Submitted:
03 January 2023
Posted:
04 January 2023
You are already at the latest version
Abstract
Keywords:
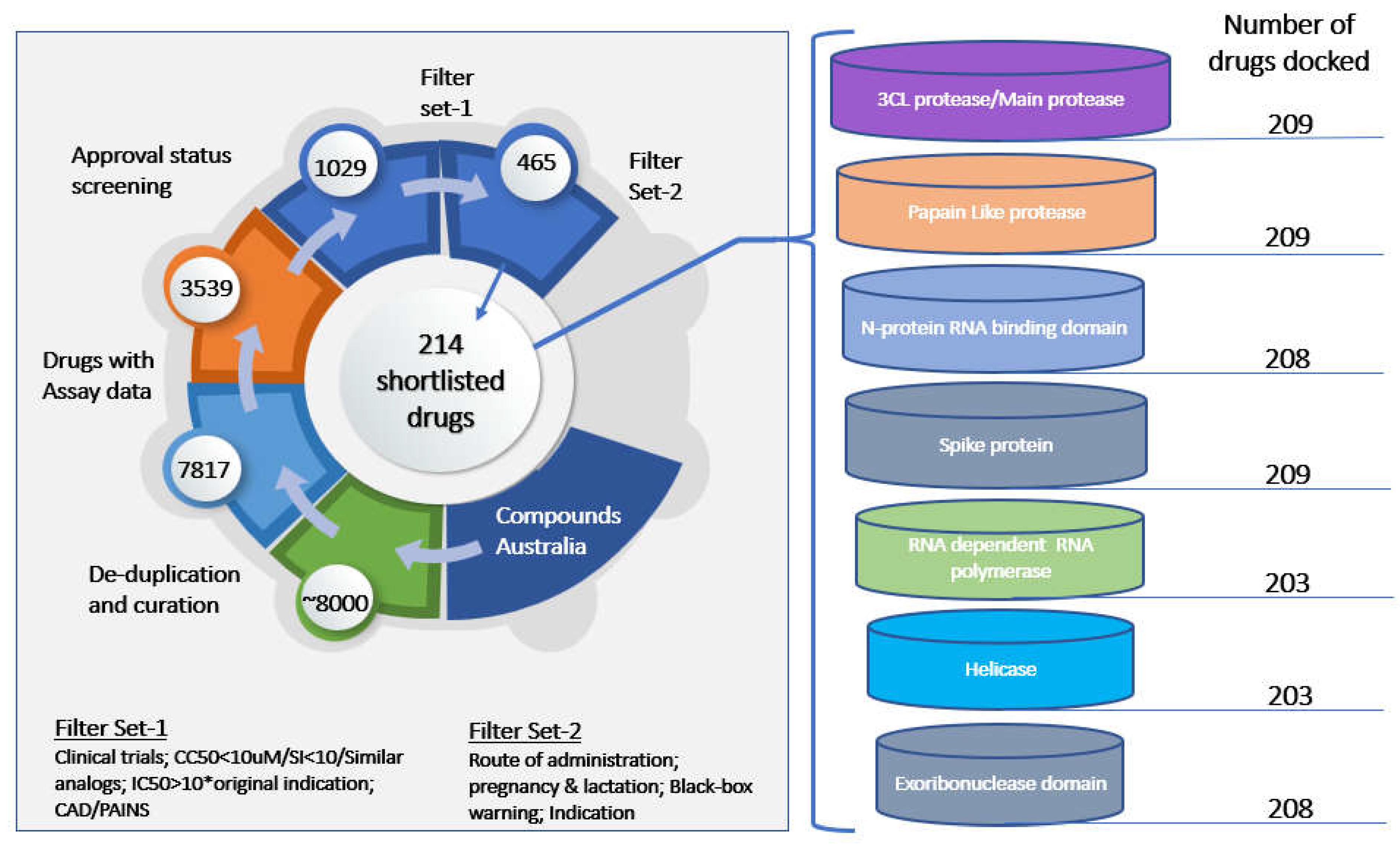
1. Introduction
2. Materials and Methodology
Protein Preparation
| S. No | Target protein | PDB ID | Resolution |
|---|---|---|---|
| 1. | Spike Receptor Binding Domain | 6M0J | 2.45 Å |
| 2. | Nucleocapsid protein RNA Binding domain | 6VYO | 2.2 Å |
| 3. | Papain Like protease (NSP3) | 6W9C | 2.7 Å |
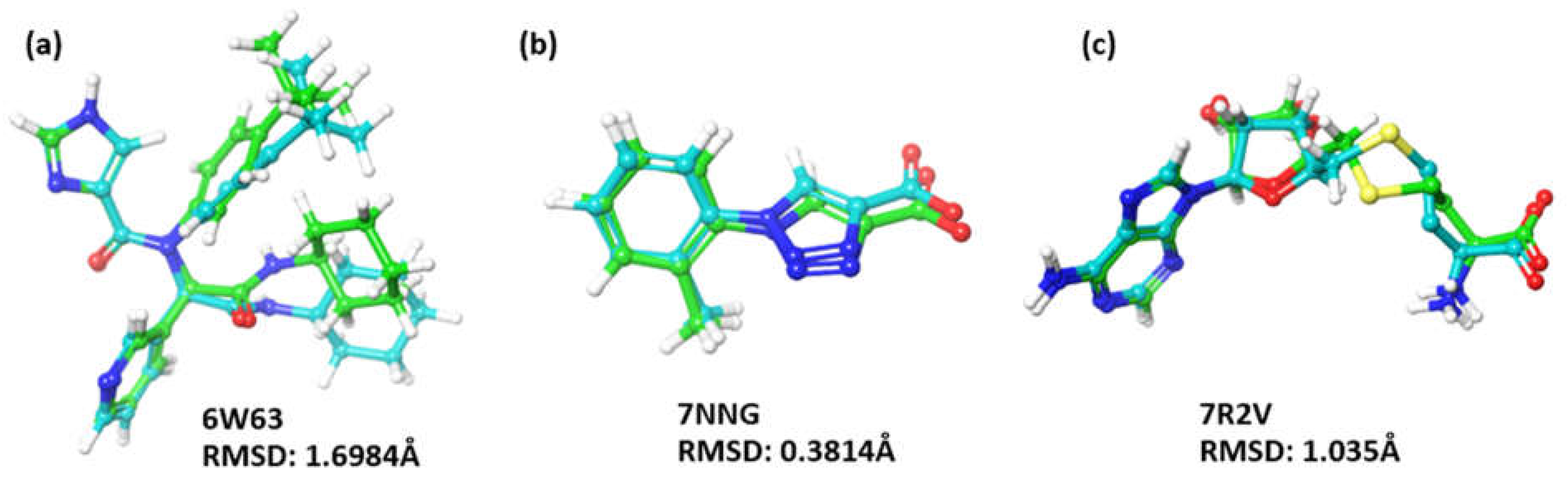
| 4. | Main protease (3CL protease) (NSP5) | 6W63 | 2.1 Å |
| 5. | RNA dependent RNA polymerase (NSP12) | 6M71 | 2.9 Å |
| 6. | Helicase (NSP13) | 7NNG | 2.38 Å |
| 7. | Exoribonuclease domain (NSP14) | 7R2V | 2.53 Å |
Ligand Preparation
Grid Generation
Molecular Docking Studies
Molecular Dynamics (MD)
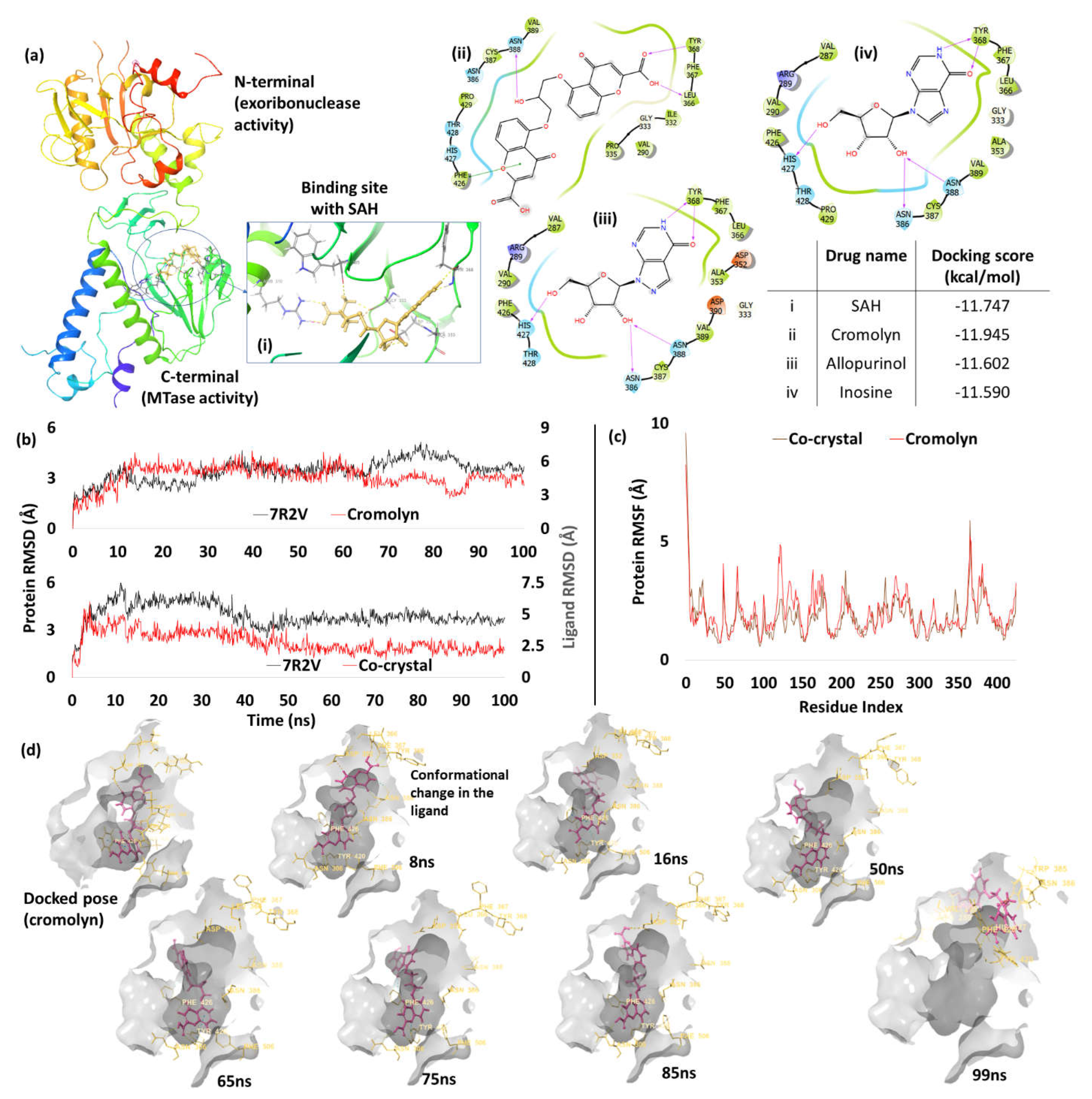
3. Results and Discussion
Docking Validation
Screening of Drugs and MD Studies
| S. No | Target code | Drug name | Dock score (kcal/mol) |
Type of interactions | Interacting residues – Bond length (in Å) |
|---|---|---|---|---|---|
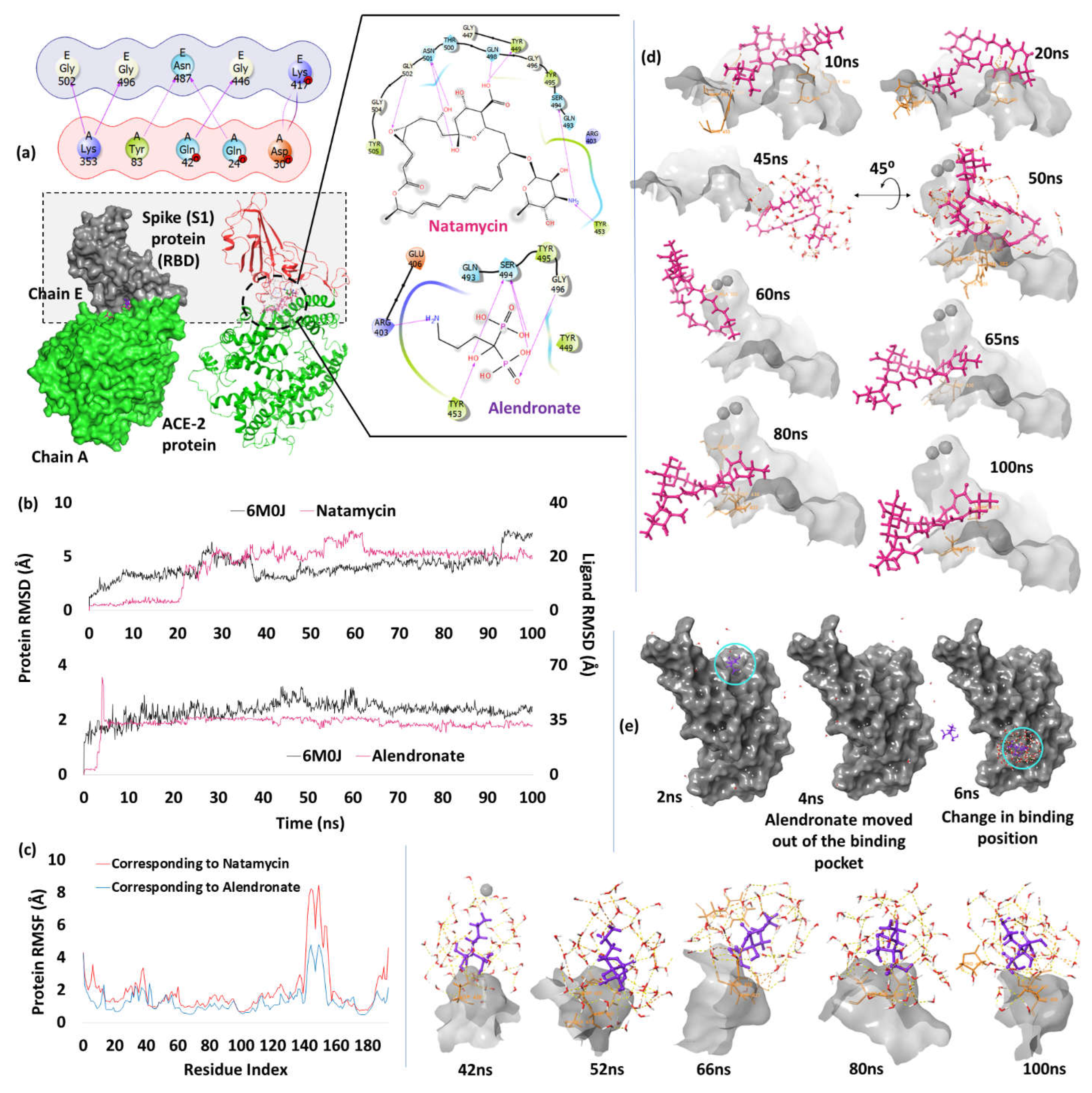
| 1. | Spike Receptor Binding Domain (6M0J) | Natamycin | -7.82 | H-bond | Tyr449: 1.97; Tyr453: 1.98; Gln498: 1.92; Asn501: 1.99, 2.46; Gly502: 2.15 |
| Alendronate | -7.69 | H-bond | Arg403: 1.95; Tyr453: 2.10; Ser494: 1.78, 1.9, 1.8; Gly496: 1.95 | ||
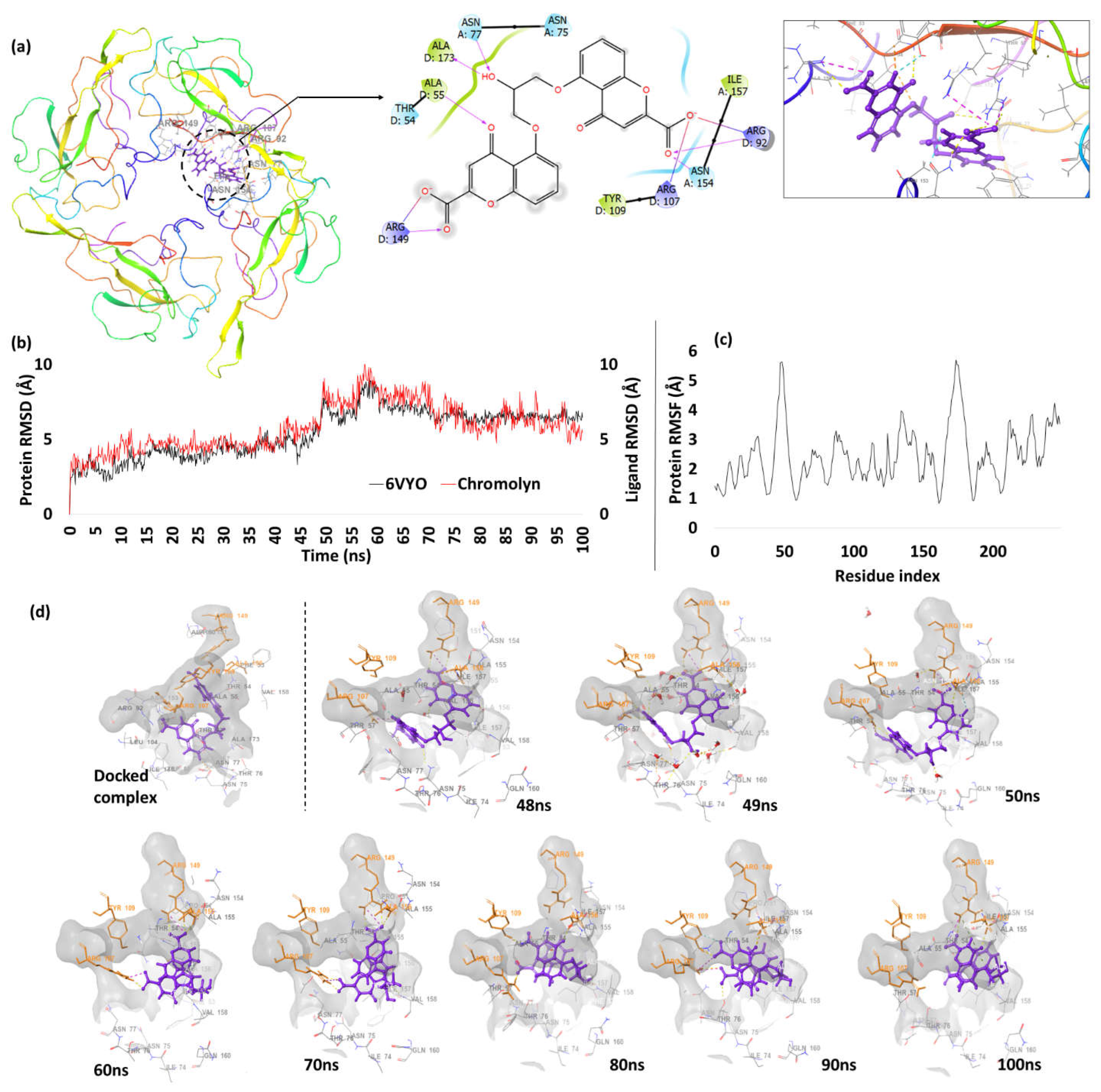
| 2. | N protein RNA Binding Domain (6VYO) | Cromolyn | -9.54 | H-bond | Ala154(A): 1.99; Asn55(D): 2.09; Ala55(D): 2.17; Arg92(D): 2.45; Ala173(D): 1.79; Arg149(D): 1.98 |
| Salt bridge | Arg107(D): 4.33; Arg149(D): 3.87 | ||||
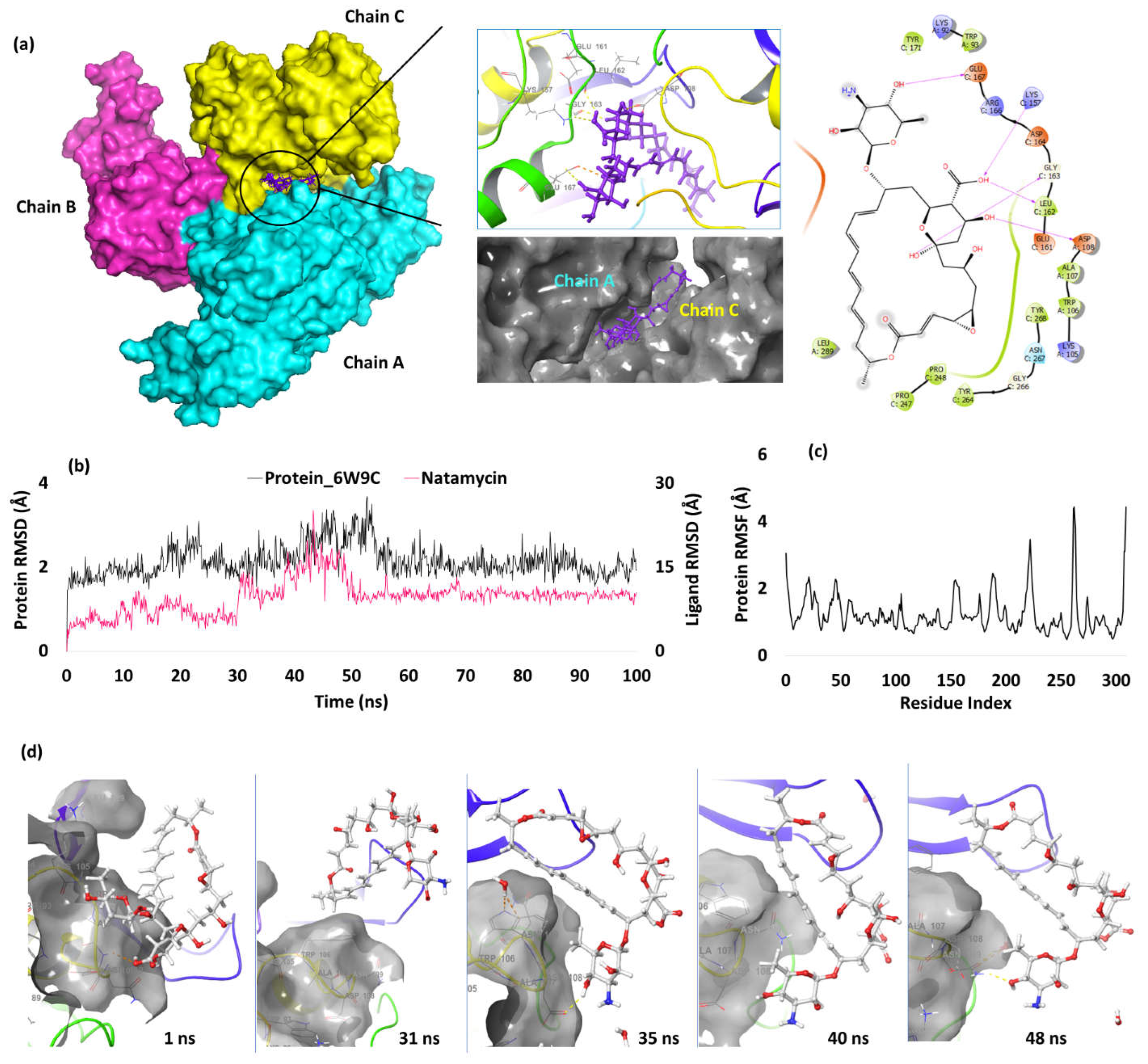
| 3. | NSP3 (6W9C) |
Natamycin | -7.39 | H-bond | Asp(A) 108: 2.52; Lys157(C): 2.08; Leu162(C): 2.10; Gly163(C): 2.57; Glu167(C): 2.06 |
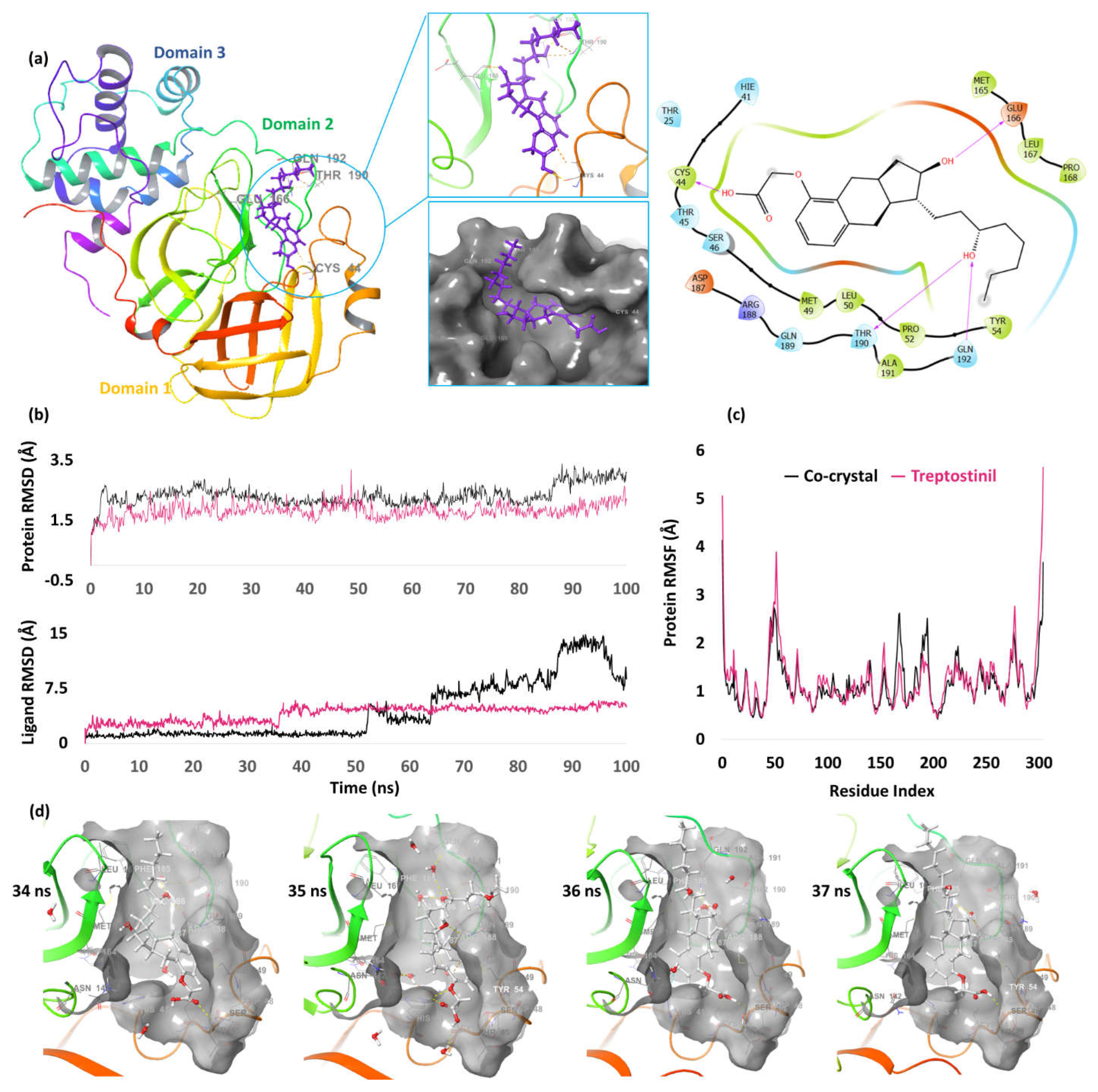
| 4. | NSP5 (6W63) |
Treprostinil | -10.76 | H-bond | Cys44: 1.63; Glu166: 1.97; Thr190: 1.87; Gln192: 1.97 |
| Co-crystal | -6.90 | H-bond | Gly143: 2.31; Asn142: 2.23; Hie163: 2.09; Glu166: 1.92 | ||
| Pi-pi stacking | Hie41: 5.14 | ||||
| 5. | NSP12 (6M71) |
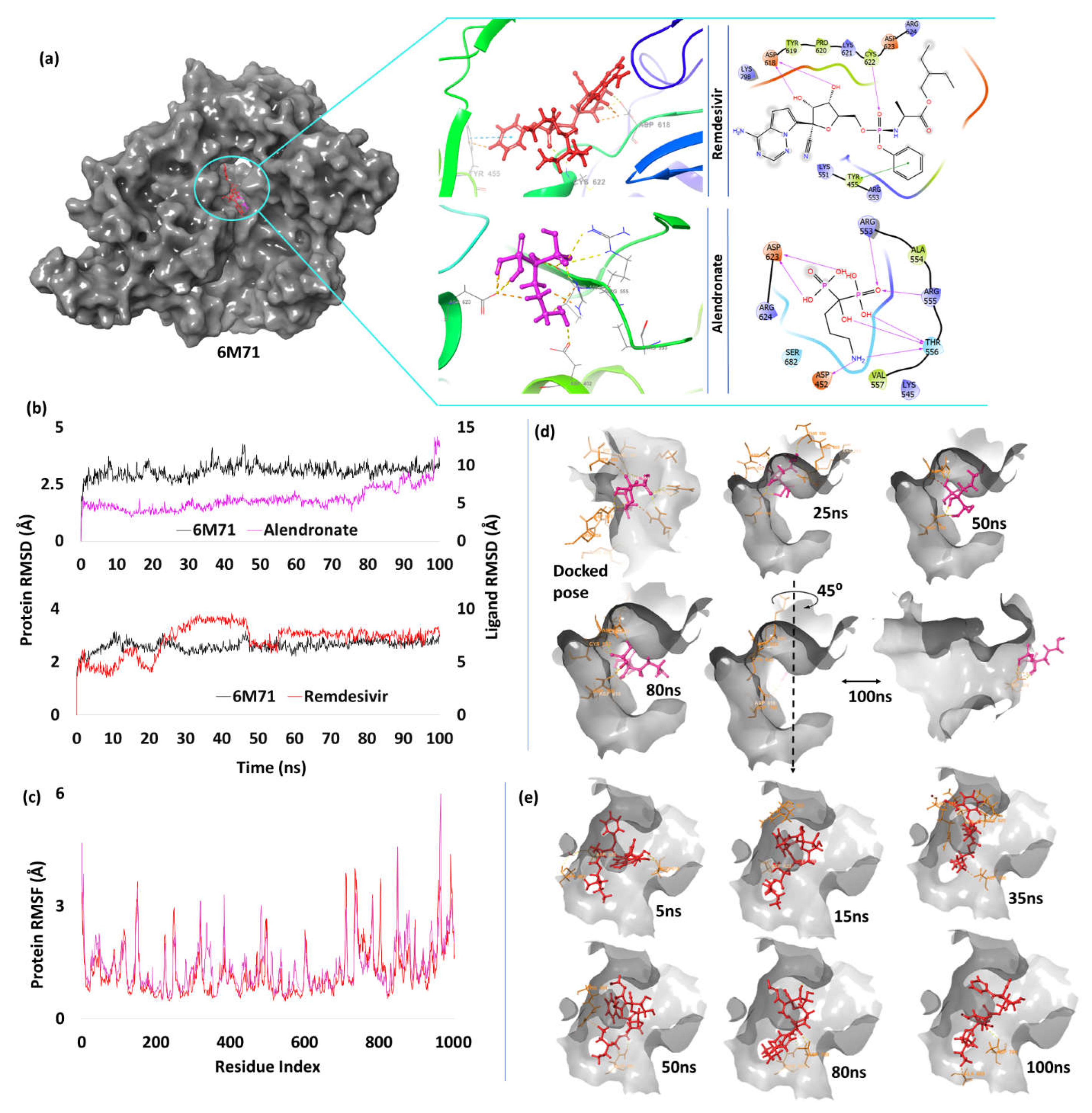
Alendronate | -7.86 | H-bond | Asp452: 1.95; Arg553: 1.94; Arg555: 2.64; Thr556: 1.61, 1.7, 1.83; Asp623: 1.76, 1.76 |
| Remdesivir | -3.27 | H-bond | Asp618: 1.81, 1.98; Cys622: 2.66 | ||
| Pi-Pi stacking | Tyr455: 5.34 | ||||
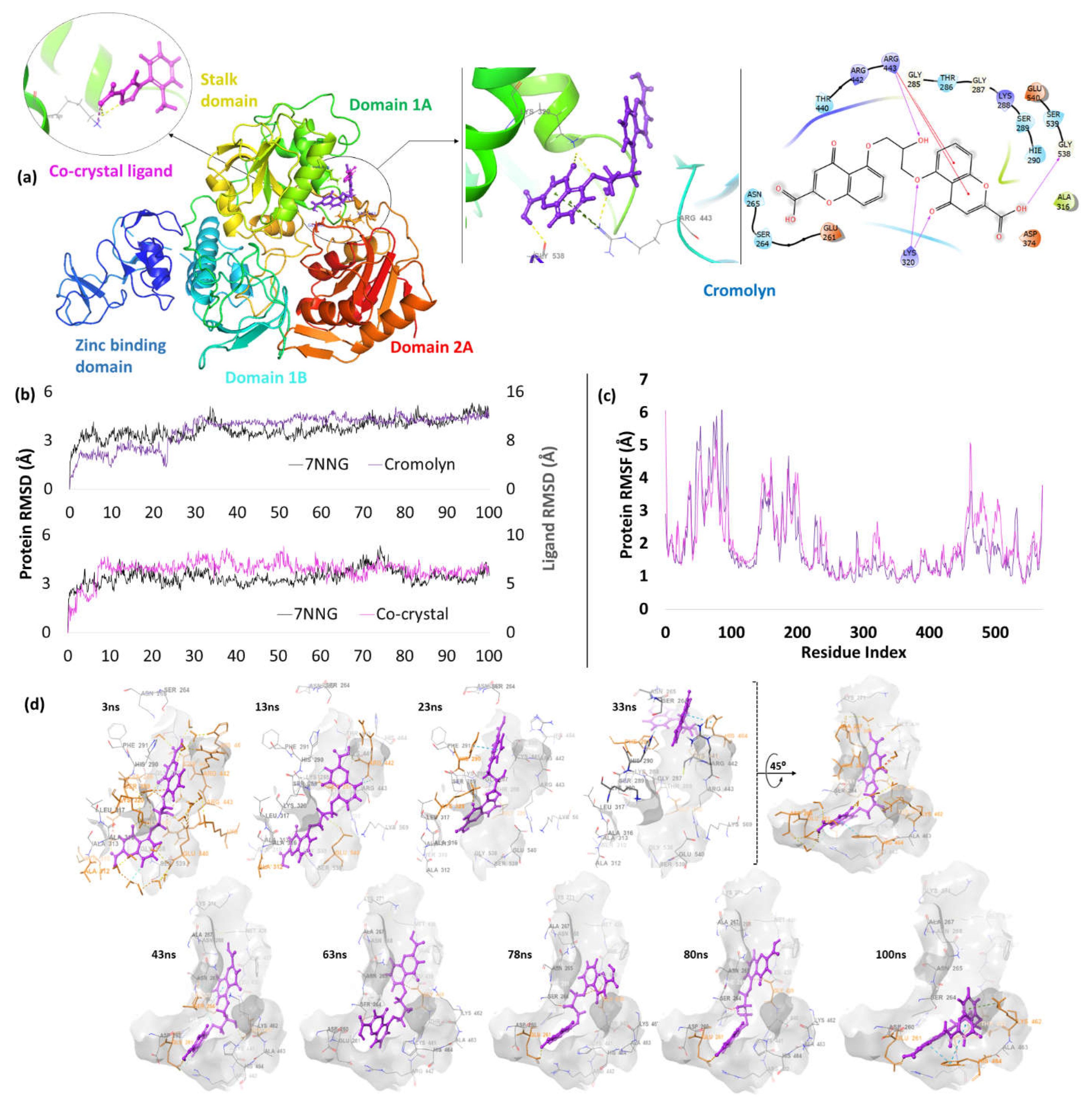
| 6. | NSP13 (7NNG) |
Cromolyn | -5.87 | H-bond | Arg443: 2.14; Lys320: 2.23, 2.54; Gly538: 2.08 |
| Pi-cation | Arg443: 4.89, 4.81 | ||||
| Co-crystal | -4.06 | H-bond | Lys320: 2.15 | ||
| Salt bridge | Lys320: 2.88; Lys323: 2.96 | ||||
| 7. | NSP14 (7R2V) |
Cromolyn | -11.95 | H-bond | Leu366: 2.13; Tyr368: 1.99; Asn388: 1.85 |
| Pi-Pi stacking | Phe426: 4.2 | ||||
| Co-crystal | -11.747 | H-bond | Arg310: 1.83; Gly333: 1.99; Asp352: 1.78, 1.99; Ala353: 2.37; Tyr368: 2.01, 2.01; Trp385: 2.10 | ||
| Salt bridge | Arg310: 2.81 |
Spike Receptor Binding Domain (RBD)
Nucleocapsid Protein RNA Binding Domain (NPRBD)
Papain Like Protease (PL pro)
3. CL Protease/Main Protease
RNA Dependent RNA Polymerase (RdRp)
Helicase
Exoribonuclease Domain
4. Conclusions and Future Studies
Institutional Review Board Statement and Informed Consent
Data Availability Statement
Acknowledgements
Conflict of Interest
Abbreviations
References
- COVID-19 Data Explorer - Our World in Data. (2022). Available online: https://ourworldindata.org/explorers/coronavirus-data-explorer (accessed on 23 November 2022).
- Wang, H.; Paulson, K.R.; Pease, S.A.; Watson, S.; Comfort, H.; et al. Estimating excess mortality due to the COVID-19 pandemic: a systematic analysis of COVID-19-related mortality, 2020-21. Lancet 2022, 399, 1513–1536. [Google Scholar] [CrossRef]
- World Health Organization. Monkeypox, COVID-19 & Other Global Health Issues Virtual Press Conference - 22 September 2022. (2022). Available online: https://www.who.int/publications/m/item/monkeypox--covid-19---other-global-health-issues-virtual-press-conference---22-september-2022 (accessed on 23 November 2022).
- World Health Organization. Weekly epidemiological update on COVID-19 - 28 December 2021. (2021). Available online: https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19---28-december-2021 (accessed on 28 November 2022).
- Mallapaty, S. China COVID wave could kill one million people, models predict. Nature 19 December 2022. [CrossRef]
- Holmes, E.C.; Zhang, Y.Z. The evolution and emergence of hantaviruses. Curr Opin Virol. 2015, 10, 27–33. [Google Scholar] [CrossRef]
- Yadav, R.; Chaudhary, J.K.; Jain, N.; Chaudhary, P.K.; Khanra, S.; et al. Role of Structural and Non-Structural Proteins and Therapeutic. Cells 2021, 10, 821. [Google Scholar] [CrossRef]
- Van Vuren, P.J.; McAuley, A.J.; Kuiper, M.J.; Singanallur, N.B.; Bruce, M.P.; et al. Highly Thermotolerant SARS-CoV-2 Vaccine Elicits Neutralising Antibodies against Delta and Omicron in Mice. Viruses 2022, 14, 800. [Google Scholar] [CrossRef] [PubMed]
- National Institutes of Health. Remdesivir. COVID-19 Treatment Guidelines. (2022). Available online: https://www.covid19treatmentguidelines.nih.gov/therapies/antiviral-therapy/remdesivir/ (accessed on 9 May 2022).
- Kozlov, M. Merck’s COVID pill loses its lustre: what that means for the pandemic. Nature 2021. [Google Scholar] [CrossRef] [PubMed]
- Butler, C.C.; Hobbs, F.D.R.; Gbinigie, O.A.; Rahman, N.M.; Hayward, G.; et al. Molnupiravir plus usual care versus usual care alone as early treatment for adults with COVID-19 at increased risk of adverse outcomes (PANORAMIC): an open-label, platform-adaptive randomised controlled trial. Lancet 2022. [Google Scholar] [CrossRef]
- Szabo, B.G.; Lenart, K.S.; Petrik, B.; Gaspar, Z.; Kiss-Dala, N.; et al. Favipiravir treatment does not influence disease progression among adult patients hospitalized with moderate-to-severe COVID-19: a prospective, sequential cohort study from Hungary. Geroscience 2021, 43, 2205–2213. [Google Scholar] [CrossRef]
- Jimenez, D.; Paxlovid: what we know about Pfizer’s Covid-19 pill. (2022). Available online: https://www.pharmaceutical-technology.com/features/paxlovid-pfizer-covid-19-pill/ (accessed on 7 February 2022).
- National Institutes of Health. Ritonavir-Boosted Nirmatrelvir (Paxlovid). (2022). Available online: https://www.covid19treatmentguidelines.nih.gov/therapies/antiviral-therapy/ritonavir-boosted-nirmatrelvir--paxlovid-/ (accessed on 9 May 2022).
- Wouters, O.J.; McKee, M.; Luyten, J. Estimated Research and Development Investment Needed to Bring a New Medicine to Market, 2009-2018. JAMA 2020, 323, 844–853. [Google Scholar] [CrossRef] [PubMed]
- Pushpakom, S.; Iorio, F.; Eyers, P.A.; Escott, K.J.; Hopper, S.; et al. Drug repurposing: Progress, challenges and recommendations. Nat. Rev. Drug Discov. 2018, 18, 41–58. [Google Scholar] [CrossRef] [PubMed]
- Jain, H.A.; Agarwal, V.; Bansal, C.; Kumar, A.; Faheem, F.; et al. CoviRx: A User-Friendly Interface for Systematic Down-Selection of Repurposed Drug Candidates for COVID-19. Data 2022, 7, 164. [Google Scholar] [CrossRef]
- MacRaild, C.A.; Mohammed, M.U.R.; Faheem Murugesan, S.; Styles, I.K.; et al. Systematic Down-Selection of Repurposed Drug Candidates for COVID-19. Int J Mol Sci 2022, 23, 11851. [Google Scholar] [CrossRef]
- McAuley, A.J.; Jansen van Vuren, P.; Mohammed, M.-U.-R.; Faheem Goldie, S.; et al. Use of Human Lung Tissue Models for Screening of Drugs against SARS-CoV-2 Infection. Viruses 2022, 14, 2417. [Google Scholar] [CrossRef] [PubMed]
- Tanimoto, T.T. An Elementary Mathematical Theory of Classification and Prediction. In: Proc. IBM Internal Report. International Business Machines Corp. pp. 1–11.
- Kushwaha, P.P.; Singh, A.K.; Bansal, T.; Yadav, A.; Prajapati, K.S.; et al. Identification of Natural Inhibitors Against SARS-CoV-2 Drugable Targets Using Molecular Docking, Molecular Dynamics Simulation, and MM-PBSA Approach. Front Cell Infect Microbiol 2021, 728. [Google Scholar] [CrossRef] [PubMed]
- Friesner, R.A.; Banks, J.L.; Murphy, R.B.; Halgren, T.A.; Klicic, J.J.; et al. Glide: A New Approach for Rapid, Accurate Docking and Scoring. 1. Method and Assessment of Docking Accuracy. J Med Chem 2004, 47, 1739–1749. [Google Scholar] [CrossRef] [PubMed]
- El-Aziz, N.; Shehata, M.; Awad, O.; El-Sohaimy, S. Inhibition of COVID-19 RNA-Dependent RNA Polymerase by Natural Bioactive Compounds: Molecular Docking Analysis. Egypt. J. Chem 2021, 1989–2001. [Google Scholar] [CrossRef]
- Li, D.; Luan, J.; Zhang, L. Molecular docking of potential SARS-CoV-2 papain-like protease inhibitors. Biochem Biophys Res Commun 2021, 538, 72–79. [Google Scholar] [CrossRef]
- Kang, S.; Yang, M.; Hong, Z.; Zhang, L.; Huang, Z.; et al. Crystal structure of SARS-CoV-2 nucleocapsid protein RNA binding domain reveals potential unique drug targeting sites. Acta Pharm Sin B 2020, 10, 1228–1238. [Google Scholar] [CrossRef]
- Shelley, J.C.; Cholleti, A.; Frye, L.L.; Greenwood, J.R.; Timlin, M.R.; et al. Epik: A software program for pKa prediction and protonation state generation for drug-like molecules. J Comput Aided Mol Des 2007, 21, 681–691. [Google Scholar] [CrossRef]
- Nutt, D.R.; Smith, J.C. Molecular dynamics simulations of proteins: Can the explicit water model be varied? J Chem Theory Comput 2007, 3, 1550–1560. [Google Scholar] [CrossRef]
- Lu, C.; Wu, C.; Ghoreishi, D.; Chen, W.; Wang, L.; et al. OPLS4: Improving force field accuracy on challenging regimes of chemical space. J Chem Theory Comput 2021, 17, 4291–4300. [Google Scholar] [CrossRef]
- Hevener, K.E.; Zhao, W.; Ball, D.M.; Babaoglu, K.; Qi, J.; et al. Validation of molecular docking programs for virtual screening against dihydropteroate synthase. J Chem Inf Model 2009, 49, 444–460. [Google Scholar] [CrossRef]
- Ahmed, S.F.; Quadeer, A.A.; McKay, M.R. Preliminary identification of potential vaccine targets for the COVID-19 Coronavirus (SARS-CoV-2) Based on SARS-CoV Immunological Studies. Viruses 2020, 12, 254. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Cheng, Y.; Wu, Y. Understanding SARS-CoV-2-Mediated Inflammatory Responses: From Mechanisms to Potential Therapeutic Tools. Virol. Sin. 2020, 35, 266–271. [Google Scholar] [CrossRef] [PubMed]
- Jawad, B.; Adhikari, P.; Podgornik, R.; Ching, W.Y. Key Interacting Residues between RBD of SARS-CoV-2 and ACE2 Receptor: Combination of Molecular Dynamics Simulation and Density Functional Calculation. J Chem Inf Model 2021, 61, 4425–4441. [Google Scholar] [CrossRef] [PubMed]
- Tatar, G.; Ozyurt, E.; Turhan, K. Computational drug repurposing study of the RNA binding domain of SARS-CoV-2 nucleocapsid protein with anti-viral agents. Biotechnol Prog 2021, 37, e3110. [Google Scholar] [CrossRef] [PubMed]
- Báez-Santos, Y.M.; St. John, S.E.; Mesecar, A.D. The SARS-coronavirus papain-like protease: Structure, function and inhibition by designed anti-viral compounds. Antivir. Res. 2015, 115, 21–38. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, B.; Batool, M.; Ain, Q.U.; Kim, M.S.; Choi, S. Exploring the binding mechanism of PF-07321332 SARS-CoV-2 protease inhibitor through molecular dynamics and binding free energy simulations. Int J Mol Sci 2021, 22, 9124. [Google Scholar] [CrossRef]
- Zhang, L.; Lin, D.; Sun, X.; Curth, U.; Drosten, C.; et al. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved a-ketoamide inhibitors. Science 2020, 368, 409–412. [Google Scholar] [CrossRef]
- Aftab, S.O.; Ghouri, M.Z.; Masood, M.U.; Haider, Z.; Khan, Z.; et al. Analysis of SARS-CoV-2 RNA-dependent RNA polymerase as a potential therapeutic drug target using a computational approach. J Transl Med 2020, 18, 1–15. [Google Scholar] [CrossRef]
- Wu, Y. Unwinding and rewinding: Double faces of helicase? J. Nucleic Acids. 2012, 2012, 140601. [Google Scholar] [CrossRef] [PubMed]
- Ambrus, A.M.; Frolov, M.V. The diverse roles of RNA helicases in RNAi. Cell Cycle 2009, 8, 3500–3505. [Google Scholar] [CrossRef]
- Adedeji, A.O.; Marchand, B.; te Velthuis, A.J.W.; Snijder, E.J.; Weiss, S.; et al. Mechanism of nucleic acid unwinding by SARS-CoV helicase. PLoS ONE 2012, 7, e36521. [Google Scholar] [CrossRef]
- Pitsillou, E.; Liang, J.; Hung, A.; Karagiannis, T.C. The SARS-CoV-2 helicase as a target for anti-viral therapy: Identification of potential small molecule inhibitors by in silico modelling. J Mol Graph Model 2022, 114, 108193. [Google Scholar] [CrossRef] [PubMed]
- Tahir, M. Coronavirus genomic nsp14-ExoN, structure, role, mechanism, and potential application as a drug target. J Med Virol. 2021, 93, 4258–4264. [Google Scholar] [CrossRef]
- Ogando, N.S.; Zevenhoven-Dobbe, J.C.; van der Meer, Y.; Bredenbeek, P.J.; Posthuma, C.C.; et al. The Enzymatic Activity of the nsp14 Exoribonuclease Is Critical for Replication of MERS-CoV and SARS-CoV-2. J Virol 2020, 94, e01246–20. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Wu, L.; Shaw, N.; Gao, Y.; Wang, J.; et al. Structural basis and functional analysis of the SARS coronavirus nsp14-nsp10 complex. Proc Natl Acad Sci USA 2015, 112, 9436–9441. [Google Scholar] [CrossRef]
- Prasetya, E.; Mulia, B.; Luke, K. Inhaled prostacyclin analogues in COVID-19 associated acute respiratory distress syndrome: scientific rationale. Egypt Heart J. 2021, 73, 82. [Google Scholar] [CrossRef]
- Nasrullah, A.; Virk, S.; Shah, A.; Jacobs, M.; Hamza, A.; et al. Acute respiratory distress syndrome and the use of inhaled pulmonary vasodilators in the COVID-19 era: A narrative review. Life 2022, 12, 1766. [Google Scholar] [CrossRef]
- Rajpal, S.; Inpatient use of inhaled pulmonary vasodilator therapy in patients infected with COVID-19 - American College of Cardiology. (2020). Available online: https://www.acc.org/latest-in-cardiology/articles/2020/05/13/08/55/inpatient-use-of-inhaled-pulmonary-vasodilator-therapy-in-patients-infected-with-covid-19 (accessed on 2 January 2023).
- Degli Esposti, L.; Perrone, V.; Sangiorgi, D.; Andretta, M.; Bartolini, F.; et al. The use of oral amino-bisphosphonates and coronavirus disease 2019 (COVID-19) Outcomes. J. Bone Miner. Res. 2021, 36, 2177–2183. [Google Scholar] [CrossRef]
- Von Andrian, U.H.; Thompson, J.; Wang, Y.; Dreischulte, T.; Barreiro, O.; et al. Association between Bisphosphonate use and COVID-19 related outcomes: a retrospective cohort study. medRxiv 2022. [Google Scholar] [CrossRef]
- Murugan, N.A.; Kumar, S.; Jeyakanthan, J.; Srivastava, V. Searching for target-specific and multi-targeting organics for Covid-19 in the Drugbank database with a double scoring approach. Sci Rep 2020, 10, 19125. [Google Scholar] [CrossRef] [PubMed]
- Staten, M. New COVID-19 treatment being explored at TTUHSC El Paso. (2022) Available online:. Available online: https://www.krwg.org/regional/2022-01-19/new-covid-19-treatment-being-explored-at-ttuhsc-el-paso (accessed on 2 January 2023).
- Michelson, E.A.; Cromolyn sodium for treatment of COVID-19 pneumonia. ClinicalTrials.gov. (2021). Available online: https://clinicaltrials.gov/ct2/show/NCT05077917 (accessed on 2 January 2023).
- Miranda-Quintana, R.A.; Bajusz, D.; Rácz, A.; Héberger, K. Extended similarity indices: the benefits of comparing more than two objects simultaneously. Part 1: Theory and characteristics. J Cheminform; 2021, 13, 32. [Google Scholar] [CrossRef] [PubMed]
- Miranda-Quintana, R.A.; Rácz, A.; Bajusz, D.; Héberger, K. Extended similarity indices: the benefits of comparing more than two objects simultaneously. Part 2: speed, consistency, diversity selection. J Cheminform 2021, 13, 33. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





