Submitted:
11 January 2023
Posted:
13 January 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
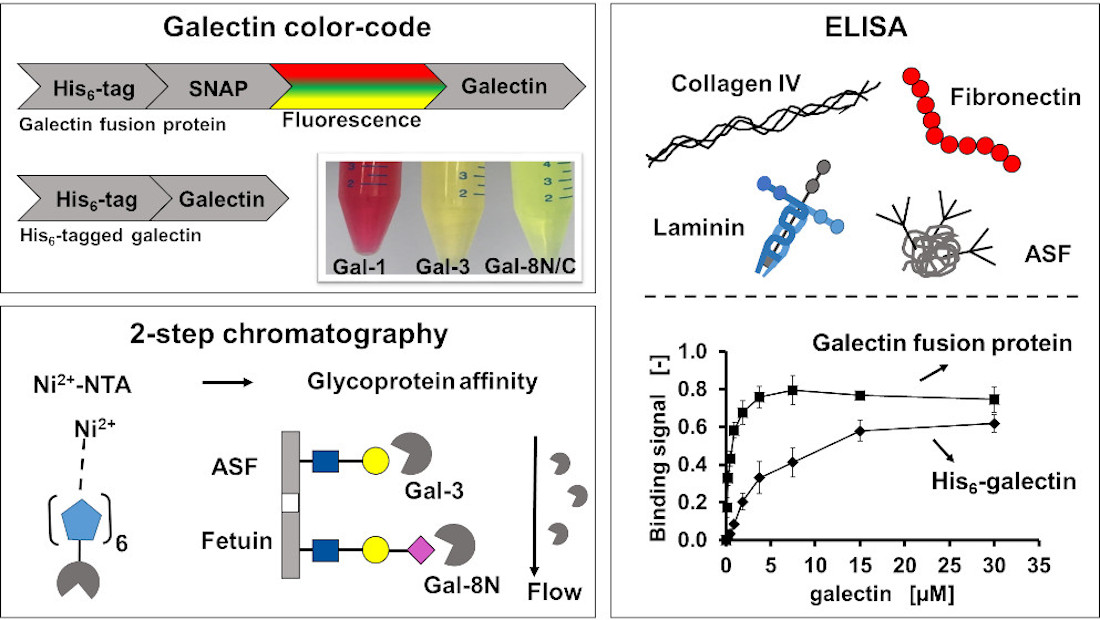
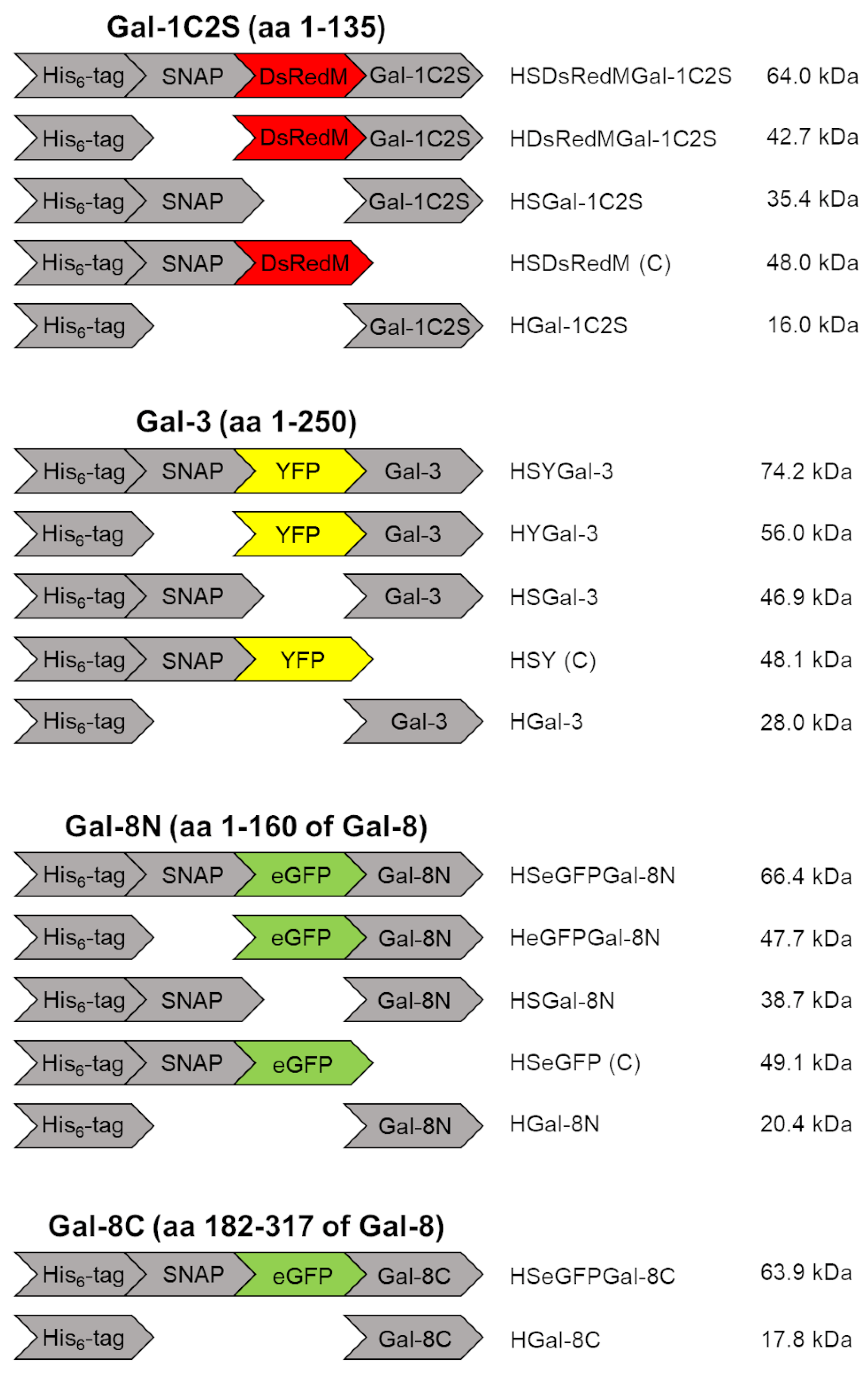
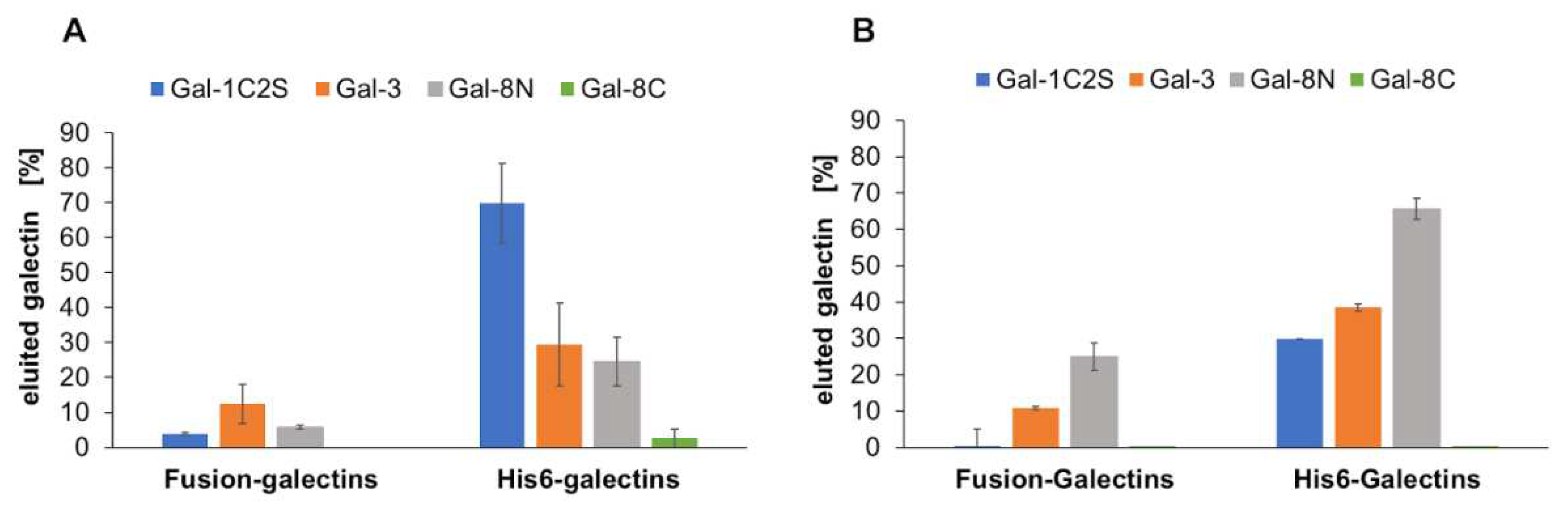
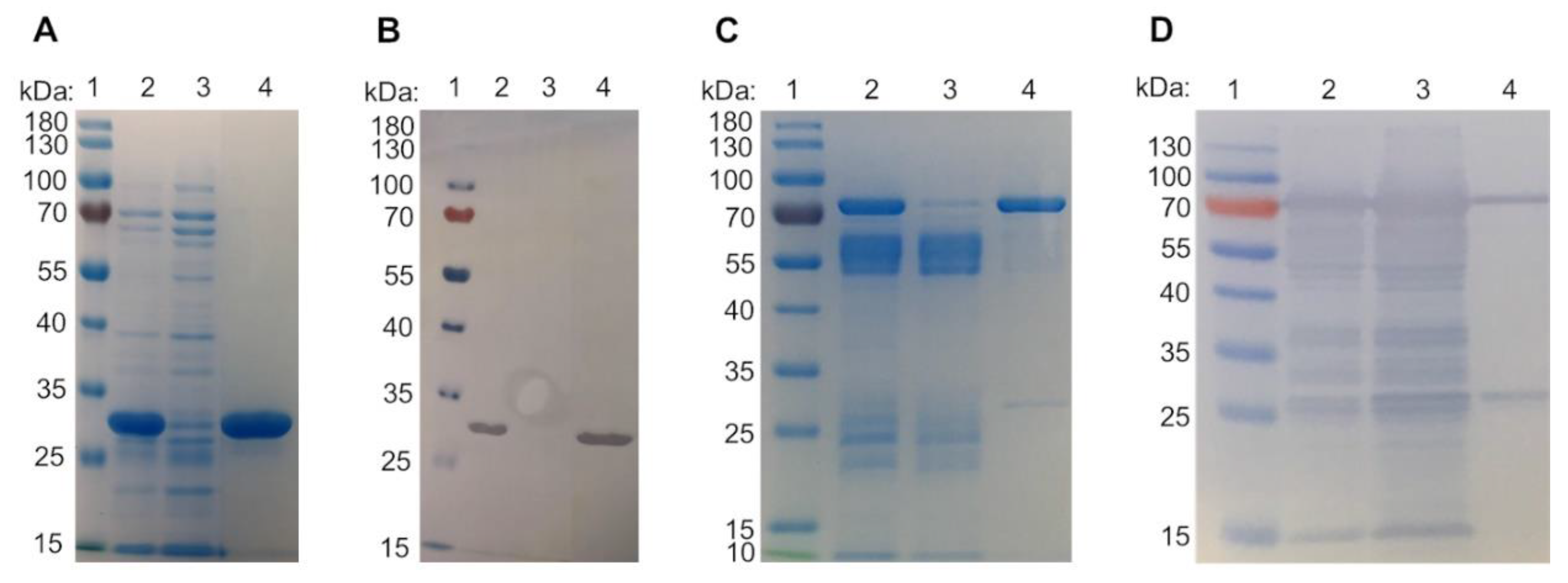
2.1. Galectin purification
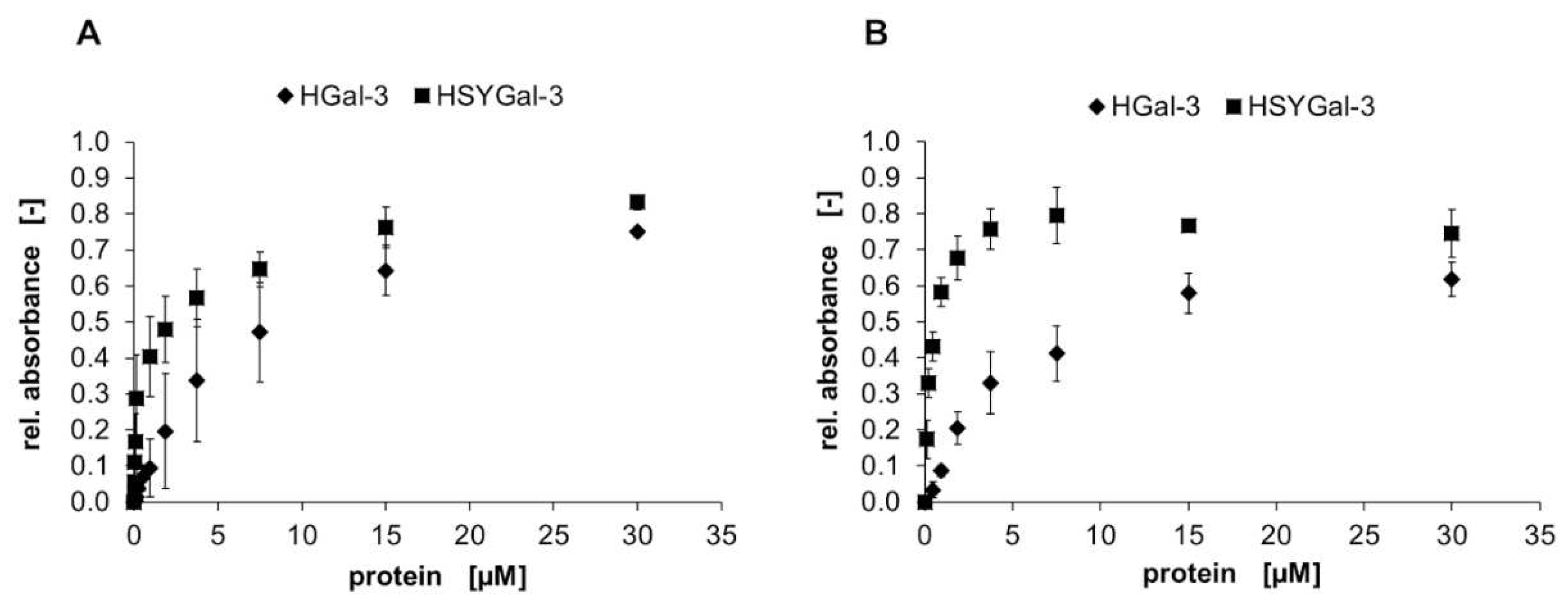
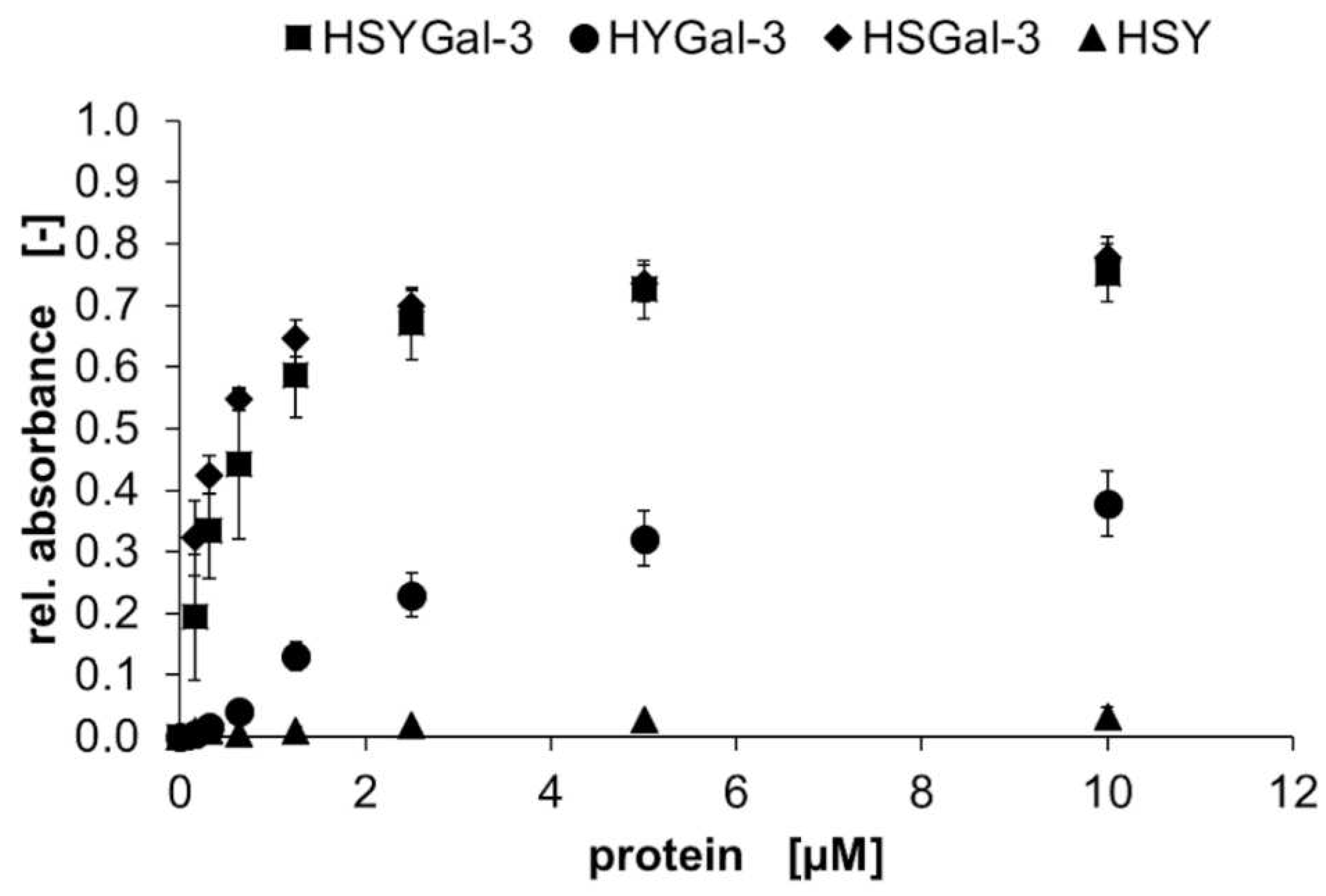
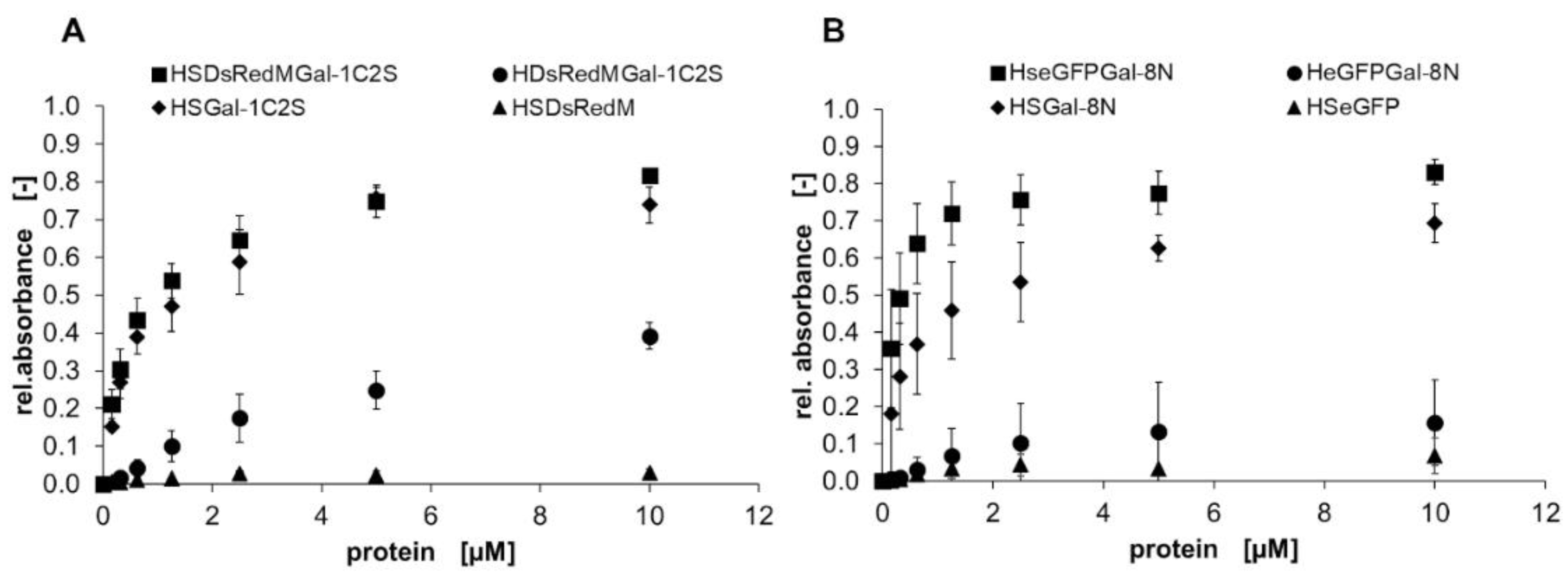
2.2. Binding characteristics of Gal-3 constructs on ASF
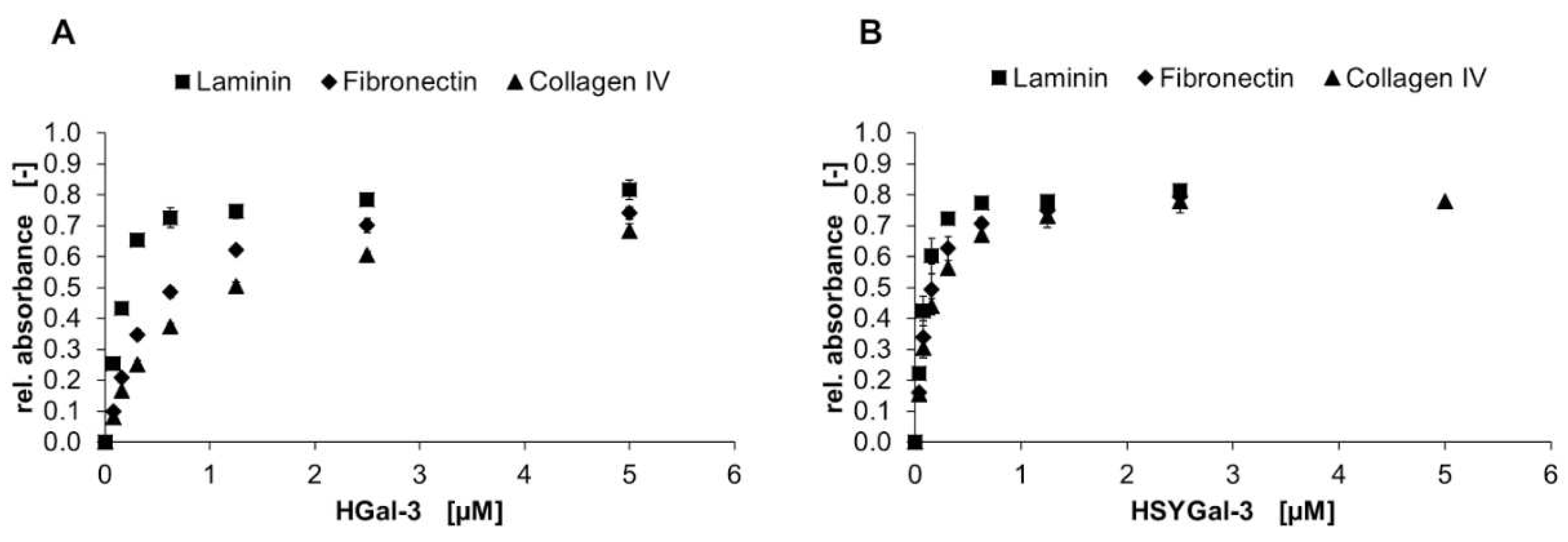
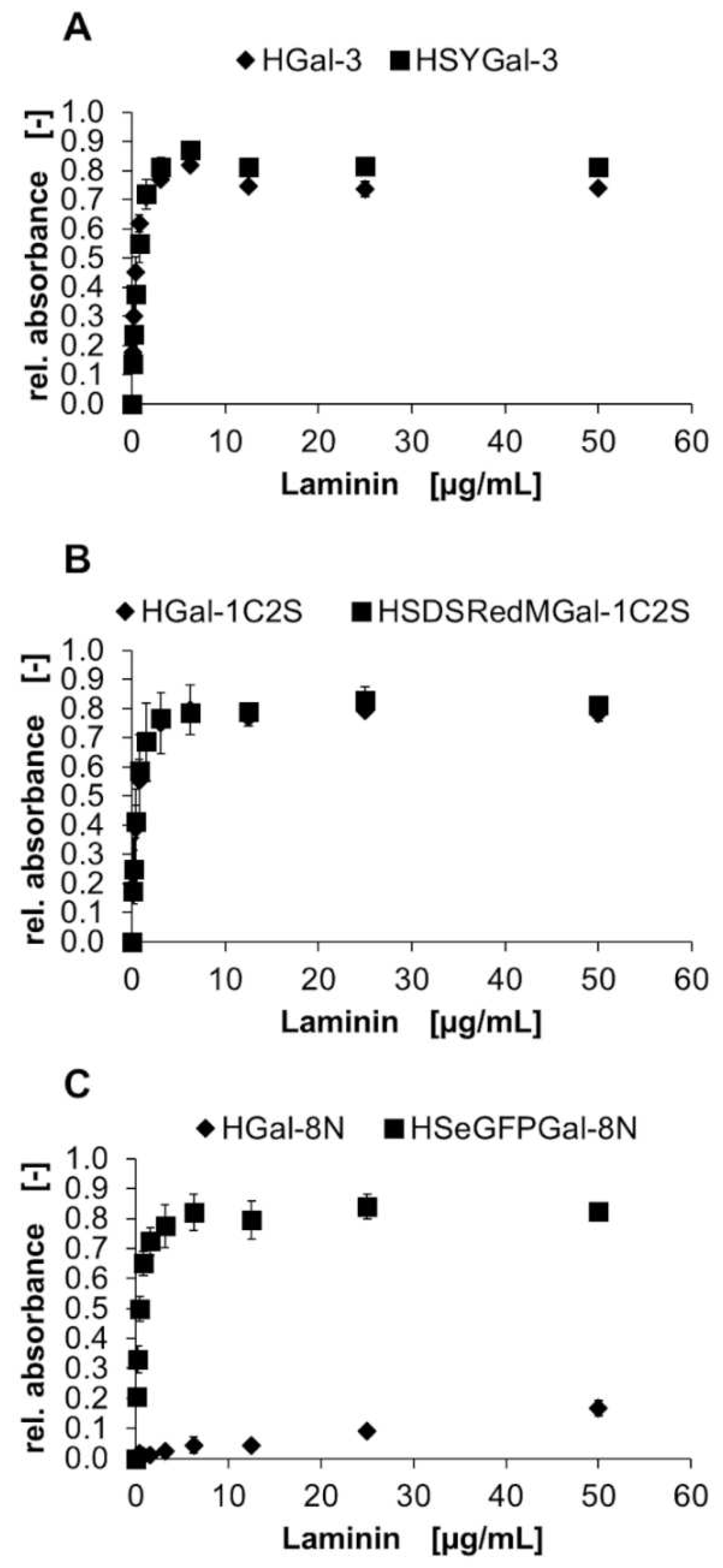
2.3. Binding of Gal-3 constructs to ECM glycoproteins
3. Discussion
3.1. Galectin purification on glycoprotein columns
3.2. Binding of galectins to ASF and ECM glycoproteins
4. Materials and Methods
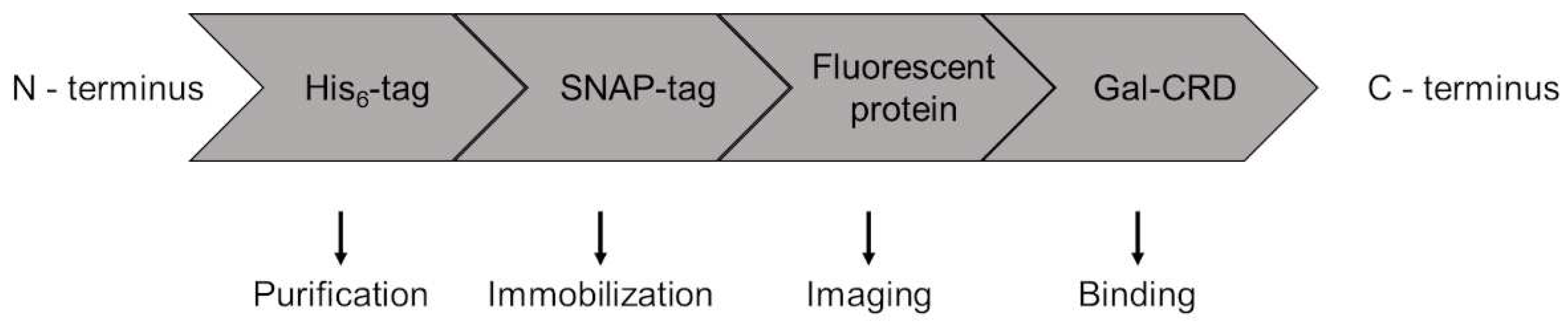
4.1. Galectin constructs
4.2. Expression and purification
4.3. Glycoprotein affinity resin and galectin purification
4.4. SDS-PAGE and Immunoblot
4.5. Size exclusion chromatography (SEC)
| Maximum pressure limit | pmax |
| Pressure limit at a maximum flow rate | ∆p |
| Pressure of the dripping column at max. flow rate (1.8 mL*min−1) | ∆pcolumn |
| Pressure with MQ at max. flow rate (not attached to the system) | ∆pbefore |
| Pressure of column at intended conditions (0.75 mL*min−1, PBS pH 7.5) | ∆pbefore(2) |
| Total system pressure | ∆ptotal |
| Average distribution constant | KAV |
| Total volume of the column | VC |
| Void volume | V0 |
| Elution volume of standard protein/sample | VE |
4.6. Galectin binding on ASF
4.7. Galectin binding on ECM glycoproteins
4.8. Galectin-mediated crosslinking of extracellular matrix glycoproteins
4.9. Enzyme-linked lectin assay (ELLA)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Cummings, R.D.; Liu, F.T.; Rabinovich, G.A.; Stowell, S.R.; Vasta, G.R. Galectins. In Essentials of Glycobiology; Varki, A., Cummings, R.D., Esko, J.D., Stanley, P., Hart, G.W., Aebi, M., Mohnen, D., Kinoshita, T., Packer, N.H., et al., Eds.; Cold Spring Harbor (NY), 2022; pp. 491–504. [Google Scholar]
- Nakahara, S.; Raz, A. On the role of galectins in signal transduction. Methods Enzymol 2006, 417, 273–289. [Google Scholar] [CrossRef]
- Viguier, M.; Advedissian, T.; Delacour, D.; Poirier, F.; Deshayes, F. Galectins in epithelial functions. Tissue Barriers 2014, 2, e29103. [Google Scholar] [CrossRef]
- Yang, R.Y.; Rabinovich, G.A.; Liu, F.T. Galectins: structure, function and therapeutic potential. Expert Rev Mol Med 2008, 10, e17. [Google Scholar] [CrossRef]
- Johannes, L.; Jacob, R.; Leffler, H. Galectins at a glance. J Cell Sci 2018, 131. [Google Scholar] [CrossRef]
- Hokama, A.; Mizoguchi, E.; Mizoguchi, A. Roles of galectins in inflammatory bowel disease. World J Gastroenterol 2008, 14, 5133–5137. [Google Scholar] [CrossRef] [PubMed]
- Girotti, M.R.; Salatino, M.; Dalotto-Moreno, T.; Rabinovich, G.A. Sweetening the hallmarks of cancer: Galectins as multifunctional mediators of tumor progression. J Exp Med 2020, 217. [Google Scholar] [CrossRef] [PubMed]
- Hara, A.; Niwa, M.; Noguchi, K.; Kanayama, T.; Niwa, A.; Matsuo, M.; Hatano, Y.; Tomita, H. Galectin-3 as a Next-Generation Biomarker for Detecting Early Stage of Various Diseases. Biomolecules 2020, 10. [Google Scholar] [CrossRef] [PubMed]
- Dong, R.; Zhang, M.; Hu, Q.; Zheng, S.; Soh, A.; Zheng, Y.; Yuan, H. Galectin-3 as a novel biomarker for disease diagnosis and a target for therapy (Review). Int J Mol Med 2018, 41, 599–614. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Bosch, N.; Barranco, L.E.; Orozco, C.A.; Moreno, M.; Visa, L.; Iglesias, M.; Oldfield, L.; Neoptolemos, J.P.; Greenhalf, W.; Earl, J.; et al. Increased plasma levels of galectin-1 in pancreatic cancer: potential use as biomarker. Oncotarget 2018, 9, 32984–32996. [Google Scholar] [CrossRef] [PubMed]
- Masoodi, M.; Shah, Z.A.; Beigh, A.H.; Ahmad, S.Z.; Mir, A.W.; Yasin, B.; Rasool, R.; Masoodi, K.Z.; Bhat, G.M. Galectin-1 as a predictive biomarker in ovarian cancer. J Ovarian Res 2021, 14, 123. [Google Scholar] [CrossRef] [PubMed]
- Yi, N.; Zhao, X.; Ji, J.; Xu, M.; Jiao, Y.; Qian, T.; Zhu, S.; Jiang, F.; Chen, J.; Xiao, M. Serum galectin-3 as a biomarker for screening, early diagnosis, prognosis and therapeutic effect evaluation of pancreatic cancer. J Cell Mol Med 2020, 24, 11583–11591. [Google Scholar] [CrossRef]
- Bojarová, P.; Tavares, M.R.; Laaf, D.; Bumba, L.; Petrásková, L.; Konefal, R.; Bláhová, M.; Pelantová, H.; Elling, L.; Etrych, T.; et al. Biocompatible glyconanomaterials based on HPMA-copolymer for specific targeting of galectin-3. J Nanobiotechnology 2018, 16, 73. [Google Scholar] [CrossRef]
- Bumba, L.; Laaf, D.; Spiwok, V.; Elling, L.; Křen, V.; Bojarová, P. Poly-N-Acetyllactosamine Neo-Glycoproteins as Nanomolar Ligands of Human Galectin-3: Binding Kinetics and Modeling. Int J Mol Sci 2018, 19. [Google Scholar] [CrossRef]
- Laaf, D.; Bojarová, P.; Pelantová, H.; Křen, V.; Elling, L. Tailored Multivalent Neo-Glycoproteins: Synthesis, Evaluation, and Application of a Library of Galectin-3-Binding Glycan Ligands. Bioconjug Chem 2017, 28, 2832–2840. [Google Scholar] [CrossRef]
- Long, F.; Li, W.; Chen, W.; Liu, D.; Chen, Y.; Zhou, R.; Li, P. An amperometric biosensor based on Cu2O@Au nanocomposites for the detection of galectin-1 via lactose-galectin interactions. Nanotechnology 2019, 30, 485706. [Google Scholar] [CrossRef]
- Martos-Maldonado, M.C.; Quesada-Soriano, I.; Garcia-Fuentes, L.; Vargas-Berenguel, A. Multivalent Lactose-Ferrocene Conjugates Based on Poly (Amido Amine) Dendrimers and Gold Nanoparticles as Electrochemical Probes for Sensing Galectin-3. Nanomaterials (Basel) 2020, 10. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Tong, L.; Li, Y.; Pan, H.; Zhang, W.; Guan, M.; Li, W.; Chen, Y.; Li, Q.; Li, Z.; et al. Lactose-Functionalized Gold Nanorods for Sensitive and Rapid Serological Diagnosis of Cancer. ACS Appl Mater Interfaces 2016, 8, 5813–5820. [Google Scholar] [CrossRef]
- Heine, V.; Dey, C.; Bojarová, P.; Křen, V.; Elling, L. Methods of in vitro study of galectin-glycomaterial interaction. Biotechnol Adv 2022, 58, 107928. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, T.J.; Lin, H.Y.; Tu, Z.; Huang, B.S.; Wu, S.C.; Lin, C.H. Structural Basis Underlying the Binding Preference of Human Galectins-1, -3 and -7 for Galbeta1-3/4GlcNAc. PLoS One 2015, 10, e0125946. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Xia, B.; Stowell, S.R.; Lasanajak, Y.; Smith, D.F.; Cummings, R.D. Novel fluorescent glycan microarray strategy reveals ligands for galectins. Chem Biol 2009, 16, 36–47. [Google Scholar] [CrossRef]
- Kupper, C.; Böcker, S.; Liu, H.; Schmidt, C.; Van de Kamp, A.; Recker, T.; Lethaus, B.; Jahnen-Dechent, W.; Neuss-Stein, S.; Müller-Newen, G.; et al. Fluorescent SNAP-Tag Galectin Fusion Proteins as Novel Tools in Glycobiology. Current pharmaceutical design 2013, 19. [Google Scholar] [CrossRef]
- Böcker, S.; Elling, L. Binding characteristics of galectin-3 fusion proteins. Glycobiology 2017, 27, 457–468. [Google Scholar] [CrossRef]
- Poland, P.A.; Kinlough, C.L.; Hughey, R.P. Cloning, Expression, and Purification of Galectins for In Vitro Studies. Methods Mol Biol 2022, 2442, 41–54. [Google Scholar] [CrossRef]
- Maller, S.M.; Cagnoni, A.J.; Bannoud, N.; Sigaut, L.; Perez Saez, J.M.; Pietrasanta, L.I.; Yang, R.Y.; Liu, F.T.; Croci, D.O.; Di Lella, S.; et al. An adipose tissue galectin controls endothelial cell function via preferential recognition of 3-fucosylated glycans. FASEB J 2020, 34, 735–753. [Google Scholar] [CrossRef] [PubMed]
- Shu, Z.; Li, J.; Mu, N.; Gao, Y.; Huang, T.; Zhang, Y.; Wang, Z.; Li, M.; Hao, Q.; Li, W.; et al. Expression, purification and characterization of galectin-1 in Escherichia coli. Protein Expr Purif 2014, 99, 58–63. [Google Scholar] [CrossRef]
- Paul, A.; Wu, S.C.; Patel, K.R.; Ho, A.D.; Allen, J.W.L.; Verkerke, H.; Arthur, C.M.; Stowell, S.R. Purification of Recombinant Galectins from Different Species Using Distinct Affinity Chromatography Methods. Methods Mol Biol 2022, 2442, 55–74. [Google Scholar] [CrossRef]
- Sakthivel, D.; Littler, D.; Shahine, A.; Troy, S.; Johnson, M.; Rossjohn, J.; Piedrafita, D.; Beddoe, T. Cloning, expression, purification and crystallographic studies of galectin-11 from domestic sheep (Ovis aries). Acta Crystallogr F Struct Biol Commun 2015, 71, 993–997. [Google Scholar] [CrossRef] [PubMed]
- Voss, P.G.; Gray, R.M.; Dickey, S.W.; Wang, W.; Park, J.W.; Kasai, K.-I.; Hirabayashi, J.; Patterson, R.J.; Wang, J.L. Dissociation of the carbohydrate-binding and splicing activities of galectin-1. Arch Biochem Biophys 2008, 478, 18–25. [Google Scholar] [CrossRef] [PubMed]
- Welch, C.J.; Kadav, P.D.; Edwards, J.L.; Krycia, J.; Talaga, M.L.; Bandyopadhyay, P.; Dam, T.K. A Rapid and Facile Purification Method for Glycan-Binding Proteins and Glycoproteins. Curr Protoc Protein Sci 2020, 101, e113. [Google Scholar] [CrossRef]
- Kadav, P.D.; Edwards, J.L.; Krycia, J.; Bandyopadhyay, P.; Dam, T.K. Rapid Detection and Purification of Galectin-3 by the Capture and Release (CaRe) Method. Methods Mol Biol 2022, 2442, 89–103. [Google Scholar] [CrossRef]
- Welch, C.J.; Talaga, M.L.; Kadav, P.D.; Edwards, J.L.; Bandyopadhyay, P.; Dam, T.K. A capture and release method based on non-covalent ligand crosslinking and facile filtration for purification of lectins and glycoproteins. J Biol Chem 2020, 295, 223–236. [Google Scholar] [CrossRef]
- Ahmed, H.; Fink, N.E.; Pohl, J.; Vasta, G.R. Galectin-1 from bovine spleen: biochemical characterization, carbohydrate specificity and tissue-specific isoform profiles. J Biochem 1996, 120, 1007–1019. [Google Scholar] [CrossRef]
- Gitt, M.A.; Colnot, C.; Poirier, F.; Nani, K.J.; Barondes, S.H.; Leffler, H. Galectin-4 and galectin-6 are two closely related lectins expressed in mouse gastrointestinal tract. J Biol Chem 1998, 273, 2954–2960. [Google Scholar] [CrossRef]
- Jones, J.L.; Saraswati, S.; Block, A.S.; Lichti, C.F.; Mahadevan, M.; Diekman, A.B. Galectin-3 is associated with prostasomes in human semen. Glycoconj J 2010, 27, 227–236. [Google Scholar] [CrossRef]
- Takeuchi, T.; Tamura, M.; Ishii, N.; Ishikida, H.; Sugimoto, S.; Suzuki, D.; Nishiyama, K.; Takahashi, H.; Natsugari, H.; Arata, Y. Purification of galectin-1 mutants using an immobilized Galactosebeta1-4Fucose affinity adsorbent. Protein Expr Purif 2015, 111, 82–86. [Google Scholar] [CrossRef]
- Ali, N.; Salahuddin, A. Isolation and some properties of mammalian hepatic membrane lectins. FEBS Lett 1989, 246, 163–165. [Google Scholar] [CrossRef]
- Beyer, E.C.; Zweig, S.E.; Barondes, S.H. Two lactose binding lectins from chicken tissues. Purified lectin from intestine is different from those in liver and muscle. J Biol Chem 1980, 255, 4236–4239. [Google Scholar] [CrossRef]
- Cerra, R.F.; Gitt, M.A.; Barondes, S.H. Three soluble rat beta-galactoside-binding lectins. J Biol Chem 1985, 260, 10474–10477. [Google Scholar] [CrossRef] [PubMed]
- Leffler, H.; Barondes, S.H. Specificity of binding of three soluble rat lung lectins to substituted and unsubstituted mammalian beta-galactosides. J Biol Chem 1986, 261, 10119–10126. [Google Scholar] [CrossRef] [PubMed]
- de Waard, A.; Hickman, S.; Kornfeld, S. Isolation and properties of beta-galactoside binding lectins of calf heart and lung. J Biol Chem 1976, 251, 7581–7587. [Google Scholar] [CrossRef] [PubMed]
- Roff, C.F.; Wang, J.L. Endogenous lectins from cultured cells. Isolation and characterization of carbohydrate-binding proteins from 3T3 fibroblasts. J Biol Chem 1983, 258, 10657–10663. [Google Scholar] [CrossRef]
- Zlocowski, N.; Grupe, V.; Garay, Y.C.; Nores, G.A.; Lardone, R.D.; Irazoqui, F.J. Purified human anti-Tn and anti-T antibodies specifically recognize carcinoma tissues. Sci Rep 2019, 9, 8097. [Google Scholar] [CrossRef]
- Dam, T.K.; Gabius, H.J.; Andre, S.; Kaltner, H.; Lensch, M.; Brewer, C.F. Galectins bind to the multivalent glycoprotein asialofetuin with enhanced affinities and a gradient of decreasing binding constants. Biochemistry 2005, 44, 12564–12571. [Google Scholar] [CrossRef]
- Ahmad, N.; Gabius, H.J.; Andre, S.; Kaltner, H.; Sabesan, S.; Roy, R.; Liu, B.; Macaluso, F.; Brewer, C.F. Galectin-3 precipitates as a pentamer with synthetic multivalent carbohydrates and forms heterogeneous crosslinked complexes. J Biol Chem 2004, 279, 10841–10847. [Google Scholar] [CrossRef] [PubMed]
- Nangia-Makker, P.; Balan, V.; Raz, A. Galectin-3 binding and metastasis. Methods Mol Biol 2012, 878, 251–266. [Google Scholar] [CrossRef]
- Kuwabara, I.; Liu, F.T. Galectin-3 promotes adhesion of human neutrophils to laminin. J Immunol 1996, 156, 3939–3944. [Google Scholar] [CrossRef] [PubMed]
- Ochieng, J.; Warfield, P. Galectin-3 binding potentials of mouse tumor EHS and human placental laminins. Biochem Biophys Res Commun 1995, 217, 402–406. [Google Scholar] [CrossRef] [PubMed]
- Ochieng, J.; Leite-Browning, M.L.; Warfield, P. Regulation of cellular adhesion to extracellular matrix proteins by galectin-3. Biochem Biophys Res Commun 1998, 246, 788–791. [Google Scholar] [CrossRef] [PubMed]
- Böcker, S.; Laaf, D.; Elling, L. Galectin Binding to Neo-Glycoproteins: LacDiNAc Conjugated BSA as Ligand for Human Galectin-3. Biomolecules 2015, 5, 1671–1696. [Google Scholar] [CrossRef] [PubMed]
- Stowell, S.R.; Arthur, C.M.; Mehta, P.; Slanina, K.A.; Blixt, O.; Leffler, H.; Smith, D.F.; Cummings, R.D. Galectin-1, -2, and -3 exhibit differential recognition of sialylated glycans and blood group antigens. J Biol Chem 2008, 283, 10109–10123. [Google Scholar] [CrossRef]
- Cuatrecasas, P.; Wilchek, M.; Anfinsen, C.B. Selective enzyme purification by affinity chromatography. Proc Natl Acad Sci U S A 1968, 61, 636–643. [Google Scholar] [CrossRef]
- Baenziger, J.U.; Fiete, D. Structure of the complex oligosaccharides of fetuin. J Biol Chem 1979, 254, 789–795. [Google Scholar] [CrossRef]
- Ahmad, N.; Gabius, H.J.; Sabesan, S.; Oscarson, S.; Brewer, C.F. Thermodynamic binding studies of bivalent oligosaccharides to galectin-1, galectin-3, and the carbohydrate recognition domain of galectin-3. Glycobiology 2004, 14, 817–825. [Google Scholar] [CrossRef]
- Brewer, C.F. Thermodynamic binding studies of galectin-1, -3 and -7. Glycoconj J 2002, 19, 459–465. [Google Scholar] [CrossRef]
- Carlsson, S.; Oberg, C.T.; Carlsson, M.C.; Sundin, A.; Nilsson, U.J.; Smith, D.; Cummings, R.D.; Almkvist, J.; Karlsson, A.; Leffler, H. Affinity of galectin-8 and its carbohydrate recognition domains for ligands in solution and at the cell surface. Glycobiology 2007, 17, 663–676. [Google Scholar] [CrossRef] [PubMed]
- Ideo, H.; Seko, A.; Ishizuka, I.; Yamashita, K. The N-terminal carbohydrate recognition domain of galectin-8 recognizes specific glycosphingolipids with high affinity. Glycobiology 2003, 13, 713–723. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Frank, M.; Schwartz-Albiez, R. Understanding the specificity of human Galectin-8C domain interactions with its glycan ligands based on molecular dynamics simulations. PLoS One 2013, 8, e59761. [Google Scholar] [CrossRef]
- Vokhmyanina, O.A.; Rapoport, E.M.; Ryzhov, I.M.; Korchagina, E.Y.; Pazynina, G.V.; Severov, V.V.; Kaltner, H.; Andre, S.; Gabius, H.J.; Bovin, N.V. Carbohydrate specificity of chicken and human tandem-repeat-type galectins-8 in composition of cells. Biochemistry (Mosc) 2011, 76, 1185–1192. [Google Scholar] [CrossRef] [PubMed]
- Vokhmyanina, O.A.; Rapoport, E.M.; Andre, S.; Severov, V.V.; Ryzhov, I.; Pazynina, G.V.; Korchagina, E.; Gabius, H.J.; Bovin, N.V. Comparative study of the glycan specificities of cell-bound human tandem-repeat-type galectin-4, -8 and -9. Glycobiology 2012, 22, 1207–1217. [Google Scholar] [CrossRef]
- Hoffmann, M.; Hayes, M.R.; Pietruszka, J.; Elling, L. Synthesis of the Thomsen-Friedenreich-antigen (TF-antigen) and binding of Galectin-3 to TF-antigen presenting neo-glycoproteins. Glycoconj J 2020, 37, 457–470. [Google Scholar] [CrossRef]
- Kremers, G.J.; Goedhart, J.; van Munster, E.B.; Gadella, T.W., Jr. Cyan and yellow super fluorescent proteins with improved brightness, protein folding, and FRET Forster radius. Biochemistry 2006, 45, 6570–6580. [Google Scholar] [CrossRef]
- Zacharias, D.A.; Violin, J.D.; Newton, A.C.; Tsien, R.Y. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science 2002, 296, 913–916. [Google Scholar] [CrossRef] [PubMed]
- Stohr, K.; Siegberg, D.; Ehrhard, T.; Lymperopoulos, K.; Oz, S.; Schulmeister, S.; Pfeifer, A.C.; Bachmann, J.; Klingmuller, U.; Sourjik, V.; et al. Quenched substrates for live-cell labeling of SNAP-tagged fusion proteins with improved fluorescent background. Anal Chem 2010, 82, 8186–8193. [Google Scholar] [CrossRef] [PubMed]
- Keppler, A.; Gendreizig, S.; Gronemeyer, T.; Pick, H.; Vogel, H.; Johnsson, K. A general method for the covalent labeling of fusion proteins with small molecules in vivo. Nat Biotechnol 2003, 21, 86–89. [Google Scholar] [CrossRef]
- Kyte, J.; Doolittle, R.F. A simple method for displaying the hydropathic character of a protein. J Mol Biol 1982, 157, 105–132. [Google Scholar] [CrossRef] [PubMed]
- Strongin, D.E.; Bevis, B.; Khuong, N.; Downing, M.E.; Strack, R.L.; Sundaram, K.; Glick, B.S.; Keenan, R.J. Structural rearrangements near the chromophore influence the maturation speed and brightness of DsRed variants. Protein Eng Des Sel 2007, 20, 525–534. [Google Scholar] [CrossRef]
- Phillips, G.N., Jr. Structure and dynamics of green fluorescent protein. Curr Opin Struct Biol 1997, 7, 821–827. [Google Scholar] [CrossRef]
- Hughes, R.C. Galectins as modulators of cell adhesion. Biochimie 2001, 83, 667–676. [Google Scholar] [CrossRef]
- Matarrese, P.; Tinari, N.; Semeraro, M.L.; Natoli, C.; Iacobelli, S.; Malorni, W. Galectin-3 overexpression protects from cell damage and death by influencing mitochondrial homeostasis. FEBS Lett 2000, 473, 311–315. [Google Scholar] [CrossRef]
- Sato, S.; Hughes, R.C. Binding specificity of a baby hamster kidney lectin for H type I and II chains, polylactosamine glycans, and appropriately glycosylated forms of laminin and fibronectin. J Biol Chem 1992, 267, 6983–6990. [Google Scholar] [CrossRef]
- Wang, J.L.; Werner, E.A.; Laing, J.G.; Patterson, R.J. Nuclear and cytoplasmic localization of a lectin-ribonucleoprotein complex. Biochem Soc Trans 1992, 20, 269–274. [Google Scholar] [CrossRef] [PubMed]
- Cecioni, S.; Imberty, A.; Vidal, S. Glycomimetics versus multivalent glycoconjugates for the design of high affinity lectin ligands. Chem Rev 2015, 115, 525–561. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, S.; Shinkai, H.; Deutzmann, R.; Paulsson, M.; Timpl, R. Structure and distribution of N-linked oligosaccharide chains on various domains of mouse tumour laminin. Biochem J 1988, 252, 453–461. [Google Scholar] [CrossRef]
- Hughes, R.C. Role of glycosylation in cell interactions with extracellular matrix. Biochem Soc Trans 1992, 20, 279–284. [Google Scholar] [CrossRef]
- Timpl, R.; Rohde, H.; Robey, P.G.; Rennard, S.I.; Foidart, J.M.; Martin, G.R. Laminin--a glycoprotein from basement membranes. J Biol Chem 1979, 254, 9933–9937. [Google Scholar] [CrossRef] [PubMed]
- Erickson, H.P. Stretching fibronectin. J Muscle Res Cell Motil 2002, 23, 575–580. [Google Scholar] [CrossRef]
- Basak, T.; Vega-Montoto, L.; Zimmerman, L.J.; Tabb, D.L.; Hudson, B.G.; Vanacore, R.M. Comprehensive Characterization of Glycosylation and Hydroxylation of Basement Membrane Collagen IV by High-Resolution Mass Spectrometry. J Proteome Res 2016, 15, 245–258. [Google Scholar] [CrossRef]
- Hennet, T. Collagen glycosylation. Curr Opin Struct Biol 2019, 56, 131–138. [Google Scholar] [CrossRef]
- Ochieng, J.; Furtak, V.; Lukyanov, P. Extracellular functions of galectin-3. Glycoconj J 2002, 19, 527–535. [Google Scholar] [CrossRef]
- Dias-Baruffi, M.; Zhu, H.; Cho, M.; Karmakar, S.; McEver, R.P.; Cummings, R.D. Dimeric galectin-1 induces surface exposure of phosphatidylserine and phagocytic recognition of leukocytes without inducing apoptosis. J Biol Chem 2003, 278, 41282–41293. [Google Scholar] [CrossRef]
- Leppanen, A.; Stowell, S.; Blixt, O.; Cummings, R.D. Dimeric galectin-1 binds with high affinity to alpha2,3-sialylated and non-sialylated terminal N-acetyllactosamine units on surface-bound extended glycans. J Biol Chem 2005, 280, 5549–5562. [Google Scholar] [CrossRef] [PubMed]
- Stowell, S.R.; Arthur, C.M.; Slanina, K.A.; Horton, J.R.; Smith, D.F.; Cummings, R.D. Dimeric Galectin-8 induces phosphatidylserine exposure in leukocytes through polylactosamine recognition by the C-terminal domain. J Biol Chem 2008, 283, 20547–20559. [Google Scholar] [CrossRef] [PubMed]
- Hirabayashi, J.; Kasai, K. Effect of amino acid substitution by sited-directed mutagenesis on the carbohydrate recognition and stability of human 14-kDa beta-galactoside-binding lectin. J Biol Chem 1991, 266, 23648–23653. [Google Scholar] [CrossRef] [PubMed]
- Cormack, B.P.; Valdivia, R.H.; Falkow, S. FACS-optimized mutants of the green fluorescent protein (GFP). Gene 1996, 173, 33–38. [Google Scholar] [CrossRef]
- Patnaik, S.K.; Potvin, B.; Carlsson, S.; Sturm, D.; Leffler, H.; Stanley, P. Complex N-glycans are the major ligands for galectin-1, -3, and -8 on Chinese hamster ovary cells. Glycobiology 2006, 16, 305–317. [Google Scholar] [CrossRef]
| IMAC | IMAC+ASF affinity resin | |||
| Elution Peak | MW (calc.) | Elution Peak | MW (calc.) | |
| HSYGal-3 (74.2 kDa) |
1 | 86.5 | 1 | 84.9 |
| 2 | 68 | 2 | 15.1 | |
| 3 | 31.7 | 3 | 1.8 | |
| HYGal-3 (56 kDa) |
1 | 47.8 | 1 | 50.6 |
| 2 | 30.3 | 2 | 2.0 | |
| HSGal-3 (46.9 kDa) |
1 | 48.5 | 1 | 48.3 |
| 2 | 28.7 | |||
| 3 | 6.2 | |||
| HGal-3 (28 kDa) |
1 | 21.0 | 1 | 28.7 |
| HSY (48.1 kDa) |
1 | 88.9 | ||
| Protein | Apparent KD [µM] | Apparent binding efficiency [µM−1] |
| IMAC | ||
| HGal-3 | 6.9 ± 1.6 | 0.1 |
| HSYGal-3 | 1.5 ± 0.8 | 0.6 |
| HSGal-3 | 0.3 ± 0.02 | 2.7 |
| HYGal-3 | 2.3 ± 0.4 | 0.3 |
| IMAC and ASF affinity chromatography | ||
| HGal-3 | 5.3 ± 1.0 | 0.1 |
| HSYGal-3 | 0.5 ± 0.1 | 1.7 |
| HSGal-3 | 0.3 ± 0.02 | 3.1 |
| HYGal-3 | 4.1 ± 1.2 | 0.1 |
| Protein | Apparent KD [µM] | Apparent binding efficiency [µM−1] |
| HSDsRedMGal-1C2S | 0.6 ± 0.1 | 1.4 |
| HSGal1-C2S | 0.8 ± 0.1 | 1.1 |
| HDsRedMGal-1C2S | 8.8 ± 3.0 | 0.1 |
| HGal-1C2S | 0.5 ± 0.1 | 1.6 |
| HSeGFPGal-8N | 0.2 ± 0.1 | 4.0 |
| HSGal-8N | 0.5 ± 0.2 | 1.3 |
| HeGFPGal-8N | 2.9 ± 3.3 | 0.1 |
| HGal-8N | 0.37 ± 0.02 | 2.2 |
| Apparent KD [µM] | |||
| Laminin | Fibronectin | Collagen IV | |
| HGal-3 | 0.13 ± 0.01 | 0.43 ± 0.02 | 0.64 ± 0.03 |
| HSYGal-3 | 0.07 ± 0.01 | 0.11 ± 0.01 | 0.13 ± 0.01 |
| HGal-1C2S | 0.42 ± 0.02 | 1.97 ± 0.38 | 1.34 ± 0.15 |
| HSDsRedMGal-1C2S | 0.11 ± 0.02 | 0.11 ± 0.02 | 0.14 ± 0.03 |
| HGal-8N | 0.18 ± 0.02 | 0.52 ± 0.03 | 0.60 ± 0.03 |
| HSeGFPGal-8N | 0.04 ± 0.04 | 0.06 ± 0.01 | 0.08 ± 0.01 |
| ASF-bound galectin | Apparent KD [µM] of laminin |
| HGal-3 | 0.27 ± 0.03 |
| HSYGal-3 | 0.45 ± 0.04 |
| HGal-1C2S | 0.38 ± 0.07 |
| HSDsRedMGal-1C2S | 0.37 ± 0.03 |
| HGal-8N | 69.5 ± 22.7 |
| HSeGFPGal-8N | 0.27 ± 0.02 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





