1. Introduction
Malaria is a life-threatening infection caused by
Plasmodium species, and it constitutes a significant burden on global health [
1]. In addition to several preventive measures and definitive diagnosis, effective antimalarial drug treatment is a key component in the control and elimination of malaria [
2,
3]. Unfortunately,
Plasmodium falciparum resistance to available antimalarial agents has been a major drawback to the success of malaria control, especially in malaria-endemic regions [
4,
5]. The development of resistance to chloroquine (CQ), historically the most widely used antimalarial drug [
6], opened a new vista into understanding the science of malaria parasite biology and its evolutionary capabilities for survival under different conditions.
Chloroquine was one of the most successful antimalarial drugs used in the treatment of uncomplicated malaria infection until it was rendered ineffective by the emergence of chloroquine-resistant
Plasmodium falciparum [
7]. However, CQ remains effective against other strains of human
plasmodium parasites including
P. vivax, the most widespread species outside sub-Saharan Africa [
8]. In sub-Saharan Africa, widespread clinical resistance of
Plasmodium falciparum to CQ [
4,
9] led to its withdrawal and replacement as first-line antimalarial regimen with artemisinin-based combination therapies (ACTs). Chloroquine withdrawal has had varying impacts on different endemic regions [
10]. For example, chloroquine gained therapeutic efficacy after its earlier withdrawal from clinical use in Malawi, Côte d’Ivoire and Uganda [
11,
12,
13,
14,
15]. Furthermore, regeneration of chloroquine-sensitive parasites was reported in some other African countries including Kenya, and Zambia after its withdrawal as first-line chemotherapy for uncomplicated malaria [
16,
17]. However, in Nigeria, evidence suggests that a strong background of the chloroquine-resistant
Plasmodium falciparum persists even after a similar period of replacement by the ACTs [
18,
19]. Despite its withdrawal as first-line antimalaria, chloroquine is still in circulation and available over-the-counter in many pharmacies in Nigeria [
18,
20,
21].
Unfortunately, reports of emergence of resistance to currently used ACTs are threatening malaria control efforts [
22]. Resistance to artemisinins originated from the Greater Mekong Subregion in Asia and has spread to the malaria-endemic areas of southeast Asia [
22,
23,
24]. Considering the emergence in East Africa and potential spread of parasites resistant to the ACTs [
25], and the inherent cost of fitness on such parasites in the absence of CQ pressure [
26,
27], there are proposals for potential re-introduction of CQ to complement ACTs for the treatment of uncomplicated malaria which appear to be gaining traction [
13,
15,
28,
29].
Resistance to CQ historically is mainly facilitated by genetic changes within the coding region of the gene encoding a transporter on the parasite food vacuole membrane; the
Plasmodium falciparum chloroquine resistance transporter (
PfCRT) gene. This transporter is responsible for the influx/efflux of some antimalarial drugs to and from the food vacuole of the parasite [
30,
31,
32]. Point mutations that translate to changes in amino acids at different positions have contributed to the parasites’ competence to resist drugs such as chloroquine and structurally related antimalarial drugs [[
30,
33,
34]. The most significant mutation implicated for chloroquine resistance is the non-synonymous substitution of lysine (K) with Threonine (T) at position 76 (K76T), which modulates efflux of protonated chloroquine from the parasite’s food vacuole [
35,
36]. This change, K76T, is also enhanced by substitutions at positions 72 – 75 in wildtype parasites, CVMN to CVIE in African isolates [
37,
38], and with slight geographical divergence in other regions, such as SVMN in Latin America and Asia [
29]. Substitutions at other positions which play significant roles in chloroquine resistance with clinical confirmation have been reviewed [
27,
39]. Interestingly, the ability of the transporters to efflux other drugs structurally related to chloroquine (amodiaquine and piperaquine) has also been reported with additional mutations at previously unchanged positions [
40]. Clinical and functional studies that investigated single nucleotide polymorphisms at other positions on the
PfCRT gene, reported reduced susceptibility to piperaquine but with increased sensitivity to chloroquine [
27,
40]. This may suggest an overriding fitness requirement for parasites to adapt resistance to structurally related antimalarial drugs [
26].
To date, previous studies on
PfCRT mutations in Nigeria have consistently reported a high prevalence of K76T changes [
6,
18]. However, the significant decreases in the prevalence of chloroquine-resistance markers across other regions in Africa [
7,
13,
28,
41,
42] may suggest an adaptation of resistance prowess against ACTs. Though, the data on prevalence of
PfCRT mutant allele across West Africa varies [
6,
18,
37,
38,
43], parasite population genetic diversity across the study region remains homogenous [
44]. With hindsight of the historical and geographical route of spread of
Plasmodium falciparum resistance to CQ, and reports of detection of ACT resistance in Uganda, east of Africa [
25], there is the fear of potential spread of ACT resistance to other sub-Saharan African countries. Continuous molecular surveillance of resistant markers in these countries is now expedient, especially in countries where chloroquine was not withdrawn but replaced. In this study, we evaluated the current prevalence of mutant
PfCRT in an endemic area of Southwest Nigeria, where chloroquine is still readily available over-the-counter. We isolated and genotyped
PfCRT from circulating parasites within a densely populated province.
2. Materials and Methods
2.1. Study design and site
The study was part of an ongoing drug therapeutic efficacy study of dihydroartemisinin/piperaquine conducted at the Malaria Clinic of the Malaria Research Laboratories, Institute of Advanced Medical Research and Training, College of Medicine, University of Ibadan. The study was an open-label single-arm prospective study. It was conducted between the period of June to November 2021. Ibadan is a malaria endemic area in southwest Nigeria, West Africa with an all-year-round malaria transmission which is more intense between April and October. The molecular analysis was conducted at the Hart Lab, Medical School, University of Minnesota, U.S.A.
2.2. Institutional Review Board Statement
The protocol for this study was reviewed and approved by both the Joint University of Ibadan/University College Hospital Ethics Committee (#UI/EC/20/0171) and Oyo State Ministry of Health Research Ethics Committee (AD 13/479/4219). The study was also registered and approved by the Pan Africa Clinical Trial Registry (PACTR) with identification number PACTR202108584742856. Written informed consent was obtained from study participants and from the parents/guardian of children. In addition, assent was also obtained directly from children ≥ 10 years.
2.3. Patients and sample collection
A total of 737 patients who visited the Malaria Clinic were screened using microscopy. Thick and thin blood films were obtained from patients on slides by finger pricking for malaria microscopic screening. The enrolment criteria included microscopically confirmed malaria infection and absence of other concomitant illnesses. Participants were enrolled into the study after clinical examination and microscopically confirmed Plasmodium falciparum infection. Finger-pricked blood samples from enrolled participants with confirmed falciparum malaria were collected on 3MM Whatman® filter paper before treatment initiation (day 0) and were allowed to dry at room temperature. The dried blood spots (DBS) were stored in sealed plastic bags at room temperature with silica gel until analyzed for molecular profiling.
2.4. Parasite DNA extraction
Plasmodium falciparum genomic DNA was extracted from the DBS using QIAamp DNA Mini blood kit (Qiagen, USA) according to the manufacturer’s protocol. The extracted genomic DNA concentration were quantified on nanodrop spectrophotometer and stored at 4oC.
2.5. Amplification of PfCRT
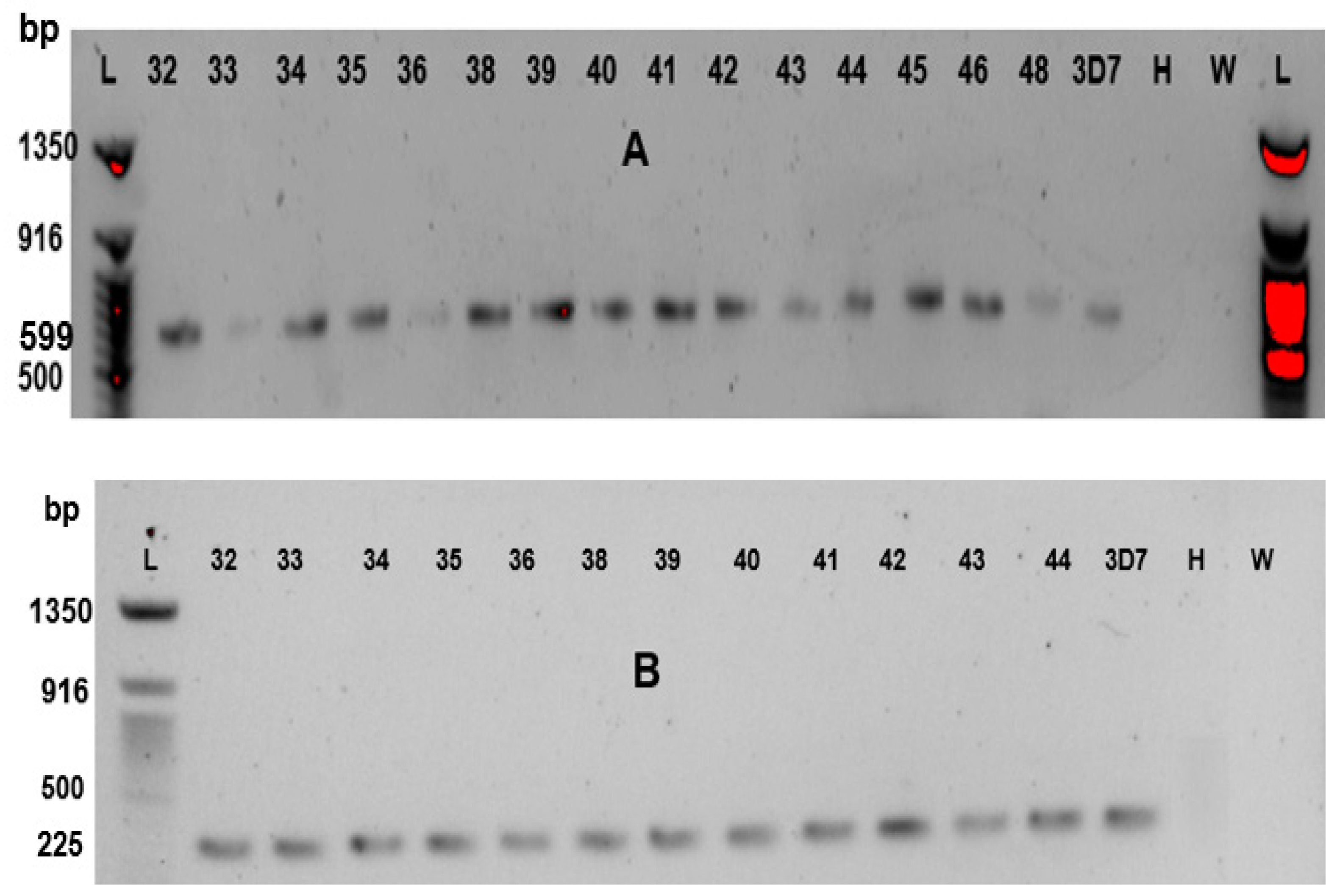
Fragments of interest of
PfCRT gene were amplified using nested polymerase chain reaction. A 1701 base pair outer fragment of
PfCRT gene (752 – 2452 bp) was amplified followed by nested amplifications of a 599 bp fragment (1144-1742) covering codons 72-76 and a 225 bp fragment (1905-2129) covering essential codons 220 and 271. Primer pairs were selected as previously reported by Mittra
et al., 2006 [
45], and were synthesized by Integrated DNA Technologies (IDT, USA). Primers for primary reaction were
PfCRT OF:5’-CCGTTAATAATAAATACAGGC-3’,
PfCRT OR: 5’-CTTTTAAAAATGGAAGGGTGT-3’ while the primers for the nested reactions were
PfCRT N72F: 5’-TGTGCTCATGTGTTTAAACTTAT-3’,
PfCRT N72R:5’-AAAATAGTATACTTACCTATATCT-3’ and
PfCRT N220F: 5’-CTTATACAATTATCTCGGAGCAG-3’,
PfCRT N220R: 5’-ATAATAAAAACAAAGTTTAAGTGT-3’.
Briefly, 10 μL reaction volume containing 5 μL iProof HF 2X master mix (Bio-rad USA), 0.25 μL (10μM) of each primer, 3.5 μL of nuclease-free water and 1 μL of Plasmodium falciparum DNA template was used for the primary amplification. For the nested amplifications, 12.5 μL reaction volume containing 6.25 μL iProof HF 2X master mix (Bio-rad, USA), 0.25 μL (10μM) of each primer, 5.25 μL of nuclease-free water and 0.5 μL of Plasmodium falciparum DNA template was used for nested amplification. Each PCR contained a Plasmodium falciparum 3D7 genomic DNA template as a positive control while a reaction containing Plasmodium falciparum-negative DNA and a no-template reaction served as negative controls. For the primary amplification, the thermal cycler (Eppendorf; USA) was programmed as follows: 94 °C for 3 min, 40 cycles of: 94 °C for 30 s, 49 °C for 1 min and 60 °C for 90 s, with a final extension at 60 °C for 3 min. Conditions for codon 72-76 nested amplifications were: 94 °C for 3 min, 25 cycles of: 94 °C for 30 s, 44 °C for 30s and 68 °C for 60 s, with a final extension at 64 °C for 3 min. Codon 220-containing fragment was amplified in a nested reaction with similar conditions except annealing temperature of 53oC.
Nested PCR products were resolved on 1.5% agarose gel stained with SYBR green dye (Invitrogen, USA) using a Quick-Load® Purple 50 bp DNA Ladder (Biolab® USA). Images of 599 and 225 base pair fragments were viewed in Gel Doc EZ Imager (Bio-Rad, USA) documentation system. The amplicons were purified by treatment with a mixture of Exonuclease 1 (ThermoFisher, USA) and Shrimp Alkaline Phosphatase (ThermoFisher, USA).
2.6. Sequence Data Analysis
Purified PCR products were subjected to Sanger dideoxy sequencing at Eurofins Genomics commercial sequencing centre (Eurofins, USA) using the forward primers of each nested reactions. The sequence data was analyzed using clustal omega alignment program. Generated sequences were aligned with the reference sequence of 3D7 (Gene ID: PF3D7_0709000) and all identified changes in nucleotides were reported.
2.7. Statistical Analysis
Data were analyzed using the statistical package for social sciences (SPSS) version 25. Fisher’s exact test was used to compare two categorical variables. A P-value of ˂ 0.05 was considered statistically significant.
3. Results
3.1. Demographic Profile of enrolled participants
Of the 737 patients who were screened at the Malaria Clinic using microscopy, 441 (59.8%) had
Plasmodium falciparum infection. One hundred and twenty-nine (129) malaria-positive patients were enrolled into the study. The baseline characteristics of the recruited participants are summarized in
Table 1. Overall, the mean age of all participants was 9.3±4.8 years (range 1-35). Twenty-eight participants were aged 1-5 years, 95 were aged >5 years to 15 years and 6 were >15 years. The mean axillary temperature was 37.3±1.1
oC (range 35.5-40.0
oC). Of the 129 participants, 48 patients presented with fever at enrolment while 81 patients had no fever. Geometric mean parasite density of enrolled participants was 14440 uL
-1 (range 181-729000).
3.2. Prevalence of PfCRT single nucleotide polymorphisms in the Plasmodium falciparum parasites
Genomic DNA samples from 52 patients were amplified while 43 were successfully sequenced. The alignment data of sequenced samples showed a uniform arrangement with the reference clone (
Pf3D7) with no synonymous change within the amplified fragments. Altogether, five non-synonymous changes were identified with three substitutions (M74I, N75E and K76T) at codons 72-76 within the 599bp fragment, and 2 substitutions (A220S and Q271E) within the 225bp fragment. At codons 72-76, changes in amino acid sequence from CVMNK (wildtype) to CVIET (mutant) were identified in 18.6% of the samples while 2.3% had CVINT mutation. The wildtype CVMNK haplotype occurred in 79.1% of the samples. All A220S changes (16.3%) occurred concurrently with the CVIET haplotype, while the Q271E was found in an isolate with the wildtype sequence (CVMNK) relative to positions 72 - 76.
Table 2 shows the position distribution and substitutions of amino acids on
PfCRT. The agarose gel resolutions for the amplified fragments are also shown in
Figure 1. The south American/southeast Asian chloroquine-resistant haplotype SVMNT was not seen in any of the samples.
3.3. Frequency of mutation between age groups
The distribution of the identified PfCRT mutations (M74I, N75E, K76T, A220S and Q271E) across the age groups are as follows: 2/43 (4.7%) were found among patients aged 1-5 years, 8/43 (18.6%) were found among patients aged >5-15 years while no mutation occurred among patients aged >15-35 years. There was no correlation between age and presence of mutant allele (P=0.84).
4. Discussion
Many studies have described widespread return of chloroquine sensitive alleles of
Plasmodium falciparum parasites in parts of Africa after at least a 10-year cessation of chloroquine use in treatment of uncomplicated malaria [
28,
41,
46,
47]. The data from the studies support calls for consideration of chloroquine as an addition to ACTs, especially in areas where chloroquine-susceptible
Plasmodium falciparum predominates [
48,
49].
From the present study, there seems to be a steady decline in the CQ resistance markers over the years in southwest Nigeria after the replacement of chloroquine as first-line treatment for uncomplicated malaria. The findings of our study revealed a lower frequency of
PfCRT 76T resistant allele (20.9%) and CVIET haplotype (18.6%) when compared to previous reports of similar studies from the same population, with 60% in 2008 and 72% in 2012, respectively [
37,
50]. Reports from other parts of Nigeria also indicate varying frequencies of CQ resistance mutant alleles [
6,
18,
38,
51,
52] which may be connected with the continued use of chloroquine due to its affordability.
While the prevalence of mutant
PfCRT haplotype we observed in south-western Nigeria is comparable to findings from samples obtained from Chinese migrants who contracted malaria in different parts of Africa [
53], it cannot be correlated with similar results from studies in East Africa where chloroquine was completely withdrawn from clinical use [
54]. However, there seem to be a slow but gradual return of the chloroquine-sensitive haplotype (CVMNK) within the study population. Mohammed
et al., 2017 reported a lower frequency of K76T in northwest Nigeria; however, the study was based on
PfCRT RNA transcripts isolated from samples. In the study, only 33.4% of samples assayed for wildtype and mutant changes at position 76 were resolved [
55]. To date, the prevalence of K76T reported in this study is the lowest that has been reported in Nigeria using direct DNA sequence analysis. The K76T
PfCRT resistant signature is gradually weaning in malaria endemic areas in Africa known to harbor strong lines of chloroquine-resistant parasites in the absence of chloroquine use. These resistant alleles may be fast disappearing within the West Africa region as similar finding has been reported in Cote de ’Ivoire [
42]. Therefore, the potential for emergence of parasites with zero chloroquine-resistant prowess in these areas should be duly considered. The ACTs are the first-line antimalarial regimen in Nigeria, while sulphadoxine-pyrimethamine is prescribed for intermittent prevention of malaria during pregnancy [
3]. The absence of chloroquine from the clinics may have contributed to the reversal of the resistant alleles of
PfCRT, probably due to fitness survival mechanisms under ACTs pressure [
26,
32,
56].
Acquisition of point mutations on
PfCRT has been reported to be directly proportional to increased levels of chloroquine resistance [
45]. The amino acid change, A220S is reported to complement the CVIET haplotype in
PfCRT [
57]. Our results indicate that all A220S changes were found within a subset of the K76T allele, suggesting that A220S may not exist in isolation to confer CQ resistance. We hypothesize that the lower prevalence of A220S and singular appearance of CVINT haplotype may suggest that parasites in the population might be undergoing gradual genetic evolution reverting to wildtype
PfCRT genotypes, by relinquishing in stepwise manner point mutations that often contribute to chloroquine resistance. This is probably supported from the observation that the CVI
NT haplotype retains asparagine amino acid contained in the wildtype CVM
NK. This is the first report of the CVINT haplotype in Nigeria, however, it has previously been reported in neighboring Niger Republic, in a study that observed a high prevalence (84.6%) of the wildtype haplotype (CVMNK) [
58] and another in Angola [
59]. Despite the time lag between our study and these other studies that reported CVINT mutations [
58,
59], the consistently low prevalence values of this mutation (≤ 4%) may be insufficient to suggest possible selection for amodiaquine resistance due to drug pressure as propounded [
58].
Plasmodium falciparum chloroquine resistance transporter has been implicated in mechanism of resistance to other drugs with associated point mutations at different positions of the gene, especially in the absence of CQ [
27,
32,
56]. The translated amino acid changes in these studies, T93S, H97Y, C101F, N326S, M343, have been associated with varying degrees of resistance to other antimalarials such as amodiaquine, piperaquine, mefloquine and lumefantrine in functional and metabolic studies on the gene activity, and in some Asian isolates. Conversely, a number of the changes resulted to chloroquine sensitivity in parasites [
32]. Interestingly, none of the highlighted changes were found in all the samples we analyzed. This information is in tandem with a previous report on African isolates [
60], despite the clinical use of the listed drugs as first-line partners to artemisinin derivatives in the study population.
5. Conclusions
There is a wide variability in the prevalence of PfCRT mutation post CQ withdrawal across the various states in Nigeria. The potential for slow but newly evolving drug resistance markers or parasites with zero-resistance prowess to CQ in the study areas should not be overlooked. Compared to the consistently high PfCRT mutations previously reported in the study area, our findings imply a reduction in the CQ resistant PfCRT mutant haplotypes in Ibadan southwest Nigeria 17 years after CQ withdrawal. However, the fact that high prevalence rates of PfCRT still occurs in some parts of Nigeria calls for urgent attention, intensified surveillance, and the need to regulate use of chloroquine in Nigeria.
Author Contributions
The authors contributed to this publication in the following ways: Conceptualization, A.I.A.; Methodology, A.I.A., G.T.H., M.H.C., and O.A.; Sample collection, A.I.A. and K.A.; Validation, A.I.A., and G.T.H., Laboratory analysis, A.I.A., G.T.H., M.H.C., J.K.D., and O.A.; Sequence analysis, A.I.A., G.T.H. and O.A.; Original draft preparation, A.I.A. and O.A.; Review and editing, G.O.G., O.A., M.H.C., J.K.D., A.I.A., G.T.H., and A.S.; Supervision, G.O.G., and G.T.H; Resources, G.T.H.,G.O.G., Funding Acquisition, A.I.A. and G.T.H. All authors have read and agreed to the published version of the manuscript.
Funding
This project was supported by Fulbright Program grant sponsored by the Bureau of Educational and Cultural Affairs of the United States Department of State and administered by the Institute of International Education. It was also supported by Hart lab through the Department of Medicine and Medical school at the University of Minnesota, U.S.A.
Informed consent statement
Written informed consent was obtained from all subjects involved in the study.
Acknowledgments
We appreciate the support offered by Mrs Alo and Mrs Ayodeji of the Malaria Research Clinic and Laboratories, Institute of Advanced Medical Research and Training, University of Ibadan, Nigeria, and Deborah Okeneye of the Department of Biological Sciences, Redeemer’s University, Ede, Osun state, Nigeria. We also thank Gavin Fuchs and Ben Zandstra of Hart’s lab, Medical School, University of Minnesota, U.S.A. We would like to thank Dr. Jianming Wu in the College of Veterinary Medicine at the University of Minnesota for gracious help in analyzing and interpreting the sequencing data and general genetics knowledge.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Moxon, C. A.; Gibbins, M. P.; McGuinness, D.; Milner, D. A.; & Marti, M. New Insights into Malaria Pathogenesis. Annual Review of Pathology: Mechanisms of Disease Rev. Pathol. Annu. 2020, 15, 315–343. [CrossRef]
- Amusan, A. I.; Akinola, O.; Akano, K.; & Gbotosho, G. O. Detection of malaria parasite protein in urine of patients with acute uncomplicated malaria using rapid diagnostic test kits. J Microbiol Infect Dis. 2022, 12, 97–106. [CrossRef]
- World Health Organisation. World malaria report. 2021. https://www.who.int/teams/global-malaria-programme/reports/world-malaria-report-2021. Accessed on 2nd November 2022.
- Happi, C. T.; Gbotosho, G. O.; Sowunmi, A.; Falade, C. O.; Akinboye, D. O.; Gerena, L.; Kyle, D. E.; Milhous, W.; Wirth, D. F.; & Oduola, A. M. J. Molecular analysis of Plasmodium falciparum recrudescent malaria infections in children treated with chloroquine in Nigeria. Am. J Trop Med Hyg. 2004, 70, 20–26.
- Paloque, L.; Coppée, R.; Stokes, B. H.; Gnädig, N. F.; Niar, K.; Augereau, J. M.; Fidock, D. A.; Clain, J.; & Benoit-Vical, F. Mutation in the Plasmodium falciparum BTB/POZ Domain of K13 Protein Confers Artemisinin Resistance. Antimicrob. Agents Chemother. 2022, 66, 1–13. [CrossRef]
- Oladipo, O. O.; Wellington, O. A.; & Sutherland, C. J. Persistence of chloroquine-resistant haplotypes of Plasmodium falciparum in children with uncomplicated Malaria in Lagos, Nigeria, four years after change of chloroquine as first-line antimalarial medicine. Diagn Pathol. 2015, 10, 4–11. [CrossRef]
- Asare, K. K.; Africa, J.; Mbata, J.; & Opoku, Y. K. The emergence of chloroquine-sensitive Plasmodium falciparum is influenced by selected communities in some parts of the Central Region of Ghana. Malar J. 2021, 20, 1–9. [CrossRef]
- Tse, E. G.; Korsik, M.; & Todd, M. H. The past, present and future of anti-malarial medicines. Malar J, 2019. 18, 93. [CrossRef]
- Sowunmi, A.; & Oduola, A. M. J. Comparative efficacy of chloroquine/chlorpheniramine combination and mefloquine for the treatment of chloroquine-resistant Plasmodium falciparum malaria in Nigerian children. Trans. R. Soc. Trop. Med. Hyg. 1997, 91, 689–693. [CrossRef]
- Ocan, M.; Akena, D.; Nsobya, S.; Kamya, M. R.; Senono, R.; Kinengyere, A. A.; & Obuku, E. A. Persistence of chloroquine resistance alleles in malaria endemic countries: A systematic review of burden and risk factors. Malar. J. 2019, 18, 1–15. [CrossRef]
- Laufer, K.; Takala-Harrison.; Dzinjalamala, K.; Taylor, T.; Plowe, C. Return of chloroquine antimalarial efficacy in Malawi. N. Engl. J. Med. 2006, 355, 11–20. [CrossRef]
- Laufer, M. K.; Takala-Harrison, S.; Dzinjalamala, F. K.; Colin Stine, O.; Taylor, T. E.; & Plowe, C. V. Return of chloroquine-susceptible falciparum malaria in malawi was a reexpansion of diverse susceptible parasites. J Infect Dis. 2010, 202, 801–808. [CrossRef]
- Dagnogo, O.; Ako, A. B.; Ouattara, L.; Dago, N. D.; Coulibaly, D. N. G.; Touré, A. O.; & Djaman, J. A. Towards a re-emergence of chloroquine sensitivity in Côte d’Ivoire? Malar J. 2018, 17, 1–7. [CrossRef]
- Balikagala, B.; Sakurai-Yatsushiro, M.; Tachibana, S.; Ikeda, M.; Yamauchi, M.; Katuro, O. T.; Ntege, E. H.; Sekihara, M.; Fukuda, N.; Takahashi, N.; Yatsushiro, S.; Mori, T.; Hirai, M.; Walter Opio, Obwoya, P. S.; Anywar, D. A.; Auma, M. A.; Palacpac.;Tsuboi, T.; Mita. Recovery and stable persistence of chloroquine sensitivity in Plasmodium falciparum parasites after its discontinued use in Northern Uganda. Malar J. 2020, 19. [CrossRef]
- Mwanza, S.; Joshi, S.; Nambozi, M.; Chileshe, J.; Malunga, P.; Kabuya, J. B. B.; Hachizovu, S.; Manyando, C.; Mulenga, M.; & Laufer, M. The return of chloroquine-susceptible Plasmodium falciparum malaria in Zambia. Malar J. 2016, 15, 1–7. [CrossRef]
- Tumwebaze, P.; Tukwasibwe, S.; Taylor, A.; Conrad, M.; Ruhamyankaka, E.; Asua, V.; Walakira, A.; Nankabirwa, J.; Yeka, A.; Staedke, S. G.; Greenhouse, B.; Nsobya, S. L.; Kamya, M. R.; Dorsey, G.; Rosenthal, P. J. Changing antimalarial drug resistance patterns identified by surveillance at three sites in Uganda. J. Infect. Dis. 2017, 215, 631–635. [CrossRef]
- Hemming-Schroeder E.; Umukoro, E.; Lo, Fung, B.; Tomas-Domingo, P.; Zhou, G.; Zhong D.; Dixit, A.; Atieli, H.; Githeko, A.; Vardo-Zalik, A.; Yan, G. Impacts of Antimalarial Drugs on Plasmodium Falciparum Drug Resistance Markers, Western Kenya, 2003-2015. Am J Trop Med and Hyg. 2018, 98, 692–99. [CrossRef]
- Kayode T. A.; Akano, K.; Fehintola V.; Ajogbasile.; Jessica N. Uwanibe.; Paul, E. O.; Bolajoko, E. B.; Sowunmi, A.; Folarin, O.A.; Volkman, S.K.; Pardis, S.; Dyann, F. W.; & Christian T. H. Polymorphisms in Plasmodium falciparum chloroquine resistance transporter (Pfcrt) and multidrug-resistant gene 1 (PfMDR1) in Nigerian children 10 years post-adoption of artemisinin-based combination treatments. Int. J. Parasitol. 2021, 51, 301–310. [CrossRef]
- Wang, X.; Zhang, X.; Chen, H.; Zhang, J.; Lu, Q.; Ruan, W.; & Linga, F. Molecular epidemiology of drug resistance genes in Plasmodium falciparum isolates imported from Nigeria between 2016 and 2020: Continued emergence of fully resistant PfDHFR-PfDHPS alleles. Microbiol Spectr. 2022, 10. [CrossRef]
- Gbotosho, G. O.; Happi, C. T., Ganiyu, A.; Ogundahunsi, O. A.; Sowunmi, A.; & Oduola, A. M. Potential contribution of prescription practices to the emergence and spread of chloroquine resistance in south-west Nigeria: Caution in the use of artemisinin combination therapy. Malar J. 2009, 8. [CrossRef]
- Efunshile, M.; Runsewe-Abiodun, T.; Ghebremedhin, B.; König, W.; & König, B. Prevalence of the molecular marker of chloroquine resistance (pfcrt 76) in Nigeria 5 years after withdrawal of the drug as first-line antimalarial: A cross-sectional study. SAJCH. 2011, 5, 39–42.
- Paloque, L.; Ramadani, A. P.; Mercereau-Puijalon, O.; Augereau, J. M.; & Benoit-Vical, F. Plasmodium falciparum: multifaceted resistance to artemisinins. Malar. J. 2016, 15. BioMed Central. [CrossRef]
- Noedl, H.; Se, Y.; Sriwichai, S.; Schaecher, K.; Teja-Isavadharm, P.; Smith, B.; Rutvisuttinunt, W.; Bethell, D.; Surasri, S.; Fukuda, M. M.; Socheat, D.; & Thap, L. C. Artemisinin resistance in Cambodia: A clinical trial designed to address an emerging problem in southeast Asia. Clin. Infect. Dis. 2010. https://doi.org/10.1086/657120.
- Imwong, M.; Hien, T. T.; Thuy-Nhien, N. T.; Dondorp, A. M.; & White, N. J. Spread of a single multidrug resistant malaria parasite lineage (PfPailin) to Vietnam. Lancet Infect Dis. 2017. 17, P1022-1023. [CrossRef]
- Balikagala, B.; Fukuda, N.; Ikeda, M.; Katuro, O. T.; Tachibana, S. I.; Yamauchi, M.; Opio, W.; Emoto, S.; Anywar, D. A.; Kimura, E.; Palacpac, Odongo-Aginya, E. I.; Ogwang, M.; Horii, T.; & Mita, T. Evidence of artemisinin-resistant malaria in Africa. N. Engl. J. Med. 2021, 385, 1163–1171. [CrossRef]
- Nair, S.; Li, X.; Arya, G. A.; McDew-White, M.; Ferrari, M.; Nosten, F.; & Anderson, T. J. C. Fitness costs and the rapid spread of kelch13-C580Y substitutions conferring artemisinin resistance. Antimicrob. Agents Chemother. 2018, 62. [CrossRef]
- Wicht, K. J.; Jennifer L. Small-Saunders, L. M. H.; Mok, S.; & Fidock, D. A. Mutant PfCRT can mediate piperaquine resistance in African Plasmodium falciparum with reduced fitness and increased susceptibility to other antimalarials. J. Infect. Dis. 2022, 22. [CrossRef]
- Achungu, C.; Nkuo-Akenji, T.; Wanji Samuel, A. T.; & Wanji, S. Re-emergence of chloroquine-sensitive Plasmodium falciparum after several years of chloroquine withdrawal in Bamenda, North West Cameroon. EC Microbiol. 2018, 831-836.
- Madkhali, A. M.; Abdulhaq, A. A.; Atroosh, W. M.; Ghzwani, A. H.; Zain, K. A.; Ghailan, K. Y.; Hamali, H. A.; Mobarki, A. A.; Eisa, Z. M., Lau, Y. L., & Al-Mekhlafi, H. M. The return of chloroquine-sensitive Plasmodium falciparum parasites in Jazan region, southwestern Saudi Arabia over a decade after the adoption of artemisinin-based combination therapy: analysis of genetic mutations in the PfCRT gene. Parasitol. Res. 2021, 120, 3771–3781. [CrossRef]
- Kim, J.; Tan, Y. Z Imwong, M.; Hien, T. T.; Thuy-Nhien, N. T.; Dondorp, A. M.; & White, N. J. Spread of a single multidrug resistant malaria parasite lineage (PfPailin) to Vietnam. Lancet Infect Dis. 2017. [CrossRef]
- Okombo, J.; Mok, S.; Tarrick Qahash, Yeo, T.; Bath, J.; Lindsey M. Orchard, E. O.; Koo, I.; Albert, I.; Llina, M.; David, & Fidock, A. Piperaquine-resistant PfCRT mutations differentially impact drug transport, hemoglobin catabolism and parasite physiology in Plasmodium falciparum asexual blood stages. PLOS Pathogens. 2022. [CrossRef]
- Shafik, S. H.; Richards, S. N.; Corry, B.; & Martin, R. E. Mechanistic basis for multidrug resistance and collateral drug sensitivity conferred to the malaria parasite by polymorphisms in PfMDR1 and PfCRT. In PLoS Biology. 2022, 20. [CrossRef]
- Dhingra, S. K.; Gabryszewski, S. J.; Small, J. L.; Yeo, T. S.; Henrich, P. P.; Sachel Mok, A.; & Fidock, D. A. Global spread of mutant PfCRT and its pleiotropic impact on plasmodium falciparum multidrug resistance and fitness. Mbio. 2019, 10. [CrossRef]
- Wicht, K. J.; K., E. S.; Dhingra, S. K.; Okombo, J.; Vendome, J.; Hagenah, L. M.; Giacometti, S. I.; Warren, A. L.; Nosol, K.; Roepe, P. D.; Potter, C. S.; Carragher, B.; Kossiakoff, A.; & Matthias Quick, David A. Fidock & Filippo Mancia. Structure and drug resistance of the Plasmodium falciparum transporter PfCRT. Nature, 2019, 576. [CrossRef]
- Kirk, A. M. L.; and Kirk, K. Chloroquine resistance-conferring mutations in PfCRT give rise to a chloroquine-associated H_ leak from the malaria parasite’s digestive vacuole. Antimicrob. Agents Chemother. 2008, 52, 4374–4380. [CrossRef]
- Chandan Patel and Dipankar Roy. A Computational Study of Molecular Mechanism of Chloroquine Resistance by Chloroquine Resistance Transporter Protein of Plasmodium falciparum via molecular modeling and molecular simulations. Physchem. 2021, 1, 232–242. [CrossRef]
- Folarin, O. A.; Gbotosho, G. O.; Sowunmi, A.; Olorunsogo, O. O.; Oduola, A. M. J.; & Happi, T. C. Chloroquine-resistant Plasmodium falciparum in Nigeria: Relationship between PfCRT and PfMDR1 polymorphisms, in-vitro resistance and treatment outcome. Open Trop. Med. J. 2008, 1, 74–82. [CrossRef]
- Agomo, C. O.; Mishra, N.; Olukosi, Y. A.; Gupta, R.; Kamlesh, K.; Aina, O. O.; & Awolola, S. T. Mutations in PfCRT and PfMDR1 genes of Plasmodium falciparum isolates from two sites in Northcentral and Southwest Nigeria. Infect. Genet. Evol. 2021, 95, 105042. [CrossRef]
- Summers, R. L.; Nash, M. N.; & Martin, R. E. Know your enemy: understanding the role of PfCRT in drug resistance could lead to new antimalarial tactics. Cell. Mol. Life Sci. 2012, 69, 1967–1995. [CrossRef]
- Shrestha, B.; Shah, Z.; Morgan, A. P.; Saingam, P.; Chaisatit, C.; Chaorattanakawee, S.; Praditpol, C.; Boonyalai, N.; Lertsethtakarn, P.; Wojnarski, M.; Deutsch-Feldman, M.; Adams, M.; Sea, D.; Chann, S.; Tyner, S. D.; Lanteri, C. A.; Spring, M. D.; Saunders, D. L.; Smith, P. L.; Takala-Harrison, S. Distribution and temporal dynamics of Plasmodium falciparum chloroquine resistance transporter mutations associated with piperaquine resistance in Northern Cambodia. J. Infect. Dis. 2021, 224, 1077–1085. [CrossRef]
- Kishoyian, G.; Njagi, E. N. M.; Orinda, G. O.; & Kimani, F. T. Chloroquine sensitivity and prevalence of chloroquine-resistant genes PfCRT and PfMDR1 in western Kenya after two decades of chloroquine withdrawal. Ann Med Health Sci Res. 2018, 331–335.
- Konaté, A.; Gnagne, P. A.; Bédia-Tanoh, V. A.; Amiah-Droh, M.; Tano, D. K.; Ignace Eby Menan, H.; & Yavo, W. Low rates of Plasmodium falciparum PfCRT K76T mutation in three sentinel sites of malaria monitoring in Côte d’Ivoire. Acta Parasitologica. 2018, 63, 795–801. [CrossRef]
- Ali, I. M.; Tchuenkam, V. P. K.; Tagomo, S. S.; Mawamba, H.; Moyeh, M. N.; Nfor, E. N.; Nji, A. M.; Fomboh, C. T.; Nana, W. D.; Kengne, J. P. C.; Niba, P. T. N.; Ekoyol, G. E.; Achu, D. F.; Bigoga, J. D.; & Mbacham, W. F. Allelic frequencies of mutants of the Plasmodium falciparum, quinoline and folate metabolizing genes in the west region of Cameroon. Heliyon. 2022, 8, e11861. [CrossRef]
- Ajogbasile, F. V.; Kayode, A. T.; Oluniyi, P. E.; Akano, K. O.; Uwanibe, J. N.; Adegboyega, B. B.; Philip, C.; John, O. G.; Eromon, P. J.; Emechebe, G.; Finimo, F.; Ogbulafor, N.; Jiya, N.; Okafor, U.; Ambe, J.; Wammanda, R. D.; Oguche, S.; Mokuolu, O. A.; Sowunmi, A.; Happi, C. T. Genetic diversity and population structure of Plasmodium falciparum in Nigeria: insights from microsatellite loci analysis. Malar. J. 2021, 20, 1–9. [CrossRef]
- Mittra, P., Vinayak, S., Chandawat, H., Das, M. K.; Singh, N.; Biswas, S.; Dev, V.; Kumar, A.; Ansari, M. A.; & Sharma, Y. D. Progressive increase in point mutations associated with chloroquine resistance in Plasmodium falciparum isolates from India. J. Infect. Dis. 2006, 193. [CrossRef]
- Frosch, A. E. P.; Laufer, M. K.; Mathanga, D. P.; Takala-Harrison, S.; Skarbinski, J.; Claassen, C. W.; Dzinjalamala, F. K.; & Plowe, C. V. Return of widespread chloroquine-sensitive Plasmodium falciparum to Malawi. J. Infect. Dis. 2014, 210, 1110–1114. [CrossRef]
- Njiro, B. J.; Mutagonda, R.; Mwakyandile, T.; Sabas, D.; & Bwire G. M. Molecular surveillance of chloroquine-resistant Plasmodium falciparum in sub-Saharan African countries after withdrawal of chloroquine for treatment of uncomplicated malaria : A systematic review. J. Infect. Public Health 2022, 15, 550-557. [CrossRef] [PubMed]
- Tyagi, R. K.; Gleeson, P. J.; Arnold, L.; Tahar, R.; Prieur, E.; Decosterd, L.; Pérignon, J. L.; Olliaro, P.; & Druilhe, P. High-level artemisinin-resistance with quinine co-resistance emerges in P. falciparum malaria under in vivo artesunate pressure. BMC Medicine. 2018, 16, 1–19. [CrossRef]
- Vydyam, P.; Dutta, X. D.; Sutram, N.; Bhattacharyya, S.; & Bhattacharyya, M. K. A small-molecule inhibitor of the DNA recombinase Rad51 from Plasmodium falciparum synergizes with the antimalarial drugs artemisinin and chloroquine. J. Biol. Chem. 2019, 294, 8171–8183. [CrossRef]
- Gbotosho, G. O.; Folarin, O. A.; Bustamante, C.; Pereira Da Silva, L. H.; Mesquita, E.; Sowunmi, A.; Zalis, M. G.; Oduola, A. M. J.; & Happi, C. T. Short report: Different patterns of PfCRT and PfMDR1 polymorphisms in P. falciparum isolates from Nigeria and Brazil: The potential role of antimalarial drug selection pressure. Am. J Trop Med Hyg. 2012, 86, 211–213.
- Ikegbunam, M.N.; Nkonganyi, C.N.; Thomas, B.N.; Esimone, C.O.; Velavan, T.P.; & Ojurongbe, O. Analysis of Plasmodium falciparum PfCRT and PfMDR1 genes in parasite isolates from asymptomatic individuals in Southeast Nigeria 11 years after withdrawal of chloroquine. Malar. J. 2019, 18. [CrossRef]
- Tola, M.; Ajibola, O.; Idowu, E. T.; Omidiji, O.; Awolola, S. T.; & Amambua-Ngwa, A.. Molecular detection of drug resistant polymorphisms in Plasmodium falciparum isolates from Southwest, Nigeria. BMC Res. Notes, 2020, 13, 1–7. [CrossRef]
- Zhao, H.; Pi, L.; Zhao, L.; Qin, Y.; Zeng, W.; Xiang, Z.; Yang, Q.; Pan, M.; Li, X.; Zou, C.; Chen, X.; Zhao, W.; Lu, Y.; Wu, Y.; Duan, M.; Wang, X.; Li, X.; Mazier, D.; Huang, Y.; & Yang, Z. First detection in west Africa of a mutation that may contribute to artemisinin resistance Plasmodium falciparum. Front Genet. 2021, 12, 1–9. [CrossRef]
- Mwai, L.; Ochong, E.; Abdirahman, A.; Kiara, S. M.; Ward, S.; Kokwaro, G.; Sasi, P.; Marsh, K.; Borrmann, S.; MacKinnon, M.; & Nzila, A. Chloroquine resistance before and after its withdrawal in Kenya. Malar. J. 2009, 8, 1–10. [CrossRef]
- Muhammad, R. H.; Nock, I. H.; Ndams, I. S.; George, J. B.; & Deeni, Y. Distribution of PfMDR1 and PfCRT chloroquine drug resistance alleles in north-western Nigeria. Malar. World J. 2017, 8, 15. http://ovidsp.ovid.com/ovidweb.cgi?T=JS&PAGE=reference&D=pmnm5&NEWS=N&AN=34532238.
- Ross, L. S.; Dhingra, S. K.; Mok, S.; Yeo, T.; Wicht, K. J.; Kümpornsin, K.; Takala-harrison, S.; Witkowski, B.; Fairhurst, R. M.; Ariey, F.; Menard, D.; & Fidock, D. A. Emerging southeast Asian PfCRT mutations confer Plasmodium falciparum resistance to the first-line. Nat Commun. 2018, 25–28. [CrossRef]
- Gabryszewski, S. J.; Modchang, C., Musset, L.; Chookajorn, T.; & Fidock, D. A. Combinatorial genetic modeling of PfCRT-mediated drug resistance evolution in Plasmodium falciparum. Mol. Biol. Evol. 2016, 33, 1554–1570. [CrossRef]
- Salissou, A.; Zamanka, H.; Biyghe Binze, B.; Rivière, T.; Tichit, M.; Ibrahim, M. L.; & Fandeur, T. Low prevalence of PfCRT resistance alleles among patients with uncomplicated falciparum malaria in niger six years after chloroquine withdrawal. Malar Res Treat. 2014. [CrossRef]
- Gama, B. E.; Al Pereira-Carvalho, G.; Ji, F., Kosi, L.; Almeida De Oliveira, N. K.; Fortes, F.; Rosenthal, P. J.; Daniel-Ribeiro, C. T.; De, M.; & Ferreira-Da-Cruz, F. Plasmodium falciparum isolates from Angola show the Stct VMNT haplotype in the PfCRT gene. Malar J. 2010, 9, 174. http://www.malariajournal.com/content/9/1/174.
- Foguim, F. T.; Bogreau, H.; Gendrot, M.; Mosnier, J.; Fonta, I.; Benoit, N.; Amalvict, R.; Madamet, M.; Wein, S.; Pradines, B.; Augis, V.; Bastien, P.; Benoit-Vical, F.; Berry, A.; Brouqui, P.; Chauvin, P.; Cividin, M.; Courtier, F.; Delaunay, P.; Wolff, A. Prevalence of mutations in the Plasmodium falciparum chloroquine resistance transporter, PfCRT, and association with ex vivo susceptibility to common anti-malarial drugs against African Plasmodium falciparum isolates. Malar. J. 2020, 19, 1–9. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).