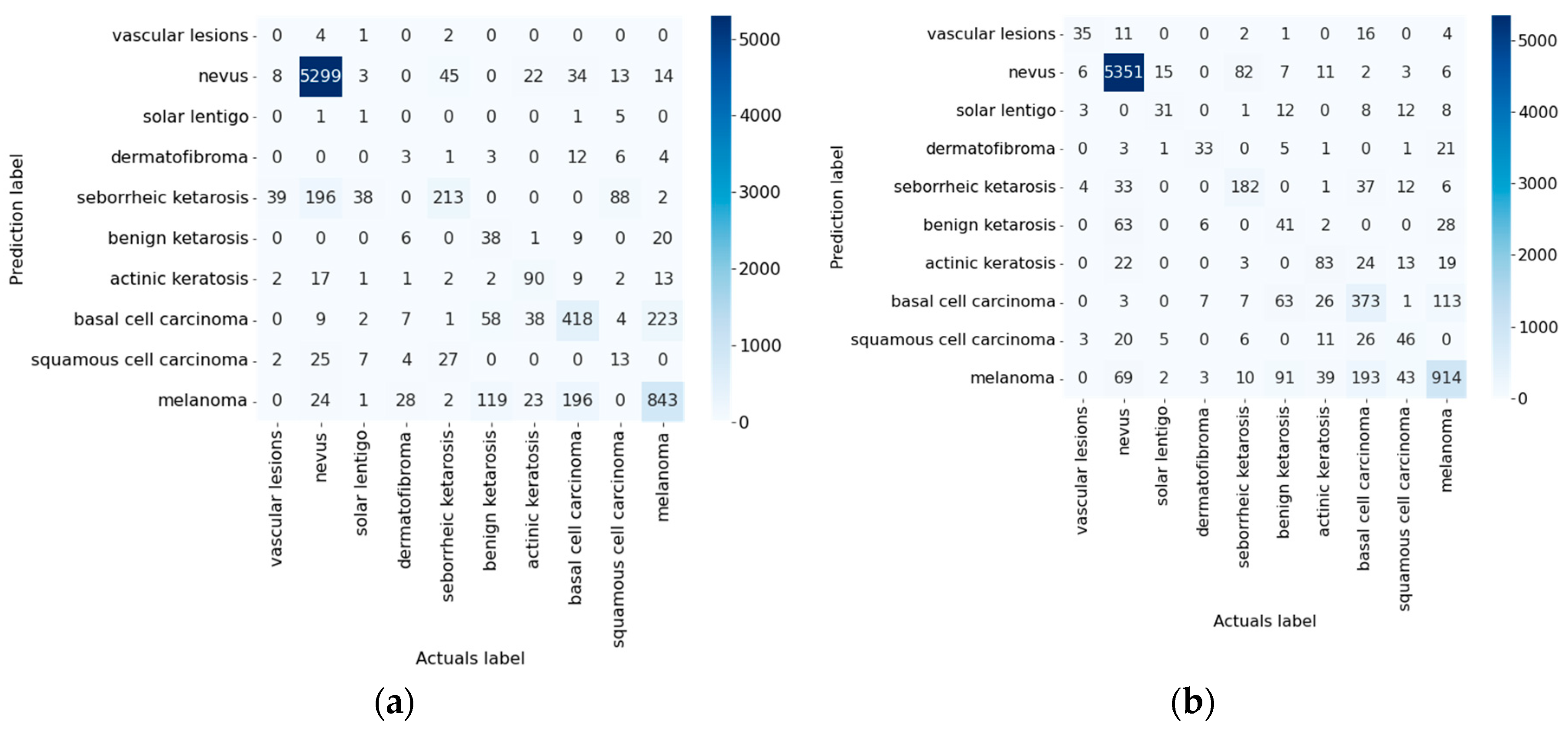
| ResNeXt_50 [78] | 83.91 | 84.93 | 1.02 |
|
CNN architecture |
Results of test | ||
| Original multimodal neural network system | Multimodal neural network system with a modified cross-entropy loss function | Different in value of the loss function between original and proposed multimodal neural network systems | |
| DenseNet_161 [76] | 0.2563 | 0.1344 | 0.1219 |
| Inception_v4 [77] | 0.2087 | 0.1964 | 0.0123 |
| ResNeXt_50 [78] | 0.1843 | 0.1475 | 0.0368 |
| CNN architecture |
Loss function weights | Specificity | Sensitivity | F-1 score | MCC | FNR | FPR | NPV | PPV | Simulation time, hh:mm:ss |
|---|---|---|---|---|---|---|---|---|---|---|
| DenseNet_161 [76] | Not used | 0.9791 | 0.8115 | 0.8115 | 0.6543 | 0.1884 | 0.0209 | 0.9791 | 0.8115 | 14:02:18 |
| Used | 0.9835 | 0.8519 | 0.8519 | 0.7169 | 0.1481 | 0.0164 | 0.9835 | 0.8519 | 13:54:55 | |
| Inception_v4 [77] | Not used | 0.9821 | 0.8397 | 0.8397 | 0.6929 | 0.1602 | 0.0178 | 0.9821 | 0.8397 | 09:28:24 |
| Used | 0.9833 | 0.8494 | 0.8494 | 0.7165 | 0.1506 | 0.0167 | 0.9833 | 0.8494 | 10:52:07 | |
| ResNeXt_50 [78] | Not used | 0.9795 | 0.8156 | 0.8156 | 0.6457 | 0.1844 | 0.0205 | 0.9795 | 0.8156 | 11:47:05 |
| Used | 0.9821 | 0.8391 | 0.8391 | 0.6846 | 0.1616 | 0.0179 | 0.9820 | 0.8391 | 10:12:15 |
| Multimodal neural network system for recognizing pigmented skin lesions | Accuracy of recognition of pigmented neoplasms of the skin, % | |
|---|---|---|
| Known neural network systems | [81] | 71.90 |
| [82] | 76.80 | |
| [83] | 80.42 | |
| The proposed multimodal neural network system based on the DenseNet_161 architecture | 85.19 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





