Introduction
Ceramide is a crucial intermediate in sphingolipid metabolism in mammalian cells (1). Ceramide is synthesized by ceramide synthases (CerS) with different acyl chain lengths. CerS share some biochemical features like their structure, intracellular localization and their catalytic mechanism (1). Additionally, different CerS display significant variability in their biological properties (2). All CerS are associated with the endoplasmic reticulum (ER), and they all contain a crucial TRAM–Lag1p–CLN8 (TLC) domain: a CerS catalytic domain necessary for ceramide synthesis (3). Originally, CerS were known as Longevity Assurance (Lass) genes due to their homology to the longevity assurance gene 1 (LAG1p) in yeast, however their name was changed after characterization of their biochemical features (4). It was found that LAG genes result in the prolonged life span of
S. cerevisiae and extend the lifespan of a cell, hence the name LAG1. The most remarkable characteristic of CerS is that each of the CerS has a specificity for an acyl CoA chain length despite all catalyzing the same reaction. It can thus be seen that CerS determine the fatty acid composition of ceramides (2).
Table 1 displays ceramides with different acyl chain lengths as synthesized by these six CerS.
Multiple investigations have been performed to determine the role of CerS in different pathologies. Although no longer true, ceramides were classically known to induce cancer cells’ growth inhibition, cell death, and senescence (11). Recent reports found that ceramides with various fatty acid chain lengths might have different functions in the pathogenesis of cancer, indicating the critical importance of CerS in sphingolipid metabolism (12-14). There are various functions of ceramides that are context dependent and are modulated by their localization in the cell and presence or absence of their targets. For instance, ceramides with various fatty-acid chain lengths have been shown to play a role in cancer cell proliferation like C16-ceramide (synthesized by CerS6) whereas C18-ceramide (synthesized by CerS1) is involved in cell death (11). Based on C18-ceramide downregulation in most head and neck squamous cell carcinoma (HNSCC) tissues compared to adjacent normal tissue, CerS1 has been found to play a role in the growth regulation of HNSCC (15). Furthermore, the balance between C16-and C18-ceramides levels could be associated with HNSCC tumor development (15, 16) related to the clinical progression of this cancer (17). CerS2 have been shown to play a role in breast cancer. In an initial study (18), it was found that the levels of total ceramide (especially C16-, C24:1- and C24:0-ceramides) were significantly increased in aggressive tumors. This was consistent with an increases in CerS2, CerS4 and CerS6 mRNA levels. In the subsequent study (19), mRNA levels of CerS2 and CerS6 were highly increased in breast tumors compared to the normal tissue, where almost half of the patients showed increased levels of CerS2 and CerS6 mRNA. Notably, there was a significant relationship between the expression of CerS6 and CerS2, and between CerS2/CerS6 and CerS4. Moreover, the sensitivity of breast cancer cells to chemotherapeutic drugs was not affected by CerS4 (unlike CerS1 and CerS5) (20). It has been suggested that CerS6 is involved in the etiology of cancer. For example, using microarray studies, CerS6 has been found to be implicated in early embryonic development and cancer differentiation (21). In contrast to this, another study showed that mRNA levels of CerS6 were increased in breast cancer tissues and regulation of CerS6 expression was dependent on estrogen receptor (18, 19, 22). This double-edge role of ceramides in cancer highlights the potential targeting of some of the enzymes in the ceramide metabolism pathway as novel therapeutic methods. For example CerS6 can be targeted in order to hamper cancer cell growth in head and neck and breast cancers while activation of CerS1 could be harnessed to develop new treatments against lung and head and neck cancers (11).
Therefore, in the current review paper we summarize different types of CerS and their products. Then, we discuss the impact of CerS and ceramides in apoptosis and autophagy which play an important role in cancer. We later explain the current methods that is being used for evaluation of lipids including ceramides in biological samples. Finally, we will provide over view for future therapeutic approaches to target CerS for developing potential new therapeutic approaches in cancer.
Ceramide Synthesis
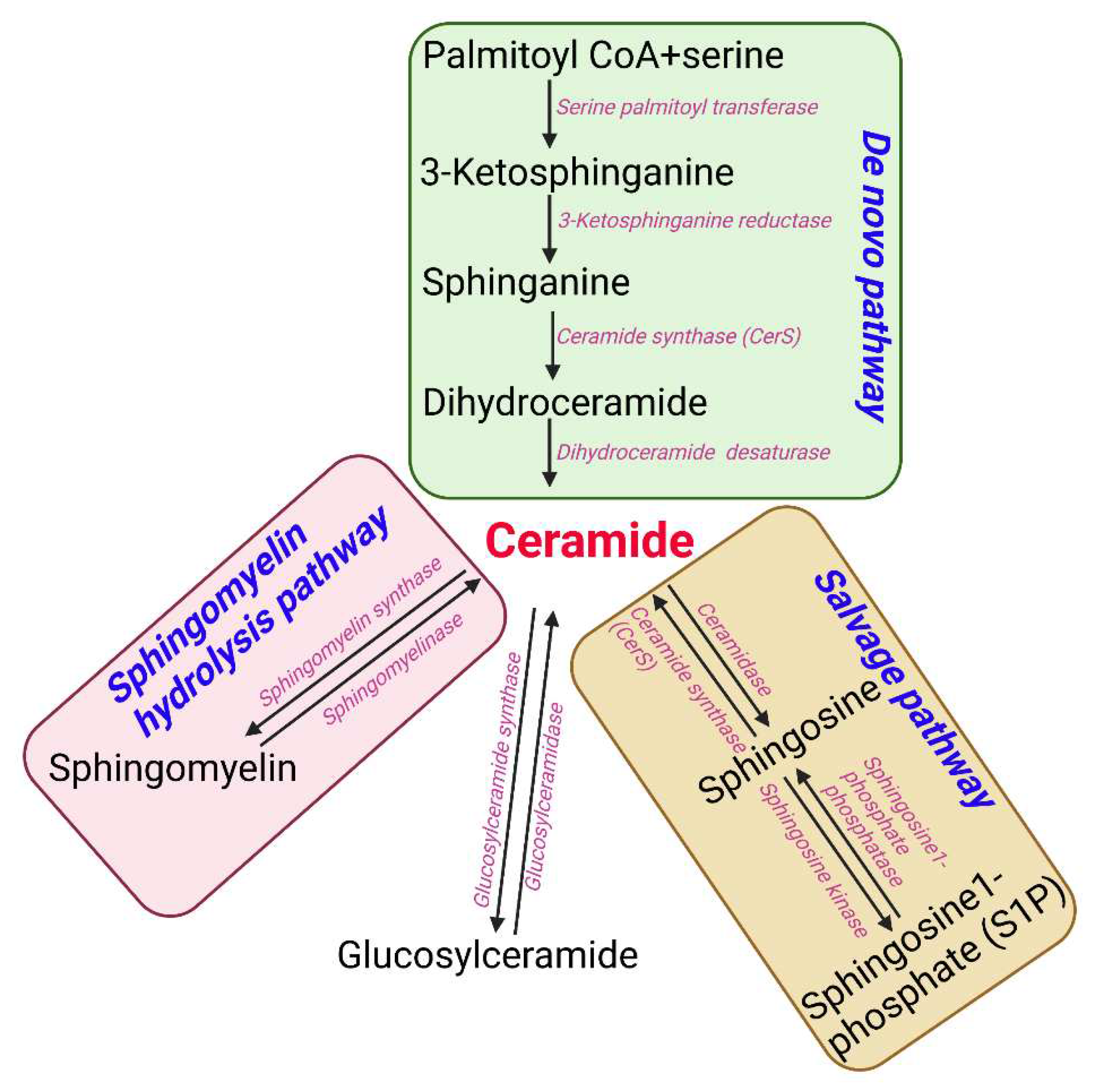
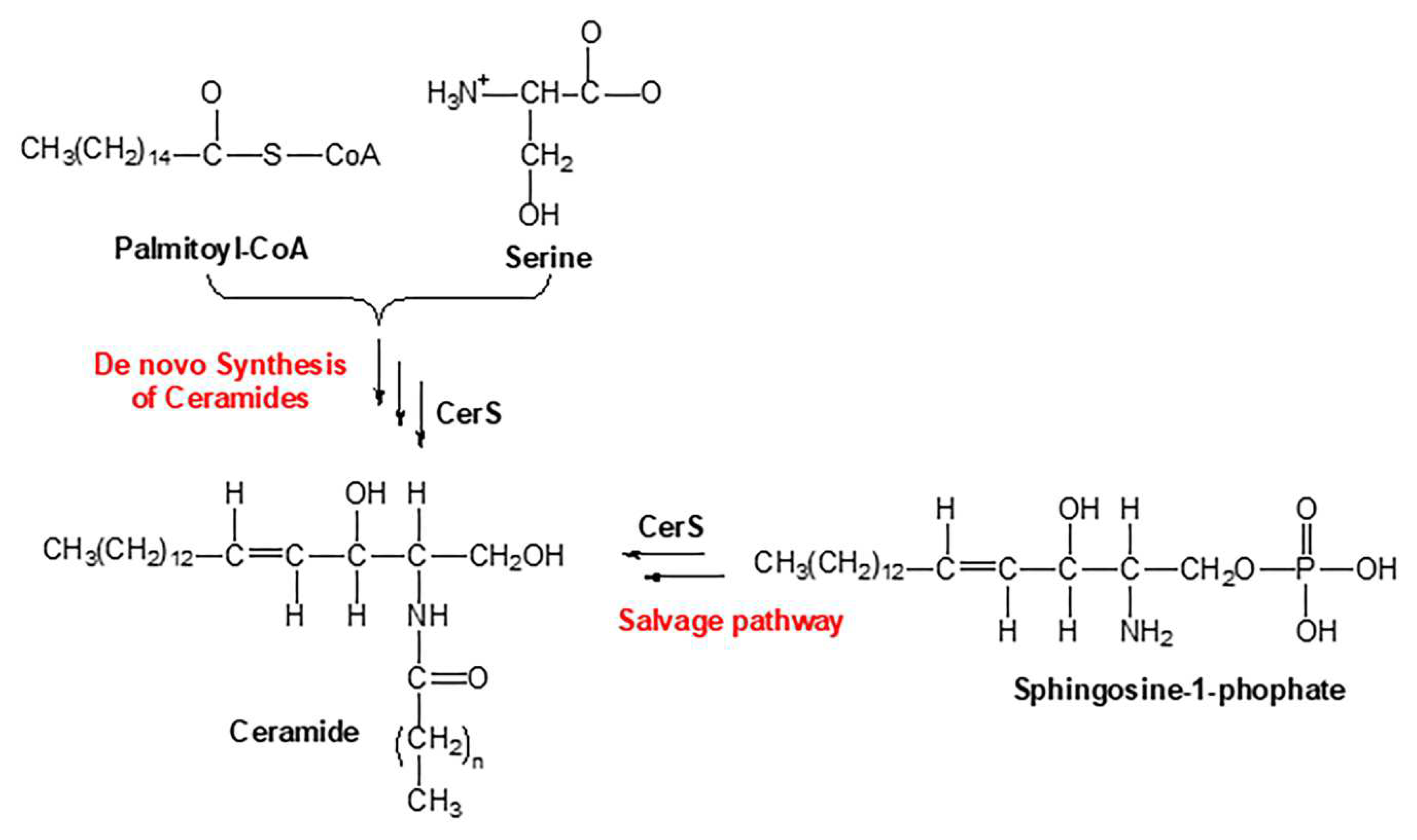
Sphingolipids are one of the main lipid types in eukaryotic cells. Ceramides are the building block of all sphingolipids. This has made ceramide the focus of recent enormous attention due to its suggested role as a crucial signaling lipid involved in the regulation of different cellular processes (23). As shown in
Figure 1, ceramide is synthesized via different pathways in the cell. Ceramide is made of a sphingosine backbone, which is a fatty acyl chain with an amide linked to it, with different lengths (24). Ceramides are used as a precursor for more complex sphingolipids, for example: glucosylceramide, ceramide-1-phosphate (C1P), sphingomyelin, and galactosyl ceramide. Ceramide is also produced through the breakdown of these complex sphingolipids using various enzymes. This type of ceramide can be used as a substrate by ceramidases to release sphingosine, which is phosphorylated to make sphingosine 1-phosphate (S1P) (11). Endogenous ceramides are synthesized
de novo in the ER through a pathway beginning with condensation of L-serine and palmitoyl-CoA followed by more enzymatic reactions leading to the production of ceramide (25, 26) (
Figure 1). D
e novo synthesis of ceramide occurs in the ER, however, the synthesis of complex sphingomyelin and glycosphingolipids occurs in the Golgi apparatus (27). During this process the ceramide is transported to Golgi apparatus from the ER via vesicular trafficking or non-vesicular transport (i.e. by ceramide transfer protein (CERT)) (28). Lastly, the salvage pathway can also generate ceramides through hydrolyzing ceramide into sphingosine which is then reacylated by CerS to regenerate ceramides (29).
CerS Activity Regulation
Mechanisms involved in the regulation of CerS activity are not fully known. Recent studies reveal that the formation of CerS dimers can regulate the ceramide synthesis which may be dependent on the interaction of CerS with each other (30). CerS is expressed in various levels in different tissues (31), however the acyl chain composition of the sphingolipid does not always correlate with CerS expression (31, 32). This has prompted the possibility of cross-regulation of CerS expression and activity (33). In fact, heterocomplexes of CerS2, -5, and -6 was recently reported in HeLa cells (34). Moreover, the activity of either monomers in this dimer form depend on, and can be regulated by, the other monomer, suggesting dimer formation as a mechanism of regulating CerS activity (30). Specifically, it was suggested that the highest level of CerS2 activity is dependent on its interaction with other CerS. CerS2 has the least activity among all CerS in vitro (35, 36), yet it has the widest tissue distribution (31). In fact, CerS monomers and dimers are in equilibrium in the ER and dimers formation and/or dissociation could be a major factor in their regulation. Dimer formation occurs rapidly which is an efficient way of increasing the ceramide levels under different conditions. This is of utmost importance in situations when de novo ceramide synthesis plays a crucial role in other signaling pathways requiring a quick mechanism of ceramide synthesis (30). The mechanisms of post translational modifications of CerS, if any, are not fully elucidated yet. There is some evidence that suggests a role for CerS phosphorylation. For example, an increase in the de novo ceramide synthesis is linked to activation of protein kinase C (PKC) and consequently up-regulation of CerS5 activity (37). It has been shown from high performance mass spectrometry that CerS might be phosphorylated (38, 39) and glycosylated (40). The rapid changes in CerS activity might be facilitated by the post-translational regulation of CerS under different conditions (41, 42).
CerS Tissue Distribution
CerS1 is mainly expressed in brain, testes, and skeletal muscle at low levels (31, 40, 43). It was shown by in situ-hybridization of brain tissue that CerS1 is expressed in most neurons and in very low levels in white matter cells (44). CerS1 is upregulated at a low level postnatally, possibly supporting the synthesis of neuronal cell membranes. The high level of CerS1 is in line with the composition of acyl chain of the neuronal sphingolipids (mainly C18-fatty acids) in neurons (44). CerS2 is more widely expressed in different human tissues compared to CerS1 with lowest expression in spleen, small intestine, colon, thymus, and peripheral blood leukocytes, moderate expression in heart, brain, placenta and lung, skeletal muscle, and highest expression in liver and kidney (31). It has been shown that CerS2 is highly expressed during active myelination in oligodendrocytes and Schwann cells in mouse brain (44). CerS3 is highly expressed in testes and skin (40, 45). The expression of CerS3 is very high in keratinocytes (46). CerS4, which is probably the least studied CerS, is highly expressed in leukocytes, heart, skin, and liver (31). CerS5 is highly expressed in lung epithelia and is likely the most studied CerS because it can synthesize C16-ceramide which is an important pro-apoptotic ceramide (47). It was found that down regulation of CerS5 in lung epithelium by siRNA or using fumonisin B1 decreased the total CerS activity (48). This shows that CerS5 is indeed involved in the ceramide synthesis in lung tissue. Lastly, CerS6 is highly expressed in intestine and immune system (2).
Role of CerS and ceramides in Regulation of Apoptosis
Apoptosis is a programmed process whereby cells are committed to death (49). There are two main apoptosic pathways namely intrinsic or extrinsic (50). Mitochondria play a crucial role in intrinsic apoptosis as the outer membrane integrity loss results in a cascade of events that leads to apoptotic death (51). Different proteins are connected to apoptosis initiation leading to the permeabilization of the mitochondrial outer membrane by the formation of different pores (52, 53), mitochondrial apoptosis-induced channel (54), Bax oligomerization into channels (55, 56), mitochondrial permeability transition pore (57, 58), and Bax/Bak hybrid channels (56, 59). Ceramides are also involved in the formation of channels in the mitochondrial outer membrane (60-62). Moreover, these events in vivo can occur at the same time resulting in a regulated and coordinated outcome.
A recent study highlighted the role of ceramides in regulation of apoptosis. It found that C16-ceramide is needed for germ cell apoptosis induced by gamma radiation in
C. elegans (63). C16-ceramide-mediated apoptosis was detected by acridine orange (AO) staining, involvement of MAPK pathway (
mpk-1; homologue of mammalian
erk and
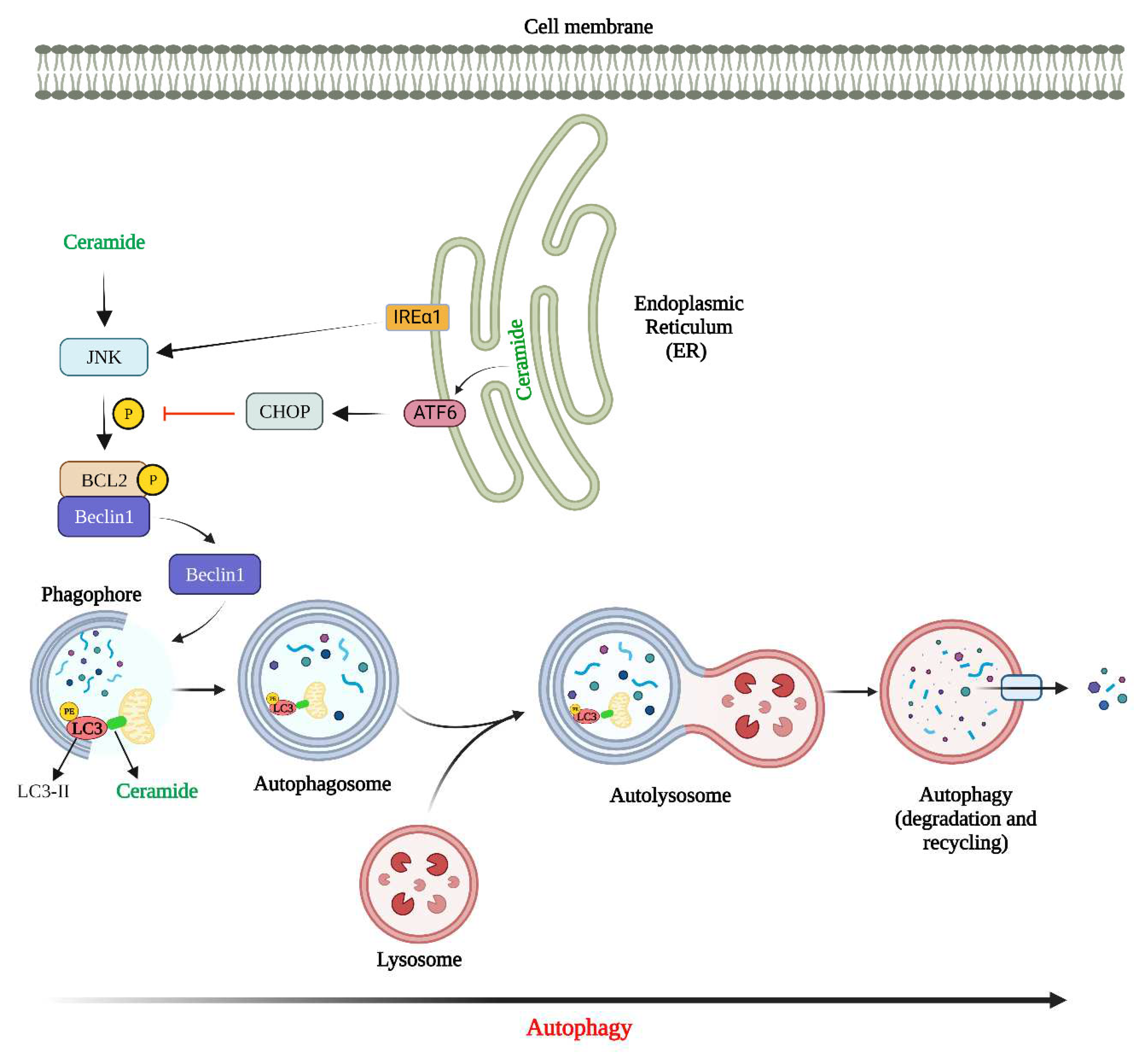
sek-1; homologue of mammalian JNK), production of ROS and reduction in mitochondrial outer membrane potential (MOMP). Most studies thus far have shown that ceramides regulate apoptosis mainly through ceramide channel formation and regulation of anti-apoptotic Bcl2 family proteins (Bcl2 and Bcl-xL), and pro-apoptotic Bcl2 family proteins (Bax and Bak) as discussed below and briefly illustrated in
Figure 2.
Regulation of Apoptosis by Ceramide Channels
Ceramides have the special ability to perforate the cell lipid bilayers through forming water-filled channels (typically 10 nm in diameter) (64, 65). Levels of mitochondrial ceramide elevates during apoptosis initiation and resulting in mitochondrial outer membrane integrity loss (66). Pro-apoptotic proteins such as cytochrome c are released from the intermembrane space into the cytosol upon the permeabilization of the mitochondrial outer membrane and at this point the cell is irreversibly committed to death (67, 68). The most recent model for the ceramide channel comprises of layers of ceramide molecules that are stacked in an anti-parallel way on top of each other forming ceramide columns in a barrel-like structure in lipid bilayer membranes (69, 70). Ceramides with various fatty acyl chain lengths such as C2-, C8-, C16- and C24-ceramides are capable of perforating the mitochondrial outer membranes (27, 71). This ceramide channel formation strongly depends on ceramide molecule structure (72). For example, dihydroceramide does not possess the 4–5 double bond in ceramide and therefore it cannot induce cell death. Furthermore, dihydroceramide cannot form ceramide channels in vitro (73). It is unsurprising that ceramides can influence each other since ceramides production with various chain lengths invariably changes the biophysical properties of membranes (74, 75). Furthermore, membranes biophysical properties alter due to the depletion of very long chain ceramides (76). Ceramide channels in the mitochondrial outer membrane are large enough to permit the release of apoptotic proteins into the cytosol (77). Previous study findings show that various ceramide species induce membrane permeability independently from one another (60). In a recent study, the egress of two intermembrane space proteins were evaluated to investigate the mechanisms by which ceramides perforate the mitochondria (78). Sulfite oxidase is a 120 KD enzyme and adenylate kinase is a 26 KD protein and both localize in the mitochondrial intermembrane space. Upon mixing C16- and C22- ceramide with isolated mitochondria, it was found that C16-ceramide induced the SOX release whereas C22-ceramide showed no release of SOX indicating that the C22-ceramide channels are significantly smaller than the ones made by C16-ceramide. Interestingly, both C16-ceramide and C22-ceramide resulted in the release of both SOX and AK. This release was possibly due to the formation of either C16-ceramide channels or C16–C22-hybrid channels as C22-ceramide channels did not release SOX. In short, these findings agree with the theory that various ceramides can form channels of varied sizes and quantities in isolated mitochondria (78). In another study by Laviad et al., it was found that CerS overexpression results in the high production of specific ceramides in HEK cells. For example, CerS2 overexpression led to the enrichment of C22-, C24- and C24:ceramide (31) while CerS5 overexpression resulted in the enrichment of C16-ceramide. Additionally, cytochrome c release was measured to examine the impact of exogenous ceramides on the membrane permeability of isolated mitochondria from HEK cells. Interestingly, C16-ceramide addition to isolated mitochondria from CerS5-transfected cells (enriched with C16-ceramide) elevated the levels of cytochrome c release while it was significantly decreased in isolated mitochondria from CerS2-transfected cells. On the other hand, C24-ceramide addition to isolated mitochondria from CerS2-transfected cells increased the release of cytochrome c whereas it was significantly reduced in isolated mitochondria from CerS5-transfected cells highlighting the existing competition ex vivo (78).
Regulation of Apoptosis by Ceramides through anti-apoptotic Bcl-2 family proteins, Bcl2 and Bcl-XL
Bcl2 phosphorylation at serine 70 is needed for its total and optimal anti-apoptotic function (79). It has been suggested that ceramide might influence the Bcl2 phosphorylation and therefore its function, since ceramide activates protein phosphatase 2A (PP2A) (a physiologic Bcl2 phosphatase) (80). In fact, C2-ceramide (and not C2-dihydroceramide) specifically activates mitochondrial PP2A that induces Bcl2 dephosphorylation and consequently ceramide-induced apoptosis in HL-60 cell line (a human leukemia cell line). These results show that ceramide-induced apoptosis might involve mitochondrial PP2A which dephosphorylates and inactivates Bcl2 in Bcl2 expressing cells (81). The fatty acyl chain lengths of the ceramide are critical in regulating the ceramide channels by Bcl-xL (82). Bcl-xL prevents the formation of ceramide channel and therefore ceramide-induced apoptosis (83). Interestingly, this prevention of ceramide channel formation is disrupted when ceramide has a truncated fatty acyl chain which shows a crucial role for the acyl chain length in binding to the Bcl-xL. Bcl-xL binding to ceramide greatly depends on the acyl chain length of ceramide due to the hydrophobic pocket in Bcl-xL structure. Bcl-xL efficiently inhibits the formation of ceramide channels formed by 16-, 18- and, 20-ceramides (82). Moreover, BH3 peptide mimetics bind to the hydrophobic grove on Bcl-xL and thus prevent its binding to Bax, inhibiting the ceramide channel formation (84).
Regulation of Apoptosis by ceramides through pro-apoptotic Bcl2 family proteins, Bax and Bak:
Bax has a crucial role in regulation of apoptosis (85). Bax translocates to mitochondria, after a variety of apoptotic stimuli, to form pores in mitochondrial outer membrane by potentiating the ceramide channels and thus inducing loss of mitochondrial membrane potential (85). This leads to the cytochrome c release from mitochondrial intermembrane spaces into the cytosol.
As opposed to Bcl-xL, formation of Bax is not dependent on nor influenced by acyl chain length of ceramide (86). It has been found that Bax, Bak, and ceramide are dependent on one another when forming ceramide channels (84). Additionally,Bak is crucial for the synthesis of long-chain ceramide by CerS during apoptosis (87). Moreover, evidence suggests that Bax and ceramide act together in a synergistic manner in the permeabilization of mitochondrial outer membrane (88, 89). It has also been reported that in Bax-transfected DU-145 cells and HL-60 cells the exogenous C2- and C6-ceramides leads to the Bax redistribution to mitochondria and causes mitochondrial permeability transition followed by cytochrome c release, then activation of caspase-3 and DNA fragmentation (90). It was found that ceramides capable of permeabilizing into the cells can promote the conformational change in the Bax N-terminus in Bax-expressing cells (91). Interestingly, loss of Bax inhibits the C2-ceramide-induced apoptosis in colorectal cancer HCT116 cells, highlighting again the role of ceramide in the regulation of apoptosis induction (92).
Endoplasmic Reticulum (ER) and Regulation of Apoptosis via Ceramides
The ER plays an important role in different cellular processes such as lipid metabolism, protein folding, protein synthesis, and calcium homeostasis and is therefore a crucial organelle within eukaryotic cells (93). When newly synthesized proteins in the ER are improperly folded or when the Ca2+ sequestration is perturbed, it leads to ER stress (94, 95). To mitigate this stress, a complicated homeostatic mechanism is activated in the ER known as the unfolded protein response (UPR) (95). Three transmembrane proteins in the ER mediate UPR namely; inositol-requiring enzyme 1 (IRE1), pancreatic ER kinase (PKR)-like ER kinase (PERK), and activating transcription factor 6 (ATF6), each of which have specific functions and targets (96, 97).
Elevated levels of ceramide in the mitochondria are linked to the induction of apoptosis. It has been found that synthesized ceramides in isolated ER vesicles transfer to the isolated mitochondria where they perforate the outer membrane leading to the release of cytochrome c and adenylate kinase (98). Interestingly, the ER-like membranes that are closely connected to mitochondria (known as mitochondria-associated membranes) synthesize ceramide that can transfer and permeabilize the mitochondrial outer membrane (98). Therefore, this mechanism of ceramide exchange between the two organelle obviates the need for a de novo pathway to synthesize the ceramide in mitochondria required for the formation of ceramide channel followed by protein release (98). In HNSCC cell lines, CerS6 overexpression followed by increased C16-ceramide has pro-survival roles whereas CerS6 knockdown induces apoptosis via activation of ATF6-CHOP arm of the UPR (99). Senkal et al. found that the induction of wild type CerS6 and not the inactive/mutant CerS6, increased the tumor growth in Severe Combined Immuno Deficient (SCID) mice. Similarly, induction of wild type-CerS (and not mutant CerS6) or expression of the mutant ATF-6 protected cells from undergoing apoptosis in response to CerS6 knockdown. Conversely, CerS6 knockdown by siRNA induced ATF-6 activation and subsequently apoptosis in multiple human squamous cell carcinomas cells (HNSCC, A549, H157, and H1650).. Altogether, this data revealed an underlying mechanism of how the alteration of CerS6 (and by extension C16-ceramide) induces the activation of ATF6 leading to the ER-stress-mediated apoptosis in human squamous cell carcinomas cells (100). Another study showed that cell-permeable C2-ceramide induces an apoptotic ER stress response in salivary adenoid cystic carcinoma cells (ACCs). Findings from this study illustrated that C2-ceramide activates pro-apoptotic factors downstream of ER stress and therefore induces apoptosis in ACCs. They found that ceramide induces CHOP which is the key transcription factor highly expressed during ER stress and is known as a crucial inducer of ER stress-mediated apoptosis. The same study found that the mRNA expression of CHOP was upregulated upon treatment with ceramide in ACC-M and ACC-2 cells (101).
Role of CerS and ceramides in Regulation of Apoptosis in Cancer
Owing to their regulatory role on apoptosis, CerS and ceramides have been implicated in the pathogenesis of various cancers. Ceramides are produced in response to a variety of stressors to initiate apoptosis. For example, levels of C16-ceramide were elevated after celecoxib (a cyclooxygenase-2 selective nonsteroidal anti-inflammatory drug for the treatment of arthritis) treatment in human colon carcinoma cells thereby mediating the anti-proliferative effects (102, 103). In another study, it was shown that overexpression of different CerS led to varied outcomes of apoptosis induced radiation in HeLa cells: overexpression of CerS5 resulted in higher levels of apoptosis in cells while overexpression of CerS2 inhibited cells from undergoing apoptosis (104). CerS1 also plays a crucial role in regulation of the sensitivity to chemotherapeutic agents. It was shown that CerS1 sensitizes human embryonic kidney cells to different anti-cancer agents such as carboplatin, cisplatin, vincristine, and doxorubicin while CerS5 sensitizes these cells to only doxorubicin and vincristine and CerS4 has no effects on the sensitivity to any of these anti-cancer agents (20). Similarly, CerS1 has a critical role in regulating apoptosis in HNSCC cells through caspase activation induced by gemcitabine/doxorubicin (105). They found that CerS1 downregulation by siRNA inhibited apoptosis about 50% in response to gemcitabine/doxorubicin. Also, CerS1 (and not CerS5) siRNA modulated caspase-3 and caspase-9, but not caspase-8, activation in response to gemcitabine/doxorubicin treatment. Moreover, gemcitabine/doxorubicin treatment led to a remarkable reduction in the growth of HNSCC tumor in severe combined immunodeficiency mice with UM-SCC-22A (a HNSCC cell line) xenografts. Interestingly, liquid chromatography and mass spectroscopy analysis showed that only the levels of C18-ceramide (synthesized by CerS1) were significantly increased in response to gemcitabine/doxorubicin in these tumors. These findings highlight a key role for C18-ceramide in gemcitabine/doxorubicin-induced apoptosis through caspase-9/3 activation in HNSCC (105). In another study on CerS1, it was found that treatment of K562 cells (a chronic myeloid leukemia cell line) with imatinib induced C18-ceramide synthesis via CerS1, which plays a role in imatinib-induced apoptosis in these cells (106). CerS1 downregulation using siRNA partly inhibited the imatinib-induced apoptosis in drug-sensitive K562 cells. Also, CerS1 overexpression (but not CerS6) triggered a significant increase in the synthesis of C18-ceramide induced by imatinib and promoted apoptosis. These results propose the involvement of C18-ceramide (synthesized by CerS1) in imatinib-induced apoptosis in K562 cells (106). As mentioned previously, de novo synthesis of ceramide is an important inducer of Bax. It has been found that localization of Bax to the mitochondria was decreased after CerS5 knockdown in NT-2 cells (a testicular embryonal carcinoma cell line) (107) indicating that CerS5 is critical in the synthesis of ceramide that leads to apoptosis in these cancer cells. CD95 (Fas) and its ligand (CD95L) are death receptors and ligands, which induce apoptosis (108). CerS6 modulates the activation of CD95 and therefore the induction of apoptosis (109). Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) is a member of the tumor necrosis factor (TNF) family able to initiate apoptosis via its death receptors (DR4 and DR5) (110, 111). This leads to caspase 8, and -10 activation. It has been found that normal cells are resistant to TRAIL-induced apoptosis because of low levels of TRAIL receptors expressed on their membrane (112). The CD95 activation and cancer cell killing were both increased after CerS6 overexpression while CerS6 knockdown abrogated CD95 activation in colon adenocarcinoma cell lines (DLD1, SW620 and SW480). Role of ceramides in the CD95-induced apoptosis in different types of cancer was investigated in another study by the same research group. Findings showed that fumonisin B1 (an inhibitor of CerS activity) inhibited the C16-ceramide increase in SW480 cells (113). Also, TRAIL-resistant SW620 cells had a lower levels of CerS6 expressed. CerS6 downregulation by siRNA led to a specific and significant reduction of C16-ceramide - enough to abrogate the TRAIL-induced apoptosis. Furthermore, only a slight increase in CerS6 expression was enough to reverse the TRAIL resistance in SW620 cells. These findings show that regulation of the CerS6 expression could potentially be a novel therapeutic approach to alter colon cancer cells’ sensitivity to apoptosis (113). In another study, it was found that the mRNA levels of CerS2 and CerS6 were highly increased in about half of the tissues obtained from patients with breast cancer in comparison to normal tissues. Findings from this study suggested that C16-ceramide (synthesized by CerS6) and C24:1- and C24-ceramide (synthesized by CerS2) may have an unusual pro-survival role and thus resist apoptosis in breast cancer pathogenesis (19). lncRNA ceramide synthase 6 antisense RNA 1 (CERS6-AS1) is a novel RNA that modulates the gene expression of ceramide synthesis 6 and is significantly overexpressed in various cancers such as gastric, hepatocellular, pancreatic and breast cancer (114). A study found that LncRNA CERS6-AS1 promotes proliferation and migration while inhibiting apoptosis of breast cancer cell lines in vitro (115). In agreement with this, flow cytometry findings on CERS6-AS1 knockdown revealed high rate of apoptosis (115). A very interesting research study showed that C16-ceramide directly binds to voltage-dependent anion channel VDAC2, a mitochondrial platform for Bax/Bak translocation, to mediate apoptosis in Human colon carcinoma HCT116 cells (91). VDAC channels are a platform for the recruitment of pro-apoptotic Bak and Bax into mitochondria, leading to cytochrome c release and permeabilization of the OMM (116). Moreover, C2-cermide has been found to induce apoptosis in lung adenocarcinoma cells (A549 and PC9) via regulation of the thioredoxin-interacting protein (Txnip) (117). Treatment of these cells with C2-ceramide caused an increase in the activity of caspase-3. Txnip has a crucial role in redox signal transmission in the cell (118). The expression of CerS6 which synthesizes C16-ceramide is significantly upregulated in breast cancer (19) and non-small-cell lung cancer (119) cells. CerS6 is a regulator of apoptosis and contributes to the metastasis and progression of cancer cells (120). Using siCerS6, Shi et al. showed that CerS6 knockdown prevented the migration and metastasis of ovarian cancer cells while inducing apoptosis in these cells (121). C6-ceramide also induces apoptosis in salivary adenoid cystic carcinoma (SACC) SACC-83 and SACC-LM cell lines in a dose dependent manner as shown by the upregulation of caspase-3, PUMA, and CHOP (122).
Role of CerS and ceramides in Regulation of Autophagy
Autophagy is a self-recycling mechanism in the cell that targets nonfunctional or misfolded proteins or damaged organelles and degrades them via lysosomes (123-127). There are three types of autophagy; macroautophagy (hereafter autophagy), microautophagy and chaperone-mediated autophagy all of which differ from one another in terms of their mechanism and function (128, 129). Autophagy not only leads to the elimination of misfolded proteins and damaged organelles, it also modulates the recycling of components within the cell thereby ensuring the cellular quality control (130-132). The primary function of autophagy is survival under stress conditions, such as starvation (133). This is accomplished through self-eating that provides the cell with required energy and metabolic precursors (134-137). Consequently, dysregulated or prolonged autophagy activation can result in cell death (138-140). Ceramides have been shown to regulate autophagy (141). Class I PI3K and Akt are two well-known regulators of autophagy. C2 or C6-ceramide have the ability to activate PP2A blocking the Akt activation thereby inducing autophagy in HT-29 (human colon) and MCF-7 (breast cancer) cell lines (142, 143). Furthermore, amino acid deprivation leads to an increase in ceramide levels. This results in inhibition of the mTOR activity and therefore autophagy induction in a PP1/PP2A dependent manner. It was shown by Edinger
et al. that ceramide rapidly and significantly downregulates amino acid transporter proteins (103). Likewise, C2-ceramide inhibited the protein expression of nutrient transporter leading to starvation and consequentlyAMPK-dependent autophagy induction in murine prolymphocytic cells (FL5. 12) (144). Beclin1 is a crucial mediator in autophagy pathway. Beclin1 expression is increased under stress and its gene is mutated or deleted in different types of cancers (145). Scarlatti
et al. demonstrated that exogenous C2-ceramide increases the expression of Beclin1 and thus autophagy induction. This effect is inhibited after using myriocin (a CerS inhibitor) which shows that ceramide regulation of Beclin1 expression occurs at either transcriptional or post-transcriptional level (143). In CNE2 and Hep3B cancer cell lines, C2-ceramide can also activate JNK and consequently c-Jun phosphorylation. C-Jun then upregulates the expression of Beclin1 mRNA (146). Moreover, elevated levels of Beclin1 could stem from the Beclin1-Bcl2 dissociation (147). In fact, ceramide triggers the JNK1 activation which leads to the phosphorylation of Bcl2 that in turn releases Beclin1 from the Beclin1-Bcl2 complex (148, 149). Also, ceramide is involved in the activation of Forkhead box protein O3 (FOXO3) that can increase the expression levels of BNIP3 (150). The increased expression of BNIP3 competitively binds to Bcl2 and Bcl-xL thereby dissociating Beclin1 from binding to Bcl2 (151). Ceramide-induced ER stress can play a role in the induction of autophagy (141). In fact, downregulation of CerS2 triggers significant accumulation of C14 and C16-ceramides and induces ER-stress-mediated autophagy (152). It should be mentioned that ER stress is also interconnected with the autophagy flux. Autophagy flux is defined as the rate of the degradation of cargos by autophagy process which shows autophagic degradation activity (153). One of the UPR arms is the dimerization of the IRE1 (154). IRE1 enhances autophagy flux through its interaction with adapter proteins such as apoptosis signal-regulating kinase 1 and tumor necrosis factor (TNF) receptor-associated factor 2 (TRAF2). This results in the formation of IRE1/TRAF2/ASK1 complex that activates JNK that will subsequently phosphorylates c-Jun, which increases Beclin1 expression (155, 156). It should be mentioned that despite the fact that IRE1 induces the protective autophagy, this could switch to autophagy-dependent cell death during prolonged ER stress (156, 157). The PERK/eIF2α arm of UPR also plays an important role in regulation of autophagy. This arm is particularly active after ER stress that leads to autophagy induction. ATF4 and CHOP (C/EBP homologous protein, a transcription factor induced by ATF4) are essential for downstream effects of PERK where they can modulate the expression of different ATG genes.
Figure 3 shows how ceramide can regulate autophagy.
Life-or-Death Dichotomy of Autophagy (Autophagy Paradox)
There is a basal level of autophagy activity the cells in the human body (158-161). This basal activity is involved in the recycling of proteins and organelles destined for degradation and highlights the antiaging role of autophagy (162, 163). Autophagy can also be induced via different stimulus like nutrient withdrawal (164). Autophagy provides the cell with required building blocks to maintain the homeostasis and metabolism for survival (165, 166). Autophagy-dependent cell death is also known as the type II programmed cell death which is defined as a highly regulated cell death that does not involve other cell death pathways but is solely dependent on the central machinery of autophagy pathway (167, 168).
Owing to its double-edged role of cell survival and death, autophagy is unsurprisingly involved in the pathology of different diseases including but not limited to cancer, liver diseases, and neurodegenerative diseases (169-171). Autophagy has advantageous and detrimental effects in all these situations (172). Furthermore, accumulating evidence implicates ceramides in the regulation of these two opposing roles of autophagy that regulate cell death and cell survival, known as the autophagy paradox (173, 174).
Regulation of Autophagy by Ceramides
Bioactive sphingolipids regulate autophagy by exerting their effect in different steps of the autophagy pathway. Research on the role of ceramides in elongation phase of autophagy is very limited and majority of studies have revealed different roles for ceramides on the initiation and degradation phases of autophagy (
Figure 3).
Ceramides regulate induction & nucleation steps of autophagy
Autophagy is regulated at the initiation step via different mechanisms. It has been demonstrated that ceramide induces the activation of JNK leading to the phosphorylation of Bcl-2 (175). Bcl-2 phosphorylation leads to its dissociation from Beclin1 and therefore lifts its inhibitory role on autophagy. Interestingly, treatments (for example glucosylceramide synthase inhibitor PDMP or tamoxifen) that elevate the long-chain ceramides levels significantly potentiate this effect of autophagy. Moreover, it was shown that treatment of nasopharyngeal carcinoma and hepatocellular carcinoma cells with ceramide enhances the Beclin1 expression through activation of JNK thereby inducing autophagy (146). Also, levels of S1P may greatly influence the formation of autophagosomes through their effect on the LC3 lipidation and thus its anchoring to the autophagosomal membrane. S1P can also modulate aBeclin1 complex although the underlying mechanism remains unknown. Additionally, it has been revealed that Sphk overexpression increases the S1P levels thereby enhancing the preautophagosomal formation in primary neurons. This remarkably enhances the overall autophagosomes formation (176). The mitochondria-associated ER membranes can be the membrane source for phagophore formation. Lipid rafts within the mitochondria-associated ER membranes are particularly important for this process. During formation of autophagosomes, ganglioside GD3 which is a component of lipid rafts and a downstream product of ceramides binds to Beclin1 complex (177, 178). GD3 contributes to the recruitment of components of the autophagic pathway. It also regulates the membrane fluidity, thus playing a role in the formation of phagophore curved membranes. Golgi-derived ATG9 vesicles donate material to the forming phagophore (179). The availability of the phosphorylated form of ceramide, ceramide-1-phosphate, (180) potentiates the formation of these vesicles which subsequently fuse to the forming phagophore (181).
Ceramides regulate fusion & degradation steps of autophagy
A direct role for sphingolipids in promoting the autophagosomes-lysosome fusion have not yet been demonstrated. It has recently been reported that C1P synthesis from sphingomyelin induces the Ca2+-dependent liposomal fusion that increases the vesicle fusion (182). Furthermore, a role for C1P has been proposed in the exocytotic and endocytic pathways (182) that are connected to the autophagic pathway (183, 184). It has also been reported that C18-ceramides regulates the degradation step of the autophagy. C18-ceramides can induce the selective targeting and degradation of mitochondria by autophagy, and mitophagy (185, 186). As stated above, localized ceramide on mitochondria binds to the lipidated LC3 on the autolysosomes and induce mitophagy (187). Interestingly, aside from their role in regulation of degradation step of autophagy, ceramides can also serve as autophagic substrates (188).
Ceramide regulates autophagy flux
Sphingolipids are also involved in the regulation of transporters which direct nutrients into the cell. These lipids are important in autophagic flux (144, 189) however, studies on their exact mechanism of actions are limited. As mentioned earlier, autophagosome fuses with lysosome to form autolysosome, thereby degrading the proteins and organelles via the acidic hydrolases in the lysosomes. Interestingly, it has been shown that the efficiency of autophagosome-lysosome fusion depends on the lipid composition in the cell such as the ceramide levels (190). Moreover, it has previously been reported that myristate enhances the autophagic flux in cardiomyocytes which is dependent on C14-ceramide and CerS5. This indicates that ceramide plays a role in the modulation of autophagic flux (189).
Cytoprotective autophagy induction by CerS and ceramide
Ceramides, and in particular long chain ceramides, are involved in regulating the cell death signaling (191). Ceramides can however trigger cytoprotective autophagy to inhibit cell death under certain conditions (192). Ceramide is involved in the down regulation of amino acid and nutrient transporters. This results in starvation which triggers survival autophagy through activating AMPK and then inhibition of mTOR signaling (193, 194). Changes in the nutrient transporters’ expression impacts cell growth and survival because these transporters regulate fuel for the cell. Moreover, survival autophagy is decreased upon downregulation of CerS2 that alters normal ceramide trafficking in the ER. This leads to the accumulation of long chain ceramides and IRE1 activation, thereby inhibiting cell death induction (152). Guenther et al. showed that C2-ceramide led to downregulation of nutrient transporters and consequently restricted the levels of nutrients in the cell leading to cell starvation and cell death (144). C2-ceramide also induced cytoprotective autophagy to inhibit cell death since autophagy inhibition by chloroquine sensitized cells to ceramide (144). In another study it was found that CerS2 downregulation led to no remarkable reduction of very long chain ceramide such as C24 ceramide. However, it increased the levels of long chain ceramide in such as C14 or C16-ceramide (~3-fold) (152). Also, a report showed that accumulation of long chain ceramide induced autophagy but not apoptosis in neuroblastoma cells (SMS-KCNR) (152). From a mechanistic standpoint, downregulation of CerS2 induced the cytoprotective autophagy and UPR. This led to the IRE1 activation which prevented the cell death induction (152). Moreover, vorinostat and sorafenib, two chemotherapeutic agents, have the ability to induce the CD95 activation through via acid sphingomyelinase activation and synthesis of C12, C22 and C26-ceramide (195). C14 and C16 ceramide activated CD95 triggered autophagy which was dependent to PERK. This indicated the cytoprotective effects since ATG5 downregulation increased the lethal effects of sorafenib and vorinostat in HEPG2 cells (195). Some studies have found that dehydroceramide (DHC) could induce protective autophagy, however its exact role is not fully elucidated (196-199). It was shown that XM462 (DHC desaturase inhibitor) delays G1/S transition in the cell cycle by ER stress activation and autophagy induction in HCG27 cells (gastric carcinoma) (196). Also, the inhibition of XM462-induced autophagy leads to a remarkable decrease in cell viability, emphasizing the role of DHC-induced autophagy in promoting cell survival (196). Interestingly, studies have shown that cytoprotective autophagy could be connected to apoptosis. It has been shown that caspases can regulate the autophagy-apoptosis crosstalk (200). Upon activation, caspases can cleave important autophagic proteins like Atg3, Atg4D, Atg5, Atg7, Beclin-1, and p62 leading to their inactivation. (201-205).It was also found that caspase-9 binds Atg7, promoting the lipidation of LC3B-I and formation of LC3B-II. This increases the autophagy activity. On the other hand, interaction between caspase-9 and Atg7 inhibits caspase-9 recruitment to the apoptosome. This inhibits caspase-9 activation and thus apoptosis (94). It was reported that in MCF-7 breast cancer cells caspase-9 knockout inhibits autophagic flux and promotes apoptosis via inhibition of cytoprotective autophagy (206). These finding highlight the intricate interconnection between autophagy and apoptosis.
Lethal autophagy induction by CerS and ceramide:
The role of ceramides in inducing apoptosis is well-known. Ceramides can induce apoptosis in response to different stimuli such as death receptor ligation, hypoxia, growth factor withdrawal, and chemotherapeutic drugs (144). Various studies have established the critical role of ceramides in regulation of lethal autophagy, however the exact mechanisms are still not fully understood. Ceramide triggers lethal autophagy through its effect on the Beclin1 expression. In fact, exogenous C2-ceramide enhances the expression of Beclin1 followed by induction of lethal autophagy in HT-29 colon cancer cells (143). This effect was inhibited when the de novo ceramide synthesis was inhibited by myriocin, suggesting the observed effects are due to C2-ceramide conversion to long chain ceramide (143). Other findings have shown that the mechanism by which C2-ceramide enhances the expression of Becloin1 is through JNK activation. JNK subsequently activates c-Jun which is a transcription factor capable of regulating Beclin1 expression (146). Additionally, chemotherapeutic drugs increase the ceramide synthesis which in turn enhances the expression of Beclin1 and therefore lethal autophagy (207). Other drugs can also result in an increase in the synthesis of ceramides. It was reported that cannabinoids trigger the production of ceramide leading to mTOR inhibition and lethal autophagy in human glioma cells (208).
Mitophagy is a selective autophagy that targets and removes damaged mitochondria. Mitophagy is therefore critical for the cell to maintain the proper homeostasis. Mitophagy involves two main steps; first the activation of general autophagy and second the selective targeting of the damaged mitochondria via different mediators (209).
Mitophagy plays an important role in tumor suppression via lethal mitophagy. Lethal mitophagy is defined as the process through which mitochondria are eliminated to the extent that cell undergoes cell death independent of the apoptosic pathway. Prolonged mitophagy results in caspase-dependent apoptosis via release of cathepsin proteases from lysosomes into the cytosol (210, 211). Lethal mitophagy is also induced by CerS1 (and also its product C18-ceramide) and is not dependent on Bax, Bak and caspase 3. It was reported that C18-ceramide synthesized by CerS1 was accumulated on the outer mitochondrial membrane. The exogenous C18-pyridinium ceramide was also localized on the outer mitochondrial membrane due to the pyridinium positive charge. This accumulated C18-ceramide interacts with LC3B-II on the forming autophagosmoe thereby facilitating the engulfment of the mitochondria into the autophagosome (212).
Role of CerS and Ceramides in Regulation of Autophagy in Cancer
CerS and Ceramides have been implicated in the pathogenesis of various cancers due to their regulatory role in autophagy. Recent reports demonstrated that under starvation condition, Bcl-2 expression inhibits autophagy (13), while C2 and C6-ceramide could trigger autophagy in HT-29 colon carcinoma cell line (8). In fact, C2-ceramide increased the BNIP3 accumulation (213) which induced autophagy via interfering with the Beclin1:Bcl2 complex and dissociating Beclin1 (151). The Beclin1:Bcl-2 dissociation is independent of Bcl-2 phosphorylation (214). Therefore, there could be two mechanisms through which ceramide dissociates this complex: 1) JNK1 activation and subsequent Bcl2 phosphorylation or 2) BNIP3 accumulation on mitochondria which interferes with the Beclin1:Bcl2 complex and dissociates Beclin1. It was previously shown that treatment of human HNSCC cells (UM-SCC 22A) with CerS1-mediated generation of C18-ceramide and C18-pyridinium-ceramide induced lethal autophagy was independent of apoptosis (212). The lethal autophagy induced by C18-ceramide was modulated by LC3B-II followed by the selective engulfment of mitochondria via the interaction between ceramide and LC3B-II (212). Dbaibo et al. reported that arsenic trioxide (As2O3) triggered cytotoxic accumulation of overall ceramide in leukemia cells (HuT-102, C91-Pl and MT-2 cells) by inhibiting the activity of GluCeramide synthase (215). Additionally, As2O3 induced both apoptosis and autophagic cell death in these cells. The lethal autophagy was associated with an increase in the Beclin1 expression. Also, this lethal autophagy was inhibited when cells were treated with 3-MA (216). C2-ceramide triggers lethal autophagy (LC3B-II lipidation, formation of autophagosomes, and acidic lysosomes) in U373-MG and T98G cells (malignant glioma) (213). It was found that the mechanism involved was the reduction of mitochondrial membrane potential and BNIP3 activation by C2- ceramide (213). Moreover, the lethal autophagy was mediated by C2-ceramide and triggers the activation of c-Jun by JNK which increases the expression of Beclin-1 in human nasopharyngeal carcinoma (CNE2) and hepatocellular carcinoma (Hep3B) cell lines (146). Treatment of cells with SP600125 (JNK inhibitor) or Beclin1 knockdown by siRNA rescued these cancer cells from C2-ceramide-induced lethal autophagy (146). Also, in leukemia cells (HL-60) and Chinese hamster ovary cells (CHO) ceramide activated protein phosphatases (CAPPs) inhibit mTOR which induces lethal autophagy. However, S1P induces mTOR thereby suppressing lethal autophagy in these cells (217). In another study, it was shown that overexpression of CerS1 triggers lethal autophagy in brain cancer cells (U251 and A172) (204). LC3B-I to LC3B-II conversion and p62 degradation indicated autophagy induction in both cells. CerS1 overexpression and exogenous C18-ceramide greatly enhanced the occurrence of LC3 punctate and formation of autophagosomes in both cells. Additionally, CerS1 induced autophagy via inhibiting the PI3K/AKT pathway in these cells (218).
A role for CerS has also been described on the outcome of patients with cancer via the regulation of autophagy. The results from a study by Hartmann
et al. displayed a correlation between strong CerS5 staining and poor prognosis in colorectal patients. Consistent with this observation, patients with a weak CerS5 staining had a better prognosis. These findings highlight that overexpression of CerS5 is connected to a more aggressive cancer that could be modulated through ceramide levels by currently unknown mechanisms. Changes in the CerS5 expression can interfere with the ceramides balance in colon cancer and therefore promote cancer progression (219). In this study, the proteomic network analysis showed a shift from apoptosis (patients with CerS5 low expression) to autophagy (patients with CerS5 high expression), indicating an association between high CerS5 expression and poor survival (220). Altogether these findings show that altering the ceramide-mediated apoptosis and autophagy activation in patients with high CerS5 expression could possibly be responsible for the link between strong CerS5 expression and poor survival (220).
Table 2 summarizes the roles of various molecules in the ceramide metabolism pathway in regulation of autophagy in different types of cancer discussed in this section.
Overall View on Lipid Analysis
Over the last decades, the detection of early cancer stages has been the focus of intensive investigation in medical research, especially non-invasive cancer screening methods based on blood analysis (225). Lipids serve a plethora of roles in human metabolism (226), acting as: cell membrane constituents, signaling molecules, energy supply, and energy storage. Changes in lipid concentrations have been reported in various types of cancer (227-233) and lipid metabolic reprogramming has become a hallmark of cancer (228, 233). However, despite the evidence highlighting its relevance to cancer, lipid metabolism remains difficult to analyze due to the lack of standard systematic quantitation methods, lipidome nomenclature, and analysis applied to cancer research (234-236).
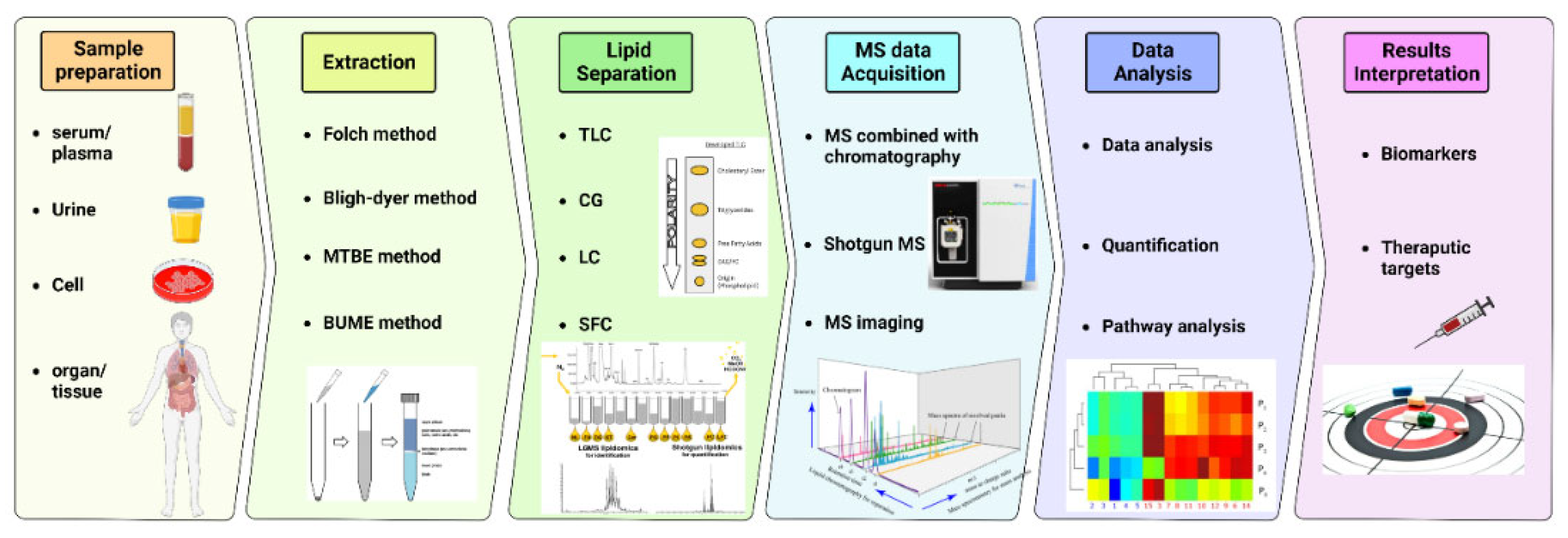
Lipidomics is the study that statistically profiles and quantitates lipid molecules on a large scale (237). The development of lipidomics followed the advances in techniques such as: mass spectrometry (MS), nuclear magnetic resonance spectrometry (NMR), and chromatography. Mass spectrometry uses mass spectrometers to study the mass-to-charge (m/z) ratio of single analytes allowing structural elucidation and quantitation, which includes steps of molecular ions and related fragment generation, ion separation based on their m/z, signal detection, and the intensity of individual ion measurement. Typically, a mass spectrometer consists of an ion source, a mass analyzer system, a detector, and a data processing system. Among the lipid separation methods, liquid high-performance chromatography (HPLC), or simply LC, combined with mass spectrometry (MS), is the primary choice for lipidomics studies due to the sensitivity and selectivity of the analysis (238). MS-based lipidomics can be categorized as non-targeting or targeted lipidomics. The former identifies and quantifies all detected lipids, whereas the latter focuses only on specified lipid classes (239). Notably, the HPLC-MS technique has been reported back to the 1970s (240, 241) as a powerful method for sphingolipids, such as ceramides, detection.. Additionally, numerous other studies have validated this method of ceramides detection over the last decades for humans, animals and plants (242-250).
Prior to LC-MS analysis, biological samples are prepared through different methods, including liquid-liquid extraction, organic solvent precipitation, and solid phase extraction, separating cellular or fluid lipids from the other constituents and preserving these lipids for further analyses (251). Most procedures extract high solubility lipids in organic solvents (252), such as Folch, Bligh and Dyer, methyl tert-butyl ether, and Butanol:Methanol.
Once lipids are extracted, samples are ready to undergo mass spectrometry lipid separation (253). A typical workflow of lipidomics analysis of biological samples consists of sample collection and storage, homogenization, internal standard addition for calibration, analysis of lipids with and without separation, data processing and lipid identification, and biomedical application, summarized in (
Figure 4).
The generation of lipid databases and the tools to cross-compare them with experimentally obtained lipidomics data has been an important development. Examples are LipidMaps, LipidBank, LipidHome, LipidBlast, and LipidSearch (254). Moreover, comprehensive mass spectrometry determination of a great range of serum lipids can reveal significant differences between studied groups of diseases, such as cancer, where selected lipid species may indicate potential prognostic biomarkers. For instance, a study by Wolrab et al., 2022, conducted lipidomic profiling of serum to screen for pancreatic cancer in 830 samples. They demonstrated differences in serum versus plasma lipidome concentrations between samples of pancreatic cancer patients and healthy controls using mass spectrometry (MS) based approaches, followed by statistical analysis. They concluded that lipid concentrations in serum were mildly higher than in plasma, yielding an enhanced sensitivity (255).
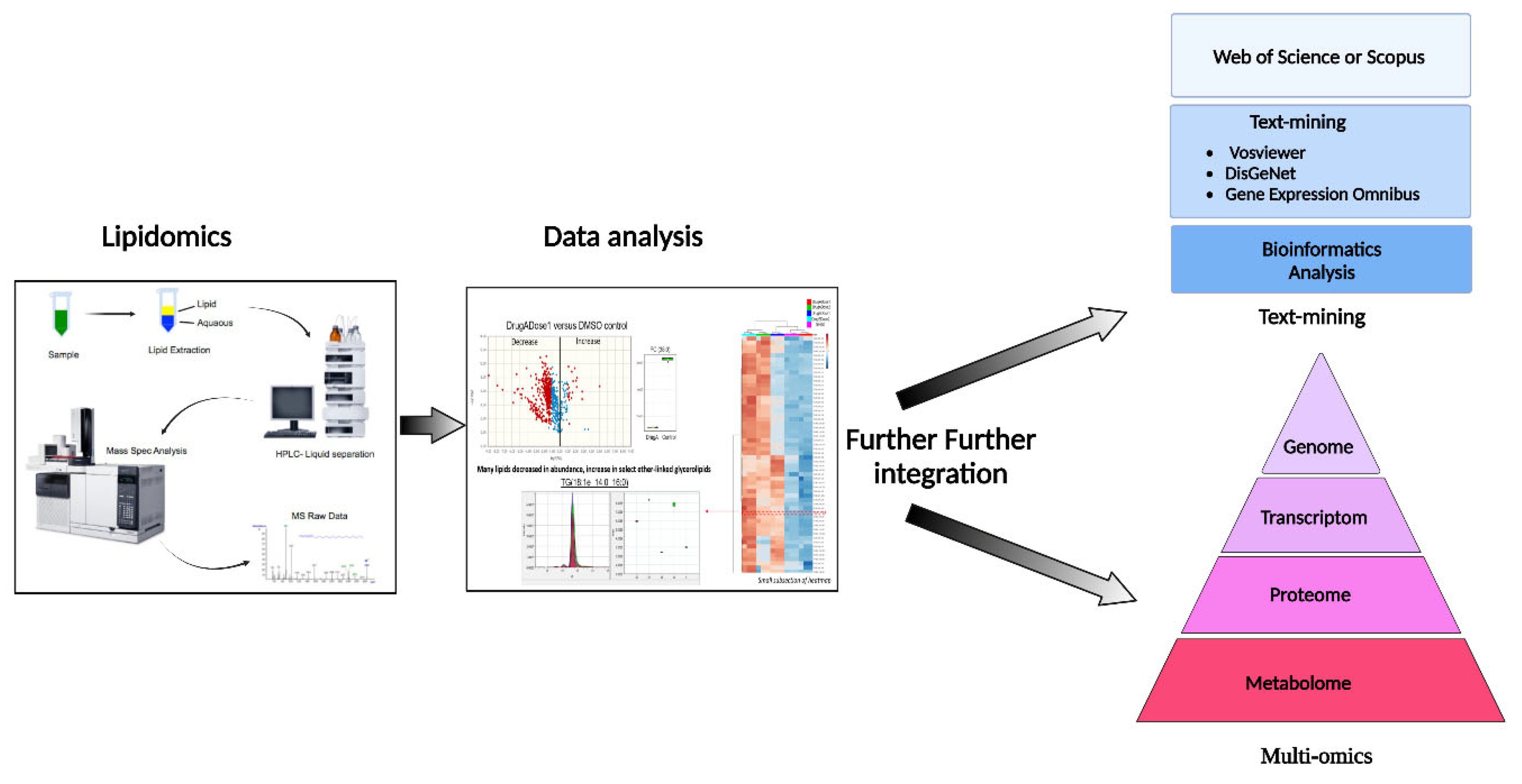
After lipidomics and identification of key targets, the next critical step in modern bioinformatics is the integration of systems biology for proteomics, genomics, regulatory genomics, and metabolomics data to provide a further systems-level overview of phenotypic responses of living systems to stimuli, (
Figure 5)
. The conventional statistical methods can be applied to each type of data, such as gene expression, proteomics or lipidomics; however, the integration across these data types, to provide mechanistically meaningful models, remains challenging (256).
To overcome this integration challenge, a new pipeline strategy developed by Lima et al., 2022, could be an interesting alternative method to validate the altered lipid levels in the samples under study. For example, the automatic text-mining feature of VOSviewer can be used to create coincidence networks of terms associated with the disease of interest, e.g., prostate cancer. These results will be complemented with DisGENET data, a repository of disease associations, and a recent bioinformatics analysis integrating all differentially expressed lipids identified in tumor tissue and serum samples from patients to improve VOSviewer's limited term selection. Later, the results can be integrated with gene expression data from the Gene Expression Omnibus database to correlate gene and protein levels (257). Additional lipid tools and resources for bioinformatic analysis are listed in
Table 3.
Despite the technological advances in lipidomics, there are still limitations to this approach. Not only does the diversity and structural complexity of many lipidomes remain a challenge, but understanding, interpreting, and integrating large data sets in conjunction with classical toxicological parameters is a major task. An integrative approach is needed to understand the principles underlying metabolic regulation of a system and how their combined interactions, in conjunction with variations in clinical phenotype, lead to pathophysiology. This challenge requires new data exploration strategies such as analysis workflows, statistical and computational algorithms for data integration, filtering, and network analysis, if we are to be able to truly transform the large multivariate data collected in such metabolomic experiments into new biological insights.
Artificial intelligence (AI) methods are advancing rapidly, and breakthrough technologies are changing the landscape of health research with high diagnostic and prognostic value. Machine learning methods (also referred to as complex AI), supervised and unsupervised, are used by AI systems to account for complex interactions, either by collecting input data, including biofluids and tissues, to predict output values based on new input samples, or by finding underlying patterns in an unlabeled data set to identify subclusters and outliers in the data.
Although AI methods have been well described in other areas of healthcare, there is little information on the value of using AI methods to understand the complex nature of phlebotomy. Machine learning has enabled the discovery of more robust biomarkers that have been approved by the Food and Drug Administration (FDA) to guide treatment, which can be very valuable in disease. Additionally, biomarkers serve as powerful clinical predictors that can be used to individualize treatment options for patients to achieve desired outcomes. The machine learning approach called Random Forest (RF) can integrate multiple data types and combine classical clinical chemistry and toxicology test results with multivariate metabolomic and lipidomic data. Several authors have previously adapted RF methods for data integration (also referred to as data fusion) that developed and evaluated a fuzzy logic combination with RF to prioritize the discriminative features of gene expression data.
In summary, to move towards standardization, minimum requirements for lipid studies should become routine. As described by McDonald et al., 2022, the nine major categories for conducting lipidomics research are 1) overall study design, 2) pre-analytics, 3) lipid extraction, 4) analytical platform, 5) lipid identification, 6) lipid quantification, 7) quality control, 8) method validation, and 9) data reporting (235).
Conclusion and Future Directions
As we discussed ceramides may play essential roles response to different cancer therapy strategies through apoptosis and autophagy pathways, therefore targeting ceramides and their biosynthesis pathway could be beneficial for developing new cancer therapy methods. It can be seen that the development of new therapeutics that interact with novel pharmacological targets is urgently needed. All anti-cancer drugs can be classified into two major categories: small molecules and monoclonal antibodies (258). Small-molecule drugs are preferable in drug development compared to antibodies due to better pharmacokinetic properties, lower preclinical and clinical costs, and large-scale production and purification (259, 260).
Modulation of the sphingolipid/ceramide-controlled apoptosis with small molecules represents an important tool and addition to the current anticancer therapies. Currently, there have been only five clinical trials of different phases that involved ceramide as the active drug for the treatment of various cancers (
Table 4). However, none of these studies explore the new small molecules as modulators of ceramide pathways.
The biosynthesis of ceramides has been done via two pathways: de novo sphingolipid pathway and sphingolipid-salvage pathway (
Figure 6) (261). Several different CerS have been identified in both pathways, and modulation of these enzymes is reported in many mechanisms including apoptosis and protein kinase inhibition (262-264). Several studies showed that decreased levels of ceramides are reported in many malignancies and tumor progression, while ceramide levels are observed to be increased by chemotherapies (265-268). Additionly, several tyrosine kinase inhibitors (TKI) increase production of CerS on a nuclear level, which in turn leads to upregulation of ceramide biosynthesis (269, 270). Recently it has been reported that the combination of a novel sphingosine kinase 2 inhibitor ABC294640, and TKI displays synergistic anti-cancer effects in multiple myeloma and cholangiocarcinoma (271, 272). Thus, the small molecules that can modulate ceramide pathways represent valid targets for anticancer drug discovery, especially in combination with currently available TKI-based anti-cancer therapies (
Figure 7) since previous studies suggests that there may be functional crosstalk between inhibiting tyrosine kinases overexpressed in certain tumors and dysregulation of ceramide synthesis in malignancies.
It should be noted, however, that the co-administration of a tyrosine kinase inhibitor and a ceramide modulator would have many drawbacks, including the possibility of drug-drug interactions
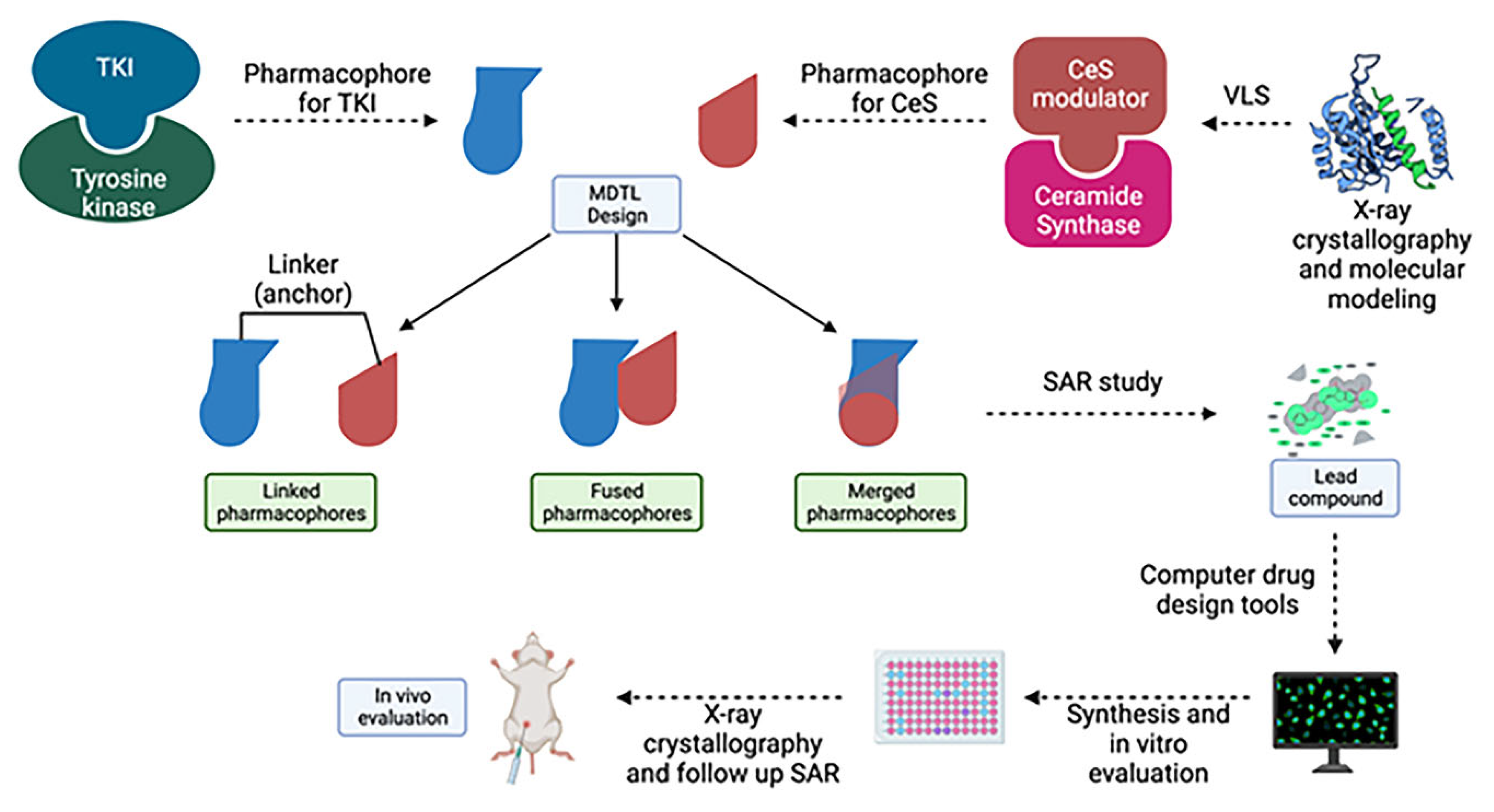
in vivo which complicates preclinical/clinical studies and the drug development process in the long term. One novel drug design strategy is the use of multi-target designed ligands (MTDLs), i.e. a polypharmacology approach. MTDLs are small molecules designed to simultaneously interact with multiple biological targets relevant for a given pathology (273). Furthermore, they have several advantages including a potential for higher efficacy and fewer side-effects compared to cocktail drugs and could avoid unpredictable pharmacokinetic and pharmacodynamics relationships (274). To develop new anticancer MTDL therapeutics, the first step will be to determine the similarities and potential overlap in the structures of already existing TKIs and ceramide modulators for targets of interest (
Figure 8). In the case that the target does not have any known modulators, drug design can be achieved using knowledge of characterized active and/or allosteric binding sites by X-ray crystallography in combination with virtual-ligand screening (VLS). A general scaffold, or core of a chemical structure, or even particular chemical groups can be used for this purpose. Once the specific structures important for the activity at the targets of interests are identified, the pharmacophore - the area responsible for the biological activity for these targets, can be defined. Next, the pharmacophores can be combined in three different fashions; they can be linked, fused or merged (275). Linked MTDLs are designed by simple linking (anchoring) of two or more individual pharmacophores via a linking group. Fused MTDLs are similar, but the pharmacophores are connected directly, without a linking group. Both, linked and fused types of multitarget compounds usually have molecular weights above 500 and increased lipophilicity, which are important for drug design but could lead to poor solubility and permeability according to Lipinski Rule of Five.(276) However, these two types of conjugated pharmacophores are a valuable tool in the early structure-activity relationship studies and the discovery of the multitarget activity. Merged pharmacophores, on the other hand, are of the greatest interest in the multitarget drug discovery and there are many successful applications of this method reported.(277-279)
Here we propose that the key pharmacophoric elements required to interact with each target of interest are combined (merged) into one single pharmacophore. Once pharmacophores are identified and combined, the next step is to preform extensive SAR studies in order to determine the best possible groups that can be tolerated on the particular pharmacophores. Next, the standard structure-based drug design approach can be followed: a) Using computer drug design tools, several additional compounds that could interact with the amino acid residues present in the binding site can be designed; b) Synthesize the second generation of lead compound(s) and test them for activity; c) Crystallographic determination of the lead compound with both targets which should identify the actual binding interactions and provide better insight in the other possible interactions. d) Finally, another more specific SAR study that will optimize the drug-target interactions should be performed.
In summary, one new rational approach is to design a single drug that would be able to interact simultaneously as an inhibitor of specific protein kinases and a modulator of certain ceramides involved in the cancer pathology. Based on the previous studies mentioned above, the novel drug should target the allosteric-binding site of the protein kinases, since the gene sequences of the allosteric sites consist of fewer homologues than those of the active sites, these drugs will be more specific. One way to identify the ceramides target involved in the cancer pathogenesis is to perform lipidomics analysis and compare lipid distribution between cancer and wild-type cells, i.e., the up- or down-regulated lipids that could represent a novel target for the MTDLs.
Author Contributions
Javad Alizadeh, Shahla Shojaei, Joadi Jacobs and Shahrokh Lorzadeh prepared the draft of general ceramide synthases, ceramides and apoptosis and autophagy. Simone C da Silva Rosa, Amir Ravandi, and Rui Vitorino participated in preparation of lipid analysis and lipidomic section. Xiaohui Weng, and Stevan Pecic prepared the drug development part. Saeid Ghavami designed and lead the whole project, and finalize the whole manuscript. .
References
- Field BC, Gordillo R, Scherer PE. The Role of Ceramides in Diabetes and Cardiovascular Disease Regulation of Ceramides by Adipokines. Frontiers in Endocrinology. 2020;11. [CrossRef]
- Levy M, Futerman AH. Mammalian ceramide synthases. IUBMB Life. 2010;62(5):347-56. [CrossRef]
- Tidhar R, Ben-Dor S, Wang E, Kelly S, Merrill AH, Jr., Futerman AH. Acyl chain specificity of ceramide synthases is determined within a region of 150 residues in the Tram-Lag-CLN8 (TLC) domain. The Journal of biological chemistry. 2012;287(5):3197-206. [CrossRef]
- Zelnik ID, Rozman B, Rosenfeld-Gur E, Ben-Dor S, Futerman AH. A Stroll Down the CerS Lane. Advances in experimental medicine and biology. 2019;1159:49-63.
- Raichur S. Ceramide Synthases Are Attractive Drug Targets for Treating Metabolic Diseases. Frontiers in Endocrinology. 2020;11. [CrossRef]
- Spassieva SD, Mullen TD, Townsend DM, Obeid LM. Disruption of ceramide synthesis by CerS2 down-regulation leads to autophagy and the unfolded protein response. Biochem J. 2009;424(2):273-83. [CrossRef]
- Jennemann R, Rabionet M, Gorgas K, Epstein S, Dalpke A, Rothermel U, et al. Loss of ceramide synthase 3 causes lethal skin barrier disruption. Human molecular genetics. 2012;21(3):586-608. [CrossRef]
- Véret J, Coant N, Berdyshev EV, Skobeleva A, Therville N, Bailbé D, et al. Ceramide synthase 4 and de novo production of ceramides with specific N-acyl chain lengths are involved in glucolipotoxicity-induced apoptosis of INS-1 β-cells. The Biochemical journal. 2011;438(1):177-89. [CrossRef]
- Gosejacob D, Jäger PS, Vom Dorp K, Frejno M, Carstensen AC, Köhnke M, et al. Ceramide Synthase 5 Is Essential to Maintain C16:0-Ceramide Pools and Contributes to the Development of Diet-induced Obesity. The Journal of biological chemistry. 2016;291(13):6989-7003. [CrossRef]
- Verlekar D, Wei S-J, Cho H, Yang S, Kang MH. Ceramide synthase-6 confers resistance to chemotherapy by binding to CD95/Fas in T-cell acute lymphoblastic leukemia. Cell death & disease. 2018;9(9):925.
- Saddoughi SA, Ogretmen B. Diverse functions of ceramide in cancer cell death and proliferation. Advances in cancer research. 2013;117:37-58.
- Zhou X, Huang F, Ma G, Wei W, Wu N, Liu Z. Dysregulated ceramides metabolism by fatty acid 2-hydroxylase exposes a metabolic vulnerability to target cancer metastasis. Signal transduction and targeted therapy. 2022;7(1):370. [CrossRef]
- Zeng J, Tan H, Huang B, Zhou Q, Ke Q, Dai Y, et al. Lipid metabolism characterization in gastric cancer identifies signatures to predict prognostic and therapeutic responses. Frontiers in genetics. 2022;13:959170. [CrossRef]
- Sheridan M, Ogretmen B. The Role of Ceramide Metabolism and Signaling in the Regulation of Mitophagy and Cancer Therapy. Cancers. 2021;13(10):2475. [CrossRef]
- Koybasi S, Senkal CE, Sundararaj K, Spassieva S, Bielawski J, Osta W, et al. Defects in cell growth regulation by C18:0-ceramide and longevity assurance gene 1 in human head and neck squamous cell carcinomas. The Journal of biological chemistry. 2004;279(43):44311-9. [CrossRef]
- Saddoughi SA, Song P, Ogretmen B. Roles of bioactive sphingolipids in cancer biology and therapeutics. Subcell Biochem. 2008;49:413-40. [CrossRef]
- Karahatay S, Thomas K, Koybasi S, Senkal CE, Elojeimy S, Liu X, et al. Clinical relevance of ceramide metabolism in the pathogenesis of human head and neck squamous cell carcinoma (HNSCC): attenuation of C(18)-ceramide in HNSCC tumors correlates with lymphovascular invasion and nodal metastasis. Cancer letters. 2007;256(1):101-11. [CrossRef]
- Schiffmann S, Sandner J, Birod K, Wobst I, Angioni C, Ruckhäberle E, et al. Ceramide synthases and ceramide levels are increased in breast cancer tissue. Carcinogenesis. 2009;30(5):745-52. [CrossRef]
- Erez-Roman R, Pienik R, Futerman AH. Increased ceramide synthase 2 and 6 mRNA levels in breast cancer tissues and correlation with sphingosine kinase expression. Biochemical and biophysical research communications. 2010;391(1):219-23. [CrossRef]
- Min J, Mesika A, Sivaguru M, Van Veldhoven PP, Alexander H, Futerman AH, et al. (Dihydro)ceramide synthase 1 regulated sensitivity to cisplatin is associated with the activation of p38 mitogen-activated protein kinase and is abrogated by sphingosine kinase 1. Molecular cancer research : MCR. 2007;5(8):801-12. [CrossRef]
- Weinmann A, Galle PR, Teufel A. LASS6, an additional member of the longevity assurance gene family. International journal of molecular medicine. 2005;16(5):905-10. [CrossRef]
- Ruckhäberle E, Rody A, Engels K, Gaetje R, von Minckwitz G, Schiffmann S, et al. Microarray analysis of altered sphingolipid metabolism reveals prognostic significance of sphingosine kinase 1 in breast cancer. Breast cancer research and treatment. 2008;112(1):41-52. [CrossRef]
- Díaz-Perales A, Escribese MM, Garrido-Arandia M, Obeso D, Izquierdo-Alvarez E, Tome-Amat J, et al. The Role of Sphingolipids in Allergic Disorders. Frontiers in allergy. 2021;2:675557. [CrossRef]
- Ponnusamy S, Meyers-Needham M, Senkal CE, Saddoughi SA, Sentelle D, Selvam SP, et al. Sphingolipids and cancer: ceramide and sphingosine-1-phosphate in the regulation of cell death and drug resistance. Future oncology (London, England). 2010;6(10):1603-24. [CrossRef]
- McCluskey G, Donaghy C, Morrison KE, McConville J, Duddy W, Duguez S. The Role of Sphingomyelin and Ceramide in Motor Neuron Diseases. Journal of personalized medicine. 2022;12(9). [CrossRef]
- Borodzicz-Jażdżyk S, Jażdżyk P, Łysik W, Cudnoch-Jȩdrzejewska A, Czarzasta K. Sphingolipid metabolism and signaling in cardiovascular diseases. Frontiers in cardiovascular medicine. 2022;9:915961. [CrossRef]
- Ho QWC, Zheng X, Ali Y. Ceramide Acyl Chain Length and Its Relevance to Intracellular Lipid Regulation. International journal of molecular sciences. 2022;23(17). [CrossRef]
- Chung LH, Liu D, Liu XT, Qi Y. Ceramide Transfer Protein (CERT): An Overlooked Molecular Player in Cancer. International journal of molecular sciences. 2021;22(24). [CrossRef]
- Taniguchi M, Okazaki T. Role of ceramide/sphingomyelin (SM) balance regulated through "SM cycle" in cancer. Cellular signalling. 2021;87:110119. [CrossRef]
- Laviad EL, Kelly S, Merrill AH, Jr., Futerman AH. Modulation of ceramide synthase activity via dimerization. The Journal of biological chemistry. 2012;287(25):21025-33. [CrossRef]
- Laviad EL, Albee L, Pankova-Kholmyansky I, Epstein S, Park H, Merrill AH, Jr., et al. Characterization of ceramide synthase 2: tissue distribution, substrate specificity, and inhibition by sphingosine 1-phosphate. The Journal of biological chemistry. 2008;283(9):5677-84.
- Ben-David O, Pewzner-Jung Y, Brenner O, Laviad EL, Kogot-Levin A, Weissberg I, et al. Encephalopathy caused by ablation of very long acyl chain ceramide synthesis may be largely due to reduced galactosylceramide levels. The Journal of biological chemistry. 2011;286(34):30022-33. [CrossRef]
- Mullen TD, Spassieva S, Jenkins RW, Kitatani K, Bielawski J, Hannun YA, et al. Selective knockdown of ceramide synthases reveals complex interregulation of sphingolipid metabolism. Journal of lipid research. 2011;52(1):68-77. [CrossRef]
- Mesicek J, Lee H, Feldman T, Jiang X, Skobeleva A, Berdyshev EV, et al. Ceramide synthases 2, 5, and 6 confer distinct roles in radiation-induced apoptosis in HeLa cells. Cellular signalling. 2010;22(9):1300-7. [CrossRef]
- Kim JL, Ben-Dor S, Rosenfeld-Gur E, Futerman AH. A novel C-terminal DxRSDxE motif in ceramide synthases involved in dimer formation. Journal of Biological Chemistry. 2022;298(2):101517. [CrossRef]
- Kim JL, Mestre B, Malitsky S, Itkin M, Kupervaser M, Futerman AH. Fatty acid transport protein 2 interacts with ceramide synthase 2 to promote ceramide synthesis. Journal of Biological Chemistry. 2022;298(4). [CrossRef]
- Wali JA, Jarzebska N, Raubenheimer D, Simpson SJ, Rodionov RN, O'Sullivan JF. Cardio-Metabolic Effects of High-Fat Diets and Their Underlying Mechanisms-A Narrative Review. Nutrients. 2020;12(5).
- Villén J, Beausoleil SA, Gerber SA, Gygi SP. Large-scale phosphorylation analysis of mouse liver. Proc Natl Acad Sci U S A. 2007;104(5):1488-93. [CrossRef]
- Sridevi P, Alexander H, Laviad EL, Pewzner-Jung Y, Hannink M, Futerman AH, et al. Ceramide synthase 1 is regulated by proteasomal mediated turnover. Biochimica et biophysica acta. 2009;1793(7):1218-27. [CrossRef]
- Mizutani Y, Kihara A, Igarashi Y. LASS3 (longevity assurance homologue 3) is a mainly testis-specific (dihydro)ceramide synthase with relatively broad substrate specificity. The Biochemical journal. 2006;398(3):531-8. [CrossRef]
- Wegner MS, Schiffmann S, Parnham MJ, Geisslinger G, Grösch S. The enigma of ceramide synthase regulation in mammalian cells. Progress in lipid research. 2016;63:93-119. [CrossRef]
- Kim JL, Ben-Dor S, Rosenfeld-Gur E, Futerman AH. A novel C-terminal DxRSDxE motif in ceramide synthases involved in dimer formation. Journal of Biological Chemistry. 2022;298(2). [CrossRef]
- Mizutani Y, Kihara A, Igarashi Y. Mammalian Lass6 and its related family members regulate synthesis of specific ceramides. The Biochemical journal. 2005;390(Pt 1):263-71. [CrossRef]
- Becker I, Wang-Eckhardt L, Yaghootfam A, Gieselmann V, Eckhardt M. Differential expression of (dihydro)ceramide synthases in mouse brain: oligodendrocyte-specific expression of CerS2/Lass2. Histochemistry and cell biology. 2008;129(2):233-41.
- Rabionet M, van der Spoel AC, Chuang C-C, von Tümpling-Radosta B, Litjens M, Bouwmeester D, et al. Male germ cells require polyenoic sphingolipids with complex glycosylation for completion of meiosis: a link to ceramide synthase-3. The Journal of biological chemistry. 2008;283(19):13357-69.
- Mizutani Y, Kihara A, Chiba H, Tojo H, Igarashi Y. 2-Hydroxy-ceramide synthesis by ceramide synthase family: enzymatic basis for the preference of FA chain length. Journal of lipid research. 2008;49(11):2356-64. [CrossRef]
- Gosejacob D, Jäger PS, Vom Dorp K, Frejno M, Carstensen AC, Köhnke M, et al. Ceramide Synthase 5 Is Essential to Maintain C16:0-Ceramide Pools and Contributes to the Development of Diet-induced Obesity. The Journal of biological chemistry. 2016;291(13):6989-7003. [CrossRef]
- Xu Z, Zhou J, McCoy DM, Mallampalli RK. LASS5 is the predominant ceramide synthase isoform involved in de novo sphingolipid synthesis in lung epithelia. Journal of lipid research. 2005;46(6):1229-38. [CrossRef]
- Martelli A, Omrani M, Zarghooni M, Citi V, Brogi S, Calderone V, et al. New Visions on Natural Products and Cancer Therapy: Autophagy and Related Regulatory Pathways. Cancers (Basel). 2022;14(23). [CrossRef]
- Ahmadi M, Amiri S, Pecic S, Machaj F, Rosik J, Los MJ, et al. Pleiotropic effects of statins: A focus on cancer. Biochim Biophys Acta Mol Basis Dis. 2020;1866(12):165968.
- Hashemi M, Aftabi S, Moazeni-Roodi A, Sarani H, Wiechec E, Ghavami S. Association of CASP8 polymorphisms and cancer susceptibility: A meta-analysis. Eur J Pharmacol. 2020;881:173201. [CrossRef]
- Suhaili SH, Karimian H, Stellato M, Lee TH, Aguilar MI. Mitochondrial outer membrane permeabilization: a focus on the role of mitochondrial membrane structural organization. Biophysical reviews. 2017;9(4):443-57. [CrossRef]
- Emami A, Shojaei S, da Silva Rosa SC, Aghaei M, Samiei E, Vosoughi AR, et al. Mechanisms of simvastatin myotoxicity: The role of autophagy flux inhibition. Eur J Pharmacol. 2019;862:172616. [CrossRef]
- Peixoto PM, Lue JK, Ryu S-Y, Wroble BN, Sible JC, Kinnally KW. Mitochondrial Apoptosis-Induced Channel (MAC) Function Triggers a Bax/Bak-Dependent Bystander Effect. The American Journal of Pathology. 2011;178(1):48-54. [CrossRef]
- Ganesan V, Walsh T, Chang K-T, Colombini M. The Dynamics of Bax Channel Formation: Influence of Ionic Strength. Biophysical Journal. 2012;103(3):483-91. [CrossRef]
- Ghavami S, Sharma P, Yeganeh B, Ojo OO, Jha A, Mutawe MM, et al. Airway mesenchymal cell death by mevalonate cascade inhibition: integration of autophagy, unfolded protein response and apoptosis focusing on Bcl2 family proteins. Biochimica et biophysica acta. 2014;1843(7):1259-71.
- Bernardi P. The mitochondrial permeability transition pore: a mystery solved? Frontiers in Physiology. 2013;4. [CrossRef]
- Iranpour M, Moghadam AR, Yazdi M, Ande SR, Alizadeh J, Wiechec E, et al. Apoptosis, autophagy and unfolded protein response pathways in Arbovirus replication and pathogenesis. Expert Rev Mol Med. 2016;18:e1. [CrossRef]
- Bleicken S, Landeta O, Landajuela A, Basañez G, García-Sáez AJ. Proapoptotic Bax and Bak Proteins Form Stable Protein-permeable Pores of Tunable Size. Journal of Biological Chemistry. 2013;288(46):33241-52. [CrossRef]
- Colombini M. Ceramide channels and mitochondrial outer membrane permeability. Journal of bioenergetics and biomembranes. 2017;49(1):57-64. [CrossRef]
- Ueda N. Ceramide-induced apoptosis in renal tubular cells: a role of mitochondria and sphingosine-1-phoshate. International journal of molecular sciences. 2015;16(3):5076-124. [CrossRef]
- Colombini M. Ceramide Channels. Advances in experimental medicine and biology. 2019;1159:33-48.
- Yang Y, Xu G, Xu Y, Cheng X, Xu S, Chen S, et al. Ceramide mediates radiation-induced germ cell apoptosis via regulating mitochondria function and MAPK factors in Caenorhabditis elegans. Ecotoxicology and Environmental Safety. 2021;208:111579. [CrossRef]
- Zhang QF, Li J, Bi FC, Liu Z, Chang ZY, Wang LY, et al. Ceramide-Induced Cell Death Depends on Calcium and Caspase-Like Activity in Rice. Frontiers in plant science. 2020;11:145. [CrossRef]
- Elsherbini A, Kirov AS, Dinkins MB, Wang G, Qin H, Zhu Z, et al. Association of Aβ with ceramide-enriched astrosomes mediates Aβ neurotoxicity. Acta neuropathologica communications. 2020;8(1):60. [CrossRef]
- Colombini M. Ceramide channels and their role in mitochondria-mediated apoptosis. Biochimica et Biophysica Acta (BBA) - Bioenergetics. 2010;1797(6):1239-44. [CrossRef]
- Parry N, Wheadon H, Copland M. The application of BH3 mimetics in myeloid leukemias. Cell death & disease. 2021;12(2):222. [CrossRef]
- Dadsena S, Zollo C, García-Sáez AJ. Mechanisms of mitochondrial cell death. Biochemical Society transactions. 2021;49(2):663-74. [CrossRef]
- Siskind LJ, Davoody A, Lewin N, Marshall S, Colombini M. Enlargement and Contracture of C2-Ceramide Channels. Biophysical Journal. 2003;85(3):1560-75. [CrossRef]
- Anishkin A, Sukharev S, Colombini M. Searching for the Molecular Arrangement of Transmembrane Ceramide Channels. Biophysical Journal. 2006;90(7):2414-26. [CrossRef]
- Fisher-Wellman KH, Hagen JT, Neufer PD, Kassai M, Cabot MC. On the nature of ceramide-mitochondria interactions - Dissection using comprehensive mitochondrial phenotyping. Cellular signalling. 2021;78:109838. [CrossRef]
- Perera MN, Ganesan V, Siskind LJ, Szulc ZM, Bielawski J, Bielawska A, et al. Ceramide channels: Influence of molecular structure on channel formation in membranes. Biochimica et Biophysica Acta (BBA) - Biomembranes. 2012;1818(5):1291-301. [CrossRef]
- Stiban J, Fistere D, Colombini M. Dihydroceramide hinders ceramide channel formation: Implications on apoptosis. Apoptosis. 2006;11(5):773-80. [CrossRef]
- Pinto SN, Laviad EL, Stiban J, Kelly SL, Merrill AH, Prieto M, et al. Changes in membrane biophysical properties induced by sphingomyelinase depend on the sphingolipid N-acyl chain. Journal of lipid research. 2014;55(1):53-61. [CrossRef]
- Pinto SN, Silva LC, Futerman AH, Prieto M. Effect of ceramide structure on membrane biophysical properties: The role of acyl chain length and unsaturation. Biochimica et Biophysica Acta (BBA) - Biomembranes. 2011;1808(11):2753-60. [CrossRef]
- Silva LC, Ben David O, Pewzner-Jung Y, Laviad EL, Stiban J, Bandyopadhyay S, et al. Ablation of ceramide synthase 2 strongly affects biophysical properties of membranes. Journal of lipid research. 2012;53(3):430-6. [CrossRef]
- Siskind LJ, Kolesnick RN, Colombini M. Ceramide Channels Increase the Permeability of the Mitochondrial Outer Membrane to Small Proteins*. Journal of Biological Chemistry. 2002;277(30):26796-803. [CrossRef]
- Stiban J, Perera M. Very long chain ceramides interfere with C16-ceramide-induced channel formation: A plausible mechanism for regulating the initiation of intrinsic apoptosis. Biochimica et biophysica acta. 2015;1848(2):561-7. [CrossRef]
- Chong SJF, Iskandar K, Lai JXH, Qu J, Raman D, Valentin R, et al. Serine-70 phosphorylated Bcl-2 prevents oxidative stress-induced DNA damage by modulating the mitochondrial redox metabolism. Nucleic acids research. 2020;48(22):12727-45. [CrossRef]
- Oaks J, Ogretmen B. Regulation of PP2A by Sphingolipid Metabolism and Signaling. Frontiers in oncology. 2014;4:388. [CrossRef]
- Deng X, Gao F, May WS. Protein phosphatase 2A inactivates Bcl2's antiapoptotic function by dephosphorylation and up-regulation of Bcl2-p53 binding. Blood. 2009;113(2):422-8. [CrossRef]
- Chang K-T, Anishkin A, Patwardhan GA, Beverly LJ, Siskind LJ, Colombini M. Ceramide channels: destabilization by Bcl-xL and role in apoptosis. Biochimica et biophysica acta. 2015;1848(10 Pt A):2374-84.
- Siskind LJ, Feinstein L, Yu T, Davis JS, Jones D, Choi J, et al. Anti-apoptotic Bcl-2 Family Proteins Disassemble Ceramide Channels*. Journal of Biological Chemistry. 2008;283(11):6622-30. [CrossRef]
- Chang KT, Anishkin A, Patwardhan GA, Beverly LJ, Siskind LJ, Colombini M. Ceramide channels: destabilization by Bcl-xL and role in apoptosis. Biochimica et biophysica acta. 2015;1848(10 Pt A):2374-84.
- Peña-Blanco A, García-Sáez AJ. Bax, Bak and beyond — mitochondrial performance in apoptosis. The FEBS Journal. 2018;285(3):416-31.
- Perera MN, Lin SH, Peterson YK, Bielawska A, Szulc ZM, Bittman R, et al. Bax and Bcl-xL exert their regulation on different sites of the ceramide channel. The Biochemical journal. 2012;445(1):81-91. [CrossRef]
- Siskind LJ, Mullen TD, Romero Rosales K, Clarke CJ, Hernandez-Corbacho MJ, Edinger AL, et al. The BCL-2 Protein BAK Is Required for Long-chain Ceramide Generation during Apoptosis*. Journal of Biological Chemistry. 2010;285(16):11818-26. [CrossRef]
- Ganesan V, Perera MN, Colombini D, Datskovskiy D, Chadha K, Colombini M. Ceramide and activated Bax act synergistically to permeabilize the mitochondrial outer membrane. Apoptosis. 2010;15(5):553-62. [CrossRef]
- Lee H, Rotolo JA, Mesicek J, Penate-Medina T, Rimner A, Liao W-C, et al. Mitochondrial Ceramide-Rich Macrodomains Functionalize Bax upon Irradiation. PLOS ONE. 2011;6(6):e19783. [CrossRef]
- Kim HJ, Mun JY, Chun YJ, Choi KH, Kim MY. Bax-dependent apoptosis induced by ceramide in HL-60 cells. FEBS letters. 2001;505(2):264-8.
- Dadsena S, Bockelmann S, Mina JGM, Hassan DG, Korneev S, Razzera G, et al. Ceramides bind VDAC2 to trigger mitochondrial apoptosis. Nature Communications. 2019;10(1):1832. [CrossRef]
- von Haefen C, Wieder T, Gillissen B, Stärck L, Graupner V, Dörken B, et al. Ceramide induces mitochondrial activation and apoptosis via a Bax-dependent pathway in human carcinoma cells. Oncogene. 2002;21(25):4009-19. [CrossRef]
- Bravo R, Parra V, Gatica D, Rodriguez AE, Torrealba N, Paredes F, et al. Endoplasmic reticulum and the unfolded protein response: dynamics and metabolic integration. Int Rev Cell Mol Biol. 2013;301:215-90.
- Szegezdi E, Logue SE, Gorman AM, Samali A. Mediators of endoplasmic reticulum stress-induced apoptosis. EMBO reports. 2006;7(9):880-5. [CrossRef]
- Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nature reviews Molecular cell biology. 2007;8(7):519-29. [CrossRef]
- Turishcheva E, Vildanova M, Onishchenko G, Smirnova E. The Role of Endoplasmic Reticulum Stress in Differentiation of Cells of Mesenchymal Origin. Biochemistry Biokhimiia. 2022;87(9):916-31. [CrossRef]
- Strzyz P. Reacting to membrane stress. Nature Reviews Molecular Cell Biology. 2017;18(8):471-.
- Stiban J, Caputo L, Colombini M. Ceramide synthesis in the endoplasmic reticulum can permeabilize mitochondria to proapoptotic proteins. Journal of lipid research. 2008;49(3):625-34. [CrossRef]
- Senkal CE, Ponnusamy S, Bielawski J, Hannun YA, Ogretmen B. Antiapoptotic roles of ceramide-synthase-6-generated C16-ceramide via selective regulation of the ATF6/CHOP arm of ER-stress-response pathways. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2010;24(1):296-308.
- Senkal CE, Ponnusamy S, Manevich Y, Meyers-Needham M, Saddoughi SA, Mukhopadyay A, et al. Alteration of ceramide synthase 6/C16-ceramide induces activating transcription factor 6-mediated endoplasmic reticulum (ER) stress and apoptosis via perturbation of cellular Ca2+ and ER/Golgi membrane network. The Journal of biological chemistry. 2011;286(49):42446-58.
- Liu Z, Xia Y, Li B, Xu H, Wang C, Liu Y, et al. Induction of ER stress-mediated apoptosis by ceramide via disruption of ER Ca(2+) homeostasis in human adenoid cystic carcinoma cells. Cell Biosci. 2014;4:71-. [CrossRef]
- Schiffmann S, Sandner J, Schmidt R, Birod K, Wobst I, Schmidt H, et al. The selective COX-2 inhibitor celecoxib modulates sphingolipid synthesiss⃞. Journal of lipid research. 2009;50(1):32-40.
- Schiffmann S, Ziebell S, Sandner J, Birod K, Deckmann K, Hartmann D, et al. Activation of ceramide synthase 6 by celecoxib leads to a selective induction of C16:0-ceramide. Biochemical Pharmacology. 2010;80(11):1632-40. [CrossRef]
- Mesicek J, Lee H, Feldman T, Jiang X, Skobeleva A, Berdyshev EV, et al. Ceramide synthases 2, 5, and 6 confer distinct roles in radiation-induced apoptosis in HeLa cells. Cellular signalling. 2010;22(9):1300-7. [CrossRef]
- Senkal CE, Ponnusamy S, Rossi MJ, Bialewski J, Sinha D, Jiang JC, et al. Role of human longevity assurance gene 1 and C18-ceramide in chemotherapy-induced cell death in human head and neck squamous cell carcinomas. Molecular cancer therapeutics. 2007;6(2):712-22. [CrossRef]
- Baran Y, Salas A, Senkal CE, Gunduz U, Bielawski J, Obeid LM, et al. Alterations of ceramide/sphingosine 1-phosphate rheostat involved in the regulation of resistance to imatinib-induced apoptosis in K562 human chronic myeloid leukemia cells. The Journal of biological chemistry. 2007;282(15):10922-34. [CrossRef]
- Jin J, Hou Q, Mullen TD, Zeidan YH, Bielawski J, Kraveka JM, et al. Ceramide generated by sphingomyelin hydrolysis and the salvage pathway is involved in hypoxia/reoxygenation-induced Bax redistribution to mitochondria in NT-2 cells. The Journal of biological chemistry. 2008;283(39):26509-17. [CrossRef]
- Peter ME, Hadji A, Murmann AE, Brockway S, Putzbach W, Pattanayak A, et al. The role of CD95 and CD95 ligand in cancer. Cell Death & Differentiation. 2015;22(4):549-59.
- Walker T, Mitchell C, Park MA, Yacoub A, Graf M, Rahmani M, et al. Sorafenib and vorinostat kill colon cancer cells by CD95-dependent and -independent mechanisms. Mol Pharmacol. 2009;76(2):342-55. [CrossRef]
- Singh D, Tewari M, Singh S, Narayan G. Revisiting the role of TRAIL/TRAIL-R in cancer biology and therapy. Future oncology (London, England). 2021;17(5):581-96. [CrossRef]
- Razeghian E, Suksatan W, Sulaiman Rahman H, Bokov DO, Abdelbasset WK, Hassanzadeh A, et al. Harnessing TRAIL-Induced Apoptosis Pathway for Cancer Immunotherapy and Associated Challenges. Frontiers in immunology. 2021;12:699746. [CrossRef]
- Hassanzadeh A, Farshdousti Hagh M, Alivand MR, Akbari AAM, Shams Asenjan K, Saraei R, et al. Down-regulation of intracellular anti-apoptotic proteins, particularly c-FLIP by therapeutic agents; the novel view to overcome resistance to TRAIL. Journal of cellular physiology. 2018;233(10):6470-85.
- White-Gilbertson S, Mullen T, Senkal C, Lu P, Ogretmen B, Obeid L, et al. Ceramide synthase 6 modulates TRAIL sensitivity and nuclear translocation of active caspase-3 in colon cancer cells. Oncogene. 2009;28(8):1132-41. [CrossRef]
- Zhang J, Lou W. A Key mRNA-miRNA-lncRNA Competing Endogenous RNA Triple Sub-network Linked to Diagnosis and Prognosis of Hepatocellular Carcinoma. Frontiers in oncology. 2020;10.
- Pan W, Chen KJ, Huang YC. Ceramide synthase 6 antisense RNA 1 contributes to the progression of breast cancer by sponging miR-16-5p to upregulate ubiquitin-conjugating enzyme E2C. Anti-cancer drugs. 2022;33(9):913-22. [CrossRef]
- Chin HS, Li MX, Tan IKL, Ninnis RL, Reljic B, Scicluna K, et al. VDAC2 enables BAX to mediate apoptosis and limit tumor development. Nature Communications. 2018;9(1):4976. [CrossRef]
- Shi Y, Jin Y, Liu F, Jiang J, Cao J, Lu Y, et al. Ceramide induces the apoptosis of non-small cell lung cancer cells through the Txnip/Trx1 complex. International journal of molecular medicine. 2021;47(5). [CrossRef]
- Wang Y, Yang J, Wang Y, Chen Y, Wang Y, Kuang H, et al. Upregulation of TXNIP contributes to granulosa cell dysfunction in polycystic ovary syndrome via activation of the NLRP3 inflammasome. Molecular and cellular endocrinology. 2022:111824.
- Suzuki M, Cao K, Kato S, Mizutani N, Tanaka K, Arima C, et al. CERS6 required for cell migration and metastasis in lung cancer. Journal of cellular and molecular medicine. 2020;24(20):11949-59. [CrossRef]
- Tang Y, Cao K, Wang Q, Chen J, Liu R, Wang S, et al. Silencing of CerS6 increases the invasion and glycolysis of melanoma WM35, WM451 and SK28 cell lines via increased GLUT1-induced downregulation of WNT5A. Oncol Rep. 2016;35(5):2907-15. [CrossRef]
- Shi Y, Zhou C, Lu H, Cui X, Li J, Jiang S, et al. Ceramide synthase 6 predicts poor prognosis and activates the AKT/mTOR/4EBP1 pathway in high-grade serous ovarian cancer. American journal of translational research. 2020;12(9):5924-39.
- Qiu L, Liu Z, Wu C, Chen W, Chen Y, Zhang B, et al. C6-ceramide induces salivary adenoid cystic carcinoma cell apoptosis via IP3R-activated UPR and UPR-independent pathways. Biochemical and biophysical research communications. 2020;525(4):997-1003. [CrossRef]
- Hewitt G, Korolchuk VI. Repair, Reuse, Recycle: The Expanding Role of Autophagy in Genome Maintenance. Trends in Cell Biology. 2017;27(5):340-51.
- Parzych KR, Klionsky DJ. An overview of autophagy: morphology, mechanism, and regulation. Antioxidants & redox signaling. 2014;20(3):460-73. [CrossRef]
- Chun Y, Kim J. Autophagy: An Essential Degradation Program for Cellular Homeostasis and Life. Cells. 2018;7(12). [CrossRef]
- Sharma P, Alizadeh J, Juarez M, Samali A, Halayko AJ, Kenyon NJ, et al. Autophagy, Apoptosis, the Unfolded Protein Response, and Lung Function in Idiopathic Pulmonary Fibrosis. Cells. 2021;10(7). [CrossRef]
- Yeganeh B, Rezaei Moghadam A, Alizadeh J, Wiechec E, Alavian SM, Hashemi M, et al. Hepatitis B and C virus-induced hepatitis: Apoptosis, autophagy, and unfolded protein response. World J Gastroenterol. 2015;21(47):13225-39. [CrossRef]
- Yang Z, Klionsky DJ. Eaten alive: a history of macroautophagy. Nature cell biology. 2010;12(9):814-22. [CrossRef]
- Klionsky DJ. The molecular machinery of autophagy: unanswered questions. Journal of cell science. 2005;118(Pt 1):7-18. [CrossRef]
- Singh R, Cuervo AM. Autophagy in the cellular energetic balance. Cell metabolism. 2011;13(5):495-504. [CrossRef]
- Rashid MU, Lorzadeh S, Gao A, Ghavami S, Coombs KM. PSMA2 knockdown impacts expression of proteins involved in immune and cellular stress responses in human lung cells. Biochim Biophys Acta Mol Basis Dis. 2023;1869(2):166617. [CrossRef]
- Dalvand A, da Silva Rosa SC, Ghavami S, Marzban H. Potential role of TGFBeta and autophagy in early crebellum development. Biochem Biophys Rep. 2022;32:101358.
- Ghavami S, Zamani M, Ahmadi M, Erfani M, Dastghaib S, Darbandi M, et al. Epigenetic regulation of autophagy in gastrointestinal cancers. Biochim Biophys Acta Mol Basis Dis. 2022;1868(11):166512. [CrossRef]
- Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132(1):27-42.
- Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451(7182):1069-75.
- Choi AM, Ryter SW, Levine B. Autophagy in human health and disease. The New England journal of medicine. 2013;368(7):651-62.
- Eshraghi M, Ahmadi M, Afshar S, Lorzadeh S, Adlimoghaddam A, Rezvani Jalal N, et al. Enhancing autophagy in Alzheimer's disease through drug repositioning. Pharmacol Ther. 2022;237:108171.
- Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122(6):927-39.
- Qu X, Zou Z, Sun Q, Luby-Phelps K, Cheng P, Hogan RN, et al. Autophagy gene-dependent clearance of apoptotic cells during embryonic development. Cell. 2007;128(5):931-46. [CrossRef]
- Alizadeh J, Kochan MM, Stewart VD, Drewnik DA, Hannila SS, Ghavami S. Inhibition of Autophagy Flux Promotes Secretion of Chondroitin Sulfate Proteoglycans in Primary Rat Astrocytes. Mol Neurobiol. 2021;58(12):6077-91. [CrossRef]
- Young MM, Kester M, Wang HG. Sphingolipids: regulators of crosstalk between apoptosis and autophagy. Journal of lipid research. 2013;54(1):5-19. [CrossRef]
- Schubert KM, Scheid MP, Duronio V. Ceramide inhibits protein kinase B/Akt by promoting dephosphorylation of serine 473. The Journal of biological chemistry. 2000;275(18):13330-5. [CrossRef]
- Scarlatti F, Bauvy C, Ventruti A, Sala G, Cluzeaud F, Vandewalle A, et al. Ceramide-mediated macroautophagy involves inhibition of protein kinase B and up-regulation of beclin 1. The Journal of biological chemistry. 2004;279(18):18384-91. [CrossRef]
- Guenther GG, Peralta ER, Rosales KR, Wong SY, Siskind LJ, Edinger AL. Ceramide starves cells to death by downregulating nutrient transporter proteins. Proc Natl Acad Sci U S A. 2008;105(45):17402-7.
- Kang R, Zeh HJ, Lotze MT, Tang D. The Beclin 1 network regulates autophagy and apoptosis. Cell death and differentiation. 2011;18(4):571-80. [CrossRef]
- Li DD, Wang LL, Deng R, Tang J, Shen Y, Guo JF, et al. The pivotal role of c-Jun NH2-terminal kinase-mediated Beclin 1 expression during anticancer agents-induced autophagy in cancer cells. Oncogene. 2009;28(6):886-98. [CrossRef]
- Bedia C, Levade T, Codogno P. Regulation of autophagy by sphingolipids. Anti-cancer agents in medicinal chemistry. 2011;11(9):844-53.
- Kravchenko G, Krasilnikova O. Jnk Activation Is Accompanied By Ceramides And Diacylglycerol Accumulation In Isolated Hepatocytes. Atherosclerosis. 2019;287:e234. [CrossRef]
- Yabu T, Shiba H, Shibasaki Y, Nakanishi T, Imamura S, Touhata K, et al. Stress-induced ceramide generation and apoptosis via the phosphorylation and activation of nSMase1 by JNK signaling. Cell Death & Differentiation. 2015;22(2):258-73. [CrossRef]
- Mammucari C, Milan G, Romanello V, Masiero E, Rudolf R, Del Piccolo P, et al. FoxO3 controls autophagy in skeletal muscle in vivo. Cell metabolism. 2007;6(6):458-71. [CrossRef]
- Zhang H, Bosch-Marce M, Shimoda LA, Tan YS, Baek JH, Wesley JB, et al. Mitochondrial autophagy is an HIF-1-dependent adaptive metabolic response to hypoxia. The Journal of biological chemistry. 2008;283(16):10892-903. [CrossRef]
- Spassieva SD, Mullen TD, Townsend DM, Obeid LM. Disruption of ceramide synthesis by CerS2 down-regulation leads to autophagy and the unfolded protein response. The Biochemical journal. 2009;424(2):273-83. [CrossRef]
- Loos B, du Toit A, Hofmeyr J-HS. Defining and measuring autophagosome flux—concept and reality. Autophagy. 2014;10(11):2087-96. [CrossRef]
- Hetz C, Zhang K, Kaufman RJ. Mechanisms, regulation and functions of the unfolded protein response. Nature reviews Molecular cell biology. 2020;21(8):421-38.
- Senft D, Ronai ZA. UPR, autophagy, and mitochondria crosstalk underlies the ER stress response. Trends in biochemical sciences. 2015;40(3):141-8. [CrossRef]
- Liu C, Yan DY, Wang C, Ma Z, Deng Y, Liu W, et al. IRE1 signaling pathway mediates protective autophagic response against manganese-induced neuronal apoptosis in vivo and in vitro. The Science of the total environment. 2020;712:136480. [CrossRef]
- Lindner P, Christensen SB, Nissen P, Møller JV, Engedal N. Cell death induced by the ER stressor thapsigargin involves death receptor 5, a non-autophagic function of MAP1LC3B, and distinct contributions from unfolded protein response components. Cell Communication and Signaling. 2020;18(1):12. [CrossRef]
- Zhou Y, Manghwar H, Hu W, Liu F. Degradation Mechanism of Autophagy-Related Proteins and Research Progress. International journal of molecular sciences. 2022;23(13). [CrossRef]
- Trojani MC, Santucci-Darmanin S, Breuil V, Carle GF, Pierrefite-Carle V. Autophagy and bone diseases. Joint bone spine. 2022;89(3):105301. [CrossRef]
- Khan A, Singh VK, Thakral D, Gupta R. Autophagy in acute myeloid leukemia: a paradoxical role in chemoresistance. Clinical & translational oncology : official publication of the Federation of Spanish Oncology Societies and of the National Cancer Institute of Mexico. 2022;24(8):1459-69.
- Gómez-Virgilio L, Silva-Lucero MD, Flores-Morelos DS, Gallardo-Nieto J, Lopez-Toledo G, Abarca-Fernandez AM, et al. Autophagy: A Key Regulator of Homeostasis and Disease: An Overview of Molecular Mechanisms and Modulators. Cells. 2022;11(15). [CrossRef]
- Aman Y, Schmauck-Medina T, Hansen M, Morimoto RI, Simon AK, Bjedov I, et al. Autophagy in healthy aging and disease. Nature Aging. 2021;1(8):634-50. [CrossRef]
- Ge Y, Zhou M, Chen C, Wu X, Wang X. Role of AMPK mediated pathways in autophagy and aging. Biochimie. 2022;195:100-13. [CrossRef]
- Habibzadeh P, Dastsooz H, Eshraghi M, Los MJ, Klionsky DJ, Ghavami S. Autophagy: The Potential Link between SARS-CoV-2 and Cancer. Cancers (Basel). 2021;13(22).
- Lum JJ, DeBerardinis RJ, Thompson CB. Autophagy in metazoans: cell survival in the land of plenty. Nature reviews Molecular cell biology. 2005;6(6):439-48. [CrossRef]
- Siri M, Behrouj H, Dastghaib S, Zamani M, Likus W, Rezaie S, et al. Casein Kinase-1-Alpha Inhibitor (D4476) Sensitizes Microsatellite Instable Colorectal Cancer Cells to 5-Fluorouracil via Authophagy Flux Inhibition. Arch Immunol Ther Exp (Warsz). 2021;69(1):26. [CrossRef]
- Galluzzi L, Vitale I, Aaronson SA, Abrams JM, Adam D, Agostinis P, et al. Molecular mechanisms of cell death: recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death & Differentiation. 2018;25(3):486-541.
- Brun P, Tarricone E, Di Stefano A, Mattiuzzo E, Mehrbod P, Ghavami S, et al. The regulatory activity of autophagy in conjunctival fibroblasts and its possible role in vernal keratoconjunctivitis. J Allergy Clin Immunol. 2020;146(5):1210-3 e9. [CrossRef]
- Yang Y, Klionsky DJ. Autophagy and disease: unanswered questions. Cell death and differentiation. 2020;27(3):858-71. [CrossRef]
- Klionsky DJ, Petroni G, Amaravadi RK, Baehrecke EH, Ballabio A, Boya P, et al. Autophagy in major human diseases. The EMBO Journal. 2021;40(19):e108863. [CrossRef]
- Lei Y, Klionsky DJ. The Emerging Roles of Autophagy in Human Diseases. Biomedicines. 2021;9(11). [CrossRef]
- Codogno P, Meijer AJ. Autophagy and signaling: their role in cell survival and cell death. Cell Death & Differentiation. 2005;12(2):1509-18. [CrossRef]
- Young MM, Kester M, Wang H-G. Sphingolipids: regulators of crosstalk between apoptosis and autophagy. Journal of lipid research. 2013;54(1):5-19. [CrossRef]
- Pattingre S, Bauvy C, Levade T, Levine B, Codogno P. Ceramide-induced autophagy: To junk or to protect cells? Autophagy. 2009;5(4):558-60. [CrossRef]
- Pattingre S, Bauvy C, Carpentier S, Levade T, Levine B, Codogno P. Role of JNK1-dependent Bcl-2 phosphorylation in ceramide-induced macroautophagy. The Journal of biological chemistry. 2009;284(5):2719-28. [CrossRef]
- Moruno Manchon JF, Uzor N-E, Dabaghian Y, Furr-Stimming EE, Finkbeiner S, Tsvetkov AS. Cytoplasmic sphingosine-1-phosphate pathway modulates neuronal autophagy. Scientific Reports. 2015;5(1):15213. [CrossRef]
- Matarrese P, Garofalo T, Manganelli V, Gambardella L, Marconi M, Grasso M, et al. Evidence for the involvement of GD3 ganglioside in autophagosome formation and maturation. Autophagy. 2014;10(5):750-65. [CrossRef]
- Garofalo T, Matarrese P, Manganelli V, Marconi M, Tinari A, Gambardella L, et al. Evidence for the involvement of lipid rafts localized at the ER-mitochondria associated membranes in autophagosome formation. Autophagy. 2016;12(6):917-35.
- Yamamoto H, Kakuta S, Watanabe TM, Kitamura A, Sekito T, Kondo-Kakuta C, et al. Atg9 vesicles are an important membrane source during early steps of autophagosome formation. Journal of Cell Biology. 2012;198(2):219-33. [CrossRef]
- Hinkovska-Galcheva V, Boxer LA, Kindzelskii A, Hiraoka M, Abe A, Goparju S, et al. Ceramide 1-Phosphate, a Mediator of Phagocytosis *. Journal of Biological Chemistry. 2005;280(28):26612-21.
- Mishra SK, Gao Y-G, Deng Y, Chalfant CE, Hinchcliffe EH, Brown RE. CPTP: A sphingolipid transfer protein that regulates autophagy and inflammasome activation. Autophagy. 2018;14(5):862-79. [CrossRef]
- Shen H, Giordano F, Wu Y, Chan J, Zhu C, Milosevic I, et al. Coupling between endocytosis and sphingosine kinase 1 recruitment. Nature cell biology. 2014;16(7):652-62. [CrossRef]
- Longatti A, Lamb CA, Razi M, Yoshimura S, Barr FA, Tooze SA. TBC1D14 regulates autophagosome formation via Rab11- and ULK1-positive recycling endosomes. The Journal of cell biology. 2012;197(5):659-75. [CrossRef]
- Szatmári Z, Kis V, Lippai M, Hegedus K, Faragó T, Lorincz P, et al. Rab11 facilitates cross-talk between autophagy and endosomal pathway through regulation of Hook localization. Molecular biology of the cell. 2014;25(4):522-31. [CrossRef]
- Dany M, Gencer S, Nganga R, Thomas RJ, Oleinik N, Baron KD, et al. Targeting FLT3-ITD signaling mediates ceramide-dependent mitophagy and attenuates drug resistance in AML. Blood. 2016;128(15):1944-58. [CrossRef]
- Mizumura K, Justice MJ, Schweitzer KS, Krishnan S, Bronova I, Berdyshev EV, et al. Sphingolipid regulation of lung epithelial cell mitophagy and necroptosis during cigarette smoke exposure. The FASEB Journal. 2018;32(4):1880-90. [CrossRef]
- Sentelle RD, Senkal CE, Jiang W, Ponnusamy S, Gencer S, Panneer Selvam S, et al. Ceramide targets autophagosomes to mitochondria and induces lethal mitophagy. Nature Chemical Biology. 2012;8(10):831-8.
- Hernandez-Diaz S, Soukup S-F. The role of lipids in autophagy and its implication in neurodegeneration. Cell Stress. 2020;4(7):167-86. [CrossRef]
- Russo SB, Baicu CF, Van Laer A, Geng T, Kasiganesan H, Zile MR, et al. Ceramide synthase 5 mediates lipid-induced autophagy and hypertrophy in cardiomyocytes. J Clin Invest. 2012;122(11):3919-30. [CrossRef]
- Koga H, Kaushik S, Cuervo AM. Altered lipid content inhibits autophagic vesicular fusion. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2010;24(8):3052-65.
- Jana A, Hogan EL, Pahan K. Ceramide and neurodegeneration: susceptibility of neurons and oligodendrocytes to cell damage and death. J Neurol Sci. 2009;278(1-2):5-15. [CrossRef]
- Pattingre S, Bauvy C, Levade T, Levine B, Codogno P. Ceramide-induced autophagy: to junk or to protect cells? Autophagy. 2009;5(4):558-60. [CrossRef]
- Edinger AL. Starvation in the midst of plenty: making sense of ceramide-induced autophagy by analysing nutrient transporter expression. Biochemical Society transactions. 2009;37(Pt 1):253-8. [CrossRef]
- Guenther GG, Edinger AL. A new take on ceramide: Starving cells by cutting off the nutrient supply. Cell Cycle. 2009;8(8):1122-6. [CrossRef]
- Park MA, Zhang G, Martin AP, Hamed H, Mitchell C, Hylemon PB, et al. Vorinostat and sorafenib increase ER stress, autophagy and apoptosis via ceramide-dependent CD95 and PERK activation. Cancer biology & therapy. 2008;7(10):1648-62. [CrossRef]
- Gagliostro V, Casas J, Caretti A, Abad JL, Tagliavacca L, Ghidoni R, et al. Dihydroceramide delays cell cycle G1/S transition via activation of ER stress and induction of autophagy. The international journal of biochemistry & cell biology. 2012;44(12):2135-43.
- Signorelli P, Munoz-Olaya JM, Gagliostro V, Casas J, Ghidoni R, Fabriàs G. Dihydroceramide intracellular increase in response to resveratrol treatment mediates autophagy in gastric cancer cells. Cancer letters. 2009;282(2):238-43. [CrossRef]
- Devlin CM, Lahm T, Hubbard WC, Van Demark M, Wang KC, Wu X, et al. Dihydroceramide-based response to hypoxia. The Journal of biological chemistry. 2011;286(44):38069-78. [CrossRef]
- Huang S, Sinicrope FA. Celecoxib-induced apoptosis is enhanced by ABT-737 and by inhibition of autophagy in human colorectal cancer cells. Autophagy. 2010;6(2):256-69. [CrossRef]
- Wu H, Che X, Zheng Q, Wu A, Pan K, Shao A, et al. Caspases: a molecular switch node in the crosstalk between autophagy and apoptosis. International journal of biological sciences. 2014;10(9):1072-83. [CrossRef]
- Matsuzawa Y, Oshima S, Nibe Y, Kobayashi M, Maeyashiki C, Nemoto Y, et al. RIPK3 regulates p62-LC3 complex formation via the caspase-8-dependent cleavage of p62. Biochemical and biophysical research communications. 2015;456(1):298-304. [CrossRef]
- Pagliarini V, Wirawan E, Romagnoli A, Ciccosanti F, Lisi G, Lippens S, et al. Proteolysis of Ambra1 during apoptosis has a role in the inhibition of the autophagic pro-survival response. Cell death and differentiation. 2012;19(9):1495-504. [CrossRef]
- You M, Savaraj N, Kuo MT, Wangpaichitr M, Varona-Santos J, Wu C, et al. TRAIL induces autophagic protein cleavage through caspase activation in melanoma cell lines under arginine deprivation. Mol Cell Biochem. 2013;374(1-2):181-90. [CrossRef]
- Wirawan E, Vande Walle L, Kersse K, Cornelis S, Claerhout S, Vanoverberghe I, et al. Caspase-mediated cleavage of Beclin-1 inactivates Beclin-1-induced autophagy and enhances apoptosis by promoting the release of proapoptotic factors from mitochondria. Cell death & disease. 2010;1(1):e18. [CrossRef]
- Oral O, Oz-Arslan D, Itah Z, Naghavi A, Deveci R, Karacali S, et al. Cleavage of Atg3 protein by caspase-8 regulates autophagy during receptor-activated cell death. Apoptosis. 2012;17(8):810-20. [CrossRef]
- Jeong HS, Choi HY, Lee ER, Kim JH, Jeon K, Lee HJ, et al. Involvement of caspase-9 in autophagy-mediated cell survival pathway. Biochimica et biophysica acta. 2011;1813(1):80-90. [CrossRef]
- Ghassan SD, Youmna K, Nadine D, Shoghag P, Lina K, Rihab N, et al. Arsenic trioxide induces accumulation of cytotoxic levels of ceramide in acute promyelocytic leukemia and adult T-cell leukemia/lymphoma cells through de novo ceramide synthesis and inhibition of glucosylceramide synthase activity. Haematologica. 2007;92(6):753-62.
- Salazar M, Carracedo A, Salanueva IJ, Hernández-Tiedra S, Lorente M, Egia A, et al. Cannabinoid action induces autophagy-mediated cell death through stimulation of ER stress in human glioma cells. J Clin Invest. 2009;119(5):1359-72. [CrossRef]
- Ding W-X, Yin X-M. Mitophagy: mechanisms, pathophysiological roles, and analysis. Biol Chem. 2012;393(7):547-64.
- Levine B, Klionsky DJ. Development by Self-Digestion: Molecular Mechanisms and Biological Functions of Autophagy. Developmental Cell. 2004;6(4):463-77. [CrossRef]
- Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451(7182):1069-75.
- Sentelle RD, Senkal CE, Jiang W, Ponnusamy S, Gencer S, Selvam SP, et al. Ceramide targets autophagosomes to mitochondria and induces lethal mitophagy. Nat Chem Biol. 2012;8(10):831-8.
- Daido S, Kanzawa T, Yamamoto A, Takeuchi H, Kondo Y, Kondo S. Pivotal role of the cell death factor BNIP3 in ceramide-induced autophagic cell death in malignant glioma cells. Cancer research. 2004;64(12):4286-93. [CrossRef]
- Levine B, Sinha S, Kroemer G. Bcl-2 family members: dual regulators of apoptosis and autophagy. Autophagy. 2008;4(5):600-6.
- Dbaibo GS, Kfoury Y, Darwiche N, Panjarian S, Kozhaya L, Nasr R, et al. Arsenic trioxide induces accumulation of cytotoxic levels of ceramide in acute promyelocytic leukemia and adult T-cell leukemia/lymphoma cells through de novo ceramide synthesis and inhibition of glucosylceramide synthase activity. Haematologica. 2007;92(6):753-62. [CrossRef]
- Qian W, Liu J, Jin J, Ni W, Xu W. Arsenic trioxide induces not only apoptosis but also autophagic cell death in leukemia cell lines via up-regulation of Beclin-1. Leukemia research. 2007;31(3):329-39. [CrossRef]
- Taniguchi M, Kitatani K, Kondo T, Hashimoto-Nishimura M, Asano S, Hayashi A, et al. Regulation of autophagy and its associated cell death by "sphingolipid rheostat": reciprocal role of ceramide and sphingosine 1-phosphate in the mammalian target of rapamycin pathway. The Journal of biological chemistry. 2012;287(47):39898-910.
- Wang Z, Wen L, Zhu F, Wang Y, Xie Q, Chen Z, et al. Overexpression of ceramide synthase 1 increases C18-ceramide and leads to lethal autophagy in human glioma. Oncotarget. 2017;8(61):104022-36. [CrossRef]
- Hartmann D, Lucks J, Fuchs S, Schiffmann S, Schreiber Y, Ferreirós N, et al. Long chain ceramides and very long chain ceramides have opposite effects on human breast and colon cancer cell growth. The international journal of biochemistry & cell biology. 2012;44(4):620-8. [CrossRef]
- Fitzgerald S, Sheehan KM, Espina V, O'Grady A, Cummins R, Kenny D, et al. High CerS5 expression levels associate with reduced patient survival and transition from apoptotic to autophagy signalling pathways in colorectal cancer. J Pathol Clin Res. 2014;1(1):54-65. [CrossRef]
- Taniguchi M, Kitatani K, Kondo T, Hashimoto-Nishimura M, Asano S, Hayashi A, et al. Regulation of autophagy and its associated cell death by "sphingolipid rheostat": reciprocal role of ceramide and sphingosine 1-phosphate in the mammalian target of rapamycin pathway. The Journal of biological chemistry. 2012;287(47):39898-910.
- Hou Q, Jin J, Zhou H, Novgorodov SA, Bielawska A, Szulc ZM, et al. Mitochondrially targeted ceramides preferentially promote autophagy, retard cell growth, and induce apoptosis. Journal of lipid research. 2011;52(2):278-88. [CrossRef]
- Lima S, Takabe K, Newton J, Saurabh K, Young MM, Leopoldino AM, et al. TP53 is required for BECN1- and ATG5-dependent cell death induced by sphingosine kinase 1 inhibition. Autophagy. 2018;14(6):942-57. [CrossRef]
- Dai L, Bai A, Smith CD, Rodriguez PC, Yu F, Qin Z. ABC294640, A Novel Sphingosine Kinase 2 Inhibitor, Induces Oncogenic Virus–Infected Cell Autophagic Death and Represses Tumor Growth. Molecular cancer therapeutics. 2017;16(12):2724-34.
- Chen X, Gole J, Gore A, He Q, Lu M, Min J, et al. Non-invasive early detection of cancer four years before conventional diagnosis using a blood test. Nat Commun. 2020;11(1):3475. [CrossRef]
- Wolrab D, Jirásko R, Chocholoušková M, Peterka O, Holčapek M. Oncolipidomics: Mass spectrometric quantitation of lipids in cancer research.. TrAC Trends in Analytical Chemistry 2019;120. [CrossRef]
- Beloribi-Djefaflia S, Vasseur S, Guillaumond F. Lipid metabolic reprogramming in cancer cells. Oncogenesis. 2016;5(1):e189.
- Butler LM, Perone Y, Dehairs J, Lupien LE, de Laat V, Talebi A, et al. Lipids and cancer: Emerging roles in pathogenesis, diagnosis and therapeutic intervention. Adv Drug Deliv Rev. 2020;159:245-93. [CrossRef]
- Fernandez LP, Gomez de Cedron M, Ramirez de Molina A. Alterations of Lipid Metabolism in Cancer: Implications in Prognosis and Treatment. Frontiers in oncology. 2020;10:577420.
- Jeong DW, Lee S, Chun YS. How cancer cells remodel lipid metabolism: strategies targeting transcription factors. Lipids Health Dis. 2021;20(1):163. [CrossRef]
- Long J, Zhang CJ, Zhu N, Du K, Yin YF, Tan X, et al. Lipid metabolism and carcinogenesis, cancer development. Am J Cancer Res. 2018;8(5):778-91.
- Vasseur S, Guillaumond F. Lipids in cancer: a global view of the contribution of lipid pathways to metastatic formation and treatment resistance. Oncogenesis. 2022;11(1):46. [CrossRef]
- Wang W, Bai L, Li W, Cui J. The Lipid Metabolic Landscape of Cancers and New Therapeutic Perspectives. Frontiers in oncology. 2020;10:605154. [CrossRef]
- Fahy E, Subramaniam S, Murphy RC, Nishijima M, Raetz CR, Shimizu T, et al. Update of the LIPID MAPS comprehensive classification system for lipids. Journal of lipid research. 2009;50 Suppl(Suppl):S9-14. [CrossRef]
- McDonald JG, Ejsing CS, Kopczynski D, Holcapek M, Aoki J, Arita M, et al. Introducing the Lipidomics Minimal Reporting Checklist. Nat Metab. 2022;4(9):1086-8. [CrossRef]
- Wood PL, Cebak JE. Lipidomics biomarker studies: Errors, limitations, and the future. Biochemical and biophysical research communications. 2018;504(3):569-75.
- Wang M, Wang C, Han RH, Han X. Novel advances in shotgun lipidomics for biology and medicine. Progress in lipid research. 2016;61:83-108. [CrossRef]
- Astarita G, Stocchero M, Paglia G. Unbiased Lipidomics and Metabolomics of Human Brain Samples. Methods Mol Biol. 2018;1750:255-69.
- Matsushita Y, Nakagawa H, Koike K. Lipid Metabolism in Oncology: Why It Matters, How to Research, and How to Treat. Cancers (Basel). 2021;13(3). [CrossRef]
- Iwamori M, Costello C, Moser HW. Analysis and quantitation of free ceramide containing nonhydroxy and 2-hydroxy fatty acids, and phytosphingosine by high-performance liquid chromatography. Journal of lipid research. 1979;20(1):86-96. [CrossRef]
- Sugita M, Iwamori M, Evans J, McCluer RH, Dulaney JT, Moser HW. High performance liquid chromatography of ceramides: application to analysis in human tissues and demonstration of ceramide excess in Farber's disease. Journal of lipid research. 1974;15(3):223-6.
- Blachnio-Zabielska AU, Persson XM, Koutsari C, Zabielski P, Jensen MD. A liquid chromatography/tandem mass spectrometry method for measuring the in vivo incorporation of plasma free fatty acids into intramyocellular ceramides in humans. Rapid Commun Mass Spectrom. 2012;26(9):1134-40. [CrossRef]
- Boiten W, Absalah S, Vreeken R, Bouwstra J, van Smeden J. Quantitative analysis of ceramides using a novel lipidomics approach with three dimensional response modelling. Biochimica et biophysica acta. 2016;1861(11):1652-61. [CrossRef]
- Cahoon RE, Solis AG, Markham JE, Cahoon EB. Mass Spectrometry-Based Profiling of Plant Sphingolipids from Typical and Aberrant Metabolism. Methods Mol Biol. 2021;2295:157-77.
- Domon B, Vath JE, Costello CE. Analysis of derivatized ceramides and neutral glycosphingolipids by high-performance tandem mass spectrometry. Anal Biochem. 1990;184(1):151-64. [CrossRef]
- Ramos P, Jenkins SM, Donato LJ, Hartman SJ, Saenger A, Baumann NA, et al. The Biological Variability of Plasma Ceramides in Healthy Subjects. J Appl Lab Med. 2022;7(4):863-70. [CrossRef]
- Sonomura K, Kudoh S, Sato TA, Matsuda F. Plasma lipid analysis by hydrophilic interaction liquid chromatography coupled with electrospray ionization tandem mass spectrometry. J Sep Sci. 2015;38(12):2033-7. [CrossRef]
- Sullards MC, Merrill AH, Jr. Analysis of sphingosine 1-phosphate, ceramides, and other bioactive sphingolipids by high-performance liquid chromatography-tandem mass spectrometry. Sci STKE. 2001;2001(67):pl1.
- Tabassum R, Ripatti S. Integrating lipidomics and genomics: emerging tools to understand cardiovascular diseases. Cell Mol Life Sci. 2021;78(6):2565-84. [CrossRef]
- Xin C, Wang Y, Chang Y, Zhang BO, Yang S. Detection and Analysis of Ceramide in Skin and Blood in a Healthy Chinese Population. J Cosmet Sci. 2020;71(6):367-75.
- Wolf C, Quinn PJ. Lipidomics: practical aspects and applications. Progress in lipid research. 2008;47(1):15-36. [CrossRef]
- Yang K, Han X. Lipidomics: Techniques, Applications, and Outcomes Related to Biomedical Sciences. Trends in biochemical sciences. 2016;41(11):954-69. [CrossRef]
- Surendran A, Atefi N, Ismail U, Shah A, Ravandi A. Impact of myocardial reperfusion on human plasma lipidome. iScience. 2022;25(2):103828. [CrossRef]
- Tumanov S, Kamphorst JJ. Recent advances in expanding the coverage of the lipidome. Curr Opin Biotechnol. 2017;43:127-33. [CrossRef]
- Wolrab D, Jirasko R, Cifkova E, Horing M, Mei D, Chocholouskova M, et al. Lipidomic profiling of human serum enables detection of pancreatic cancer. Nat Commun. 2022;13(1):124. [CrossRef]
- Subramaniam S, Fahy E, Gupta S, Sud M, Byrnes RW, Cotter D, et al. Bioinformatics and systems biology of the lipidome. Chem Rev. 2011;111(10):6452-90. [CrossRef]
- Lima T, Ferreira R, Freitas M, Henrique R, Vitorino R, Fardilha M. Integration of Automatic Text Mining and Genomic and Proteomic Analysis to Unravel Prostate Cancer Biomarkers. J Proteome Res. 2022;21(2):447-58. [CrossRef]
- Baudino TA. Targeted Cancer Therapy: The Next Generation of Cancer Treatment. Curr Drug Discov Technol. 2015;12(1):3-20. [CrossRef]
- Zhong L, Li Y, Xiong L, Wang W, Wu M, Yuan T, et al. Small molecules in targeted cancer therapy: advances, challenges, and future perspectives. Signal Transduct Target Ther. 2021;6(1):201. [CrossRef]
- Zhong L, Li Y, Xiong L, Wang W, Wu M, Yuan T, et al. Small molecules in targeted cancer therapy: advances, challenges, and future perspectives. Signal Transduction and Targeted Therapy. 2021;6(1):201. [CrossRef]
- Lewis AC, Wallington-Beddoe CT, Powell JA, Pitson SM. Targeting sphingolipid metabolism as an approach for combination therapies in haematological malignancies. Cell Death Discovery. 2018;4(1):72. [CrossRef]
- Raichur S. Ceramide Synthases Are Attractive Drug Targets for Treating Metabolic Diseases. Front Endocrinol (Lausanne). 2020;11:483. [CrossRef]
- Park JW, Park WJ, Futerman AH. Ceramide synthases as potential targets for therapeutic intervention in human diseases. Biochim Biophys Acta. 2014;1841(5):671-81. [CrossRef]
- Mullen TD, Hannun YA, Obeid LM. Ceramide synthases at the centre of sphingolipid metabolism and biology. Biochem J. 2012;441(3):789-802. [CrossRef]
- Coant N, Sakamoto W, Mao C, Hannun YA. Ceramidases, roles in sphingolipid metabolism and in health and disease. Adv Biol Regul. 2017;63:122-31. [CrossRef]
- Wang K, Wei Y, Xu R, Li Y, Mao C. Manifold Roles of Ceramide Metabolism in Non-Alcoholic Fatty Liver Disease and Liver Cancer. Adv Exp Med Biol. 2022;1372:157-68.
- Xu R, Antwi Boasiako P, Mao C. Alkaline ceramidase family: The first two decades. Cell Signal. 2021;78:109860. [CrossRef]
- Truman JP, García-Barros M, Obeid LM, Hannun YA. Evolving concepts in cancer therapy through targeting sphingolipid metabolism. Biochim Biophys Acta. 2014;1841(8):1174-88. [CrossRef]
- Gencer EB, Ural AU, Avcu F, Baran Y. A novel mechanism of dasatinib-induced apoptosis in chronic myeloid leukemia; ceramide synthase and ceramide clearance genes. Ann Hematol. 2011;90(11):1265-75. [CrossRef]
- Camgoz A, Gencer EB, Ural AU, Avcu F, Baran Y. Roles of ceramide synthase and ceramide clearence genes in nilotinib-induced cell death in chronic myeloid leukemia cells. Leuk Lymphoma. 2011;52(8):1574-84.
- Sundaramoorthy P, Gasparetto C, Kang Y. The combination of a sphingosine kinase 2 inhibitor (ABC294640) and a Bcl-2 inhibitor (ABT-199) displays synergistic anti-myeloma effects in myeloma cells without a t(11;14) translocation. Cancer Med. 2018;7(7):3257-68. [CrossRef]
- Ding X, Zhang Y, Huang T, Xu G, Peng C, Chen G, et al. Targeting sphingosine kinase 2 suppresses cell growth and synergizes with BCL2/BCL-XL inhibitors through NOXA-mediated MCL1 degradation in cholangiocarcinoma. Am J Cancer Res. 2019;9(3):546-61.
- Cavalli A, Bolognesi ML, Minarini A, Rosini M, Tumiatti V, Recanatini M, et al. Multi-target-directed ligands to combat neurodegenerative diseases. J Med Chem. 2008;51(3):347-72.
- Zhou J, Jiang X, He S, Jiang H, Feng F, Liu W, et al. Rational Design of Multitarget-Directed Ligands: Strategies and Emerging Paradigms. Journal of Medicinal Chemistry. 2019;62(20):8881-914. [CrossRef]
- Proschak E, Stark H, Merk D. Polypharmacology by Design: A Medicinal Chemist’s Perspective on Multitargeting Compounds. Journal of Medicinal Chemistry. 2019;62(2):420-44. [CrossRef]
- Benet LZ, Hosey CM, Ursu O, Oprea TI. BDDCS, the Rule of 5 and drugability. Adv Drug Deliv Rev. 2016;101:89-98. [CrossRef]
- Ma H, Huang B, Zhang Y. Recent advances in multitarget-directed ligands targeting G-protein-coupled receptors. Drug Discov Today. 2020;25(9):1682-92. [CrossRef]
- Stelitano G, Sammartino JC, Chiarelli LR. Multitargeting Compounds: A Promising Strategy to Overcome Multi-Drug Resistant Tuberculosis. Molecules. 2020;25(5):1239. [CrossRef]
- Wilt S, Kodani S, Valencia L, Hudson PK, Sanchez S, Quintana T, et al. Further exploration of the structure-activity relationship of dual soluble epoxide hydrolase/fatty acid amide hydrolase inhibitors. Bioorganic & Medicinal Chemistry. 2021;51:116507. [CrossRef]
Figure 1.
Main metabolic pathways of ceramide metabolism. Three pathways help synthesize ceramide in the cell namely, sphingomyelin hydrolysis pathway, de novo pathway, and salvage pathway. De novo sphingolipid biosynthesis begins with the condensation of serine and palmitoyl-CoA, which then is catalyzed by serine palmitoyltransferase enzyme. Its product, 3-ketosphinganine, is reduced to sphinganine by 3-ketosphinganine reductase. Next, dihydroceramide synthases contribute in synthesis of dihydroceramides using sphinganine and fatty acyls, which are then reduced by dihydroceramide desaturase to produce ceramides with different acyl chain lengths. Ceramide is also synthesized by other metabolic pathways (salvage and sphingomyelin hydrolysis). The ceramide produced via these pathways will be used as a mediator in the regulation of subsequent downstream pathways (enzymes shown in red italics).
Figure 1.
Main metabolic pathways of ceramide metabolism. Three pathways help synthesize ceramide in the cell namely, sphingomyelin hydrolysis pathway, de novo pathway, and salvage pathway. De novo sphingolipid biosynthesis begins with the condensation of serine and palmitoyl-CoA, which then is catalyzed by serine palmitoyltransferase enzyme. Its product, 3-ketosphinganine, is reduced to sphinganine by 3-ketosphinganine reductase. Next, dihydroceramide synthases contribute in synthesis of dihydroceramides using sphinganine and fatty acyls, which are then reduced by dihydroceramide desaturase to produce ceramides with different acyl chain lengths. Ceramide is also synthesized by other metabolic pathways (salvage and sphingomyelin hydrolysis). The ceramide produced via these pathways will be used as a mediator in the regulation of subsequent downstream pathways (enzymes shown in red italics).
Figure 2.
Role of ceramide in regulation of apoptosis via different mechanisms. Figure shows how ceramides regulate the apoptosis via formation of ceramide channels and interaction with anti-apoptotic proteins like Bcl-xL and pro-apoptotic proteins such as Bax. Ceramides with various fatty acyl chain lengths can perforate into the mitochondrial outer membranes. These channels are layers of ceramide molecules that are stacked in an anti-parallel way on top of each other forming ceramide columns in a barrel-like structure. Ceramide channels contribute to the permeabilization of the mitochondrial outer membrane. This leads to the release of pro-apoptotic proteins such as cytochrome c from the intermembrane space into the cytosol and thus apoptosis. Additionally, ceramides help in the oligomerization of Bax molecules during apoptosis. Furthermore, ceramide can influence the Bcl2 phosphorylation and therefore its anti-apoptotic function. Ceramide activates mitochondrial protein phosphatase 2A (PP2A) (a physiologic Bcl2 phosphatase) that induces Bcl2 dephosphorylation and therefore ceramide-induced apoptosis.
Figure 2.
Role of ceramide in regulation of apoptosis via different mechanisms. Figure shows how ceramides regulate the apoptosis via formation of ceramide channels and interaction with anti-apoptotic proteins like Bcl-xL and pro-apoptotic proteins such as Bax. Ceramides with various fatty acyl chain lengths can perforate into the mitochondrial outer membranes. These channels are layers of ceramide molecules that are stacked in an anti-parallel way on top of each other forming ceramide columns in a barrel-like structure. Ceramide channels contribute to the permeabilization of the mitochondrial outer membrane. This leads to the release of pro-apoptotic proteins such as cytochrome c from the intermembrane space into the cytosol and thus apoptosis. Additionally, ceramides help in the oligomerization of Bax molecules during apoptosis. Furthermore, ceramide can influence the Bcl2 phosphorylation and therefore its anti-apoptotic function. Ceramide activates mitochondrial protein phosphatase 2A (PP2A) (a physiologic Bcl2 phosphatase) that induces Bcl2 dephosphorylation and therefore ceramide-induced apoptosis.
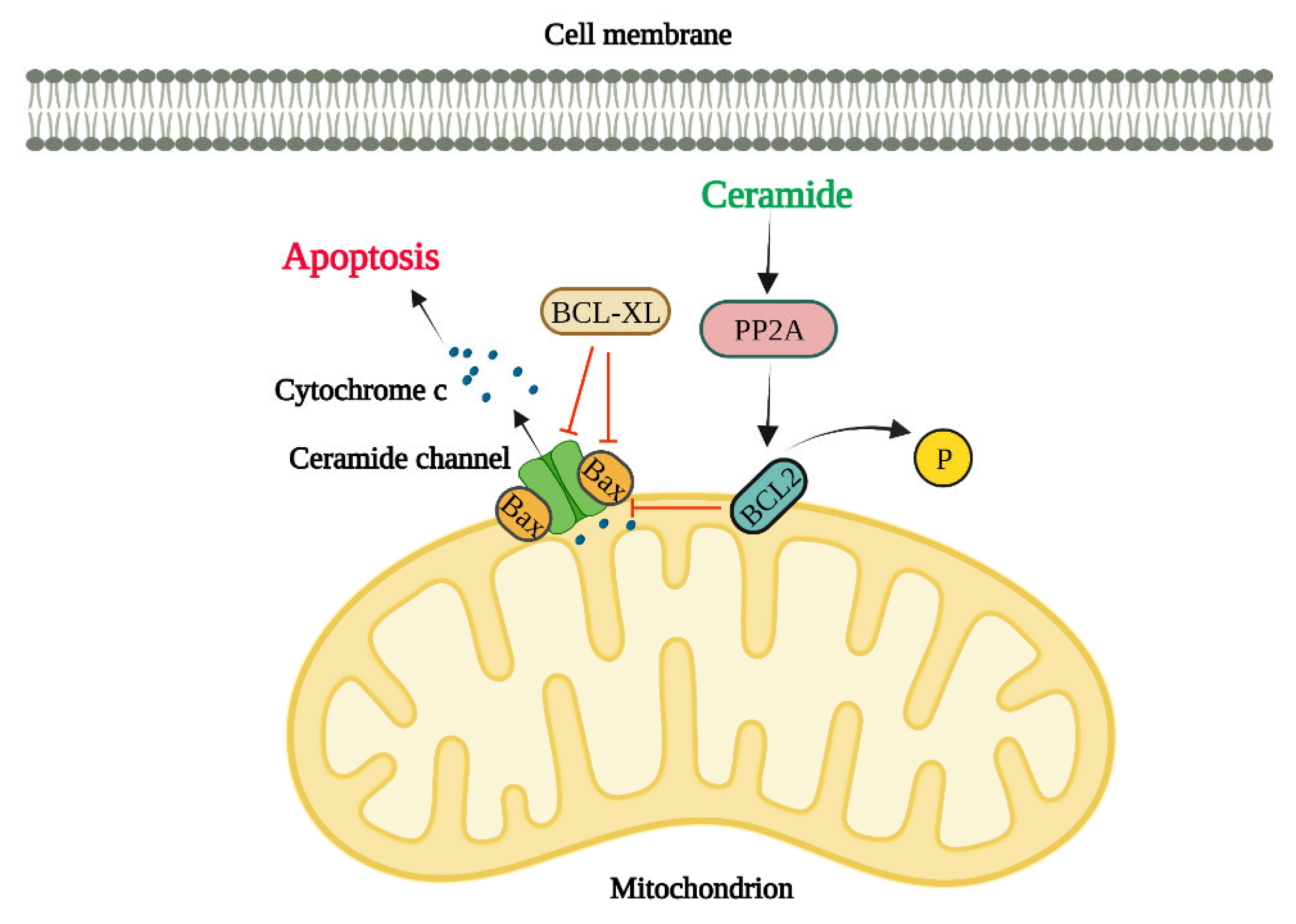
Figure 3.
Role of ceramide in regulation of autophagy by different mechanisms. Ceramide activates JNK which leads to the phosphorylation of BCL2 resulting in the dissociation of Beclin1 from the Beclin1-Bcl2 complex. Beclin1 then can participate in the formation of autophagosomes. Also, localized ceramide on the outer mitochondrial membrane can act as a LC3II-receptor, thereby bringing the mitochondria into the forming autophagosome. Figure also shows the cross talk between ER and autophagy pathway. Ceramide activates the ATF6 in the ER membrane which subsequently activates CHOP. Activation of CHOP inhibits the phosphorylation of BCL2 by JNK. .
Figure 3.
Role of ceramide in regulation of autophagy by different mechanisms. Ceramide activates JNK which leads to the phosphorylation of BCL2 resulting in the dissociation of Beclin1 from the Beclin1-Bcl2 complex. Beclin1 then can participate in the formation of autophagosomes. Also, localized ceramide on the outer mitochondrial membrane can act as a LC3II-receptor, thereby bringing the mitochondria into the forming autophagosome. Figure also shows the cross talk between ER and autophagy pathway. Ceramide activates the ATF6 in the ER membrane which subsequently activates CHOP. Activation of CHOP inhibits the phosphorylation of BCL2 by JNK. .
Figure 4.
Major steps in lipid preparation and analysis involve 1) sample collection (serum, plasma, urine, cell, organ/tissue); 2) lipid extraction (folch method, blingh-dyer method, MTBE method, BUME method); 3) Lipid separation (TLC, GC, LC, SFC); 4) mass spectrometry data acquisition (MS combined with chromatography, shot gun MS, MS imaging); 5) data analysis (identification, quantification, pathway analysis); 6) interpretation (biomarkers, therapeutic targets).
Figure 4.
Major steps in lipid preparation and analysis involve 1) sample collection (serum, plasma, urine, cell, organ/tissue); 2) lipid extraction (folch method, blingh-dyer method, MTBE method, BUME method); 3) Lipid separation (TLC, GC, LC, SFC); 4) mass spectrometry data acquisition (MS combined with chromatography, shot gun MS, MS imaging); 5) data analysis (identification, quantification, pathway analysis); 6) interpretation (biomarkers, therapeutic targets).
Figure 5.
Bioinformatics guide to lipidomics integrative analysis.
Figure 5.
Bioinformatics guide to lipidomics integrative analysis.
Figure 7.
Drug design strategies summary and potential binding sites on Bcr-Abl tyrosine kinase. Tyrosine kinases use ATP as a source of phosphate group for the phosphorylation reaction of the phenol groups on the tyrosine residues. A) Substrate (orange) binding to substrate binding site. Huang et al. used peptide inhibitors that binds to the substrate-binding region and were able to inhibit in vitro Bcr-Abl kinase in the micromolar range.Ref However, peptides have several disadvantages to be used as the therapeutics in the clinical settings, and the small molecule drug that binds and significantly inhibits substrate-based region for Bcr-Abl kinase has not yet been reported. B) The binding site for ATP is usually located close to the substrate binding site. Imatinib (shown in red) binds to ATP-binding region. A known T315I mutation, located in ATP-binding site, causes a drug resistance to imatinib, and makes this binding site less attractive target for drug design. C) Allosteric-binding site with asciminib shown in blue. The first discovered allosteric site on Bcr-Abl has been named myristoyl-binding region, since myristoyl acid binds in this binding region and produce inactivation of the kinase. This highly hydrophobic pocket represents a novel target for the site-specific drug design strategy. More drug-like small molecules that could mimic myristate could theoretically bind there and inhibit the kinase. D) A potential, novel dual inhibitor strategy: substrate-binding ligand (L1) is anchored to the ATP-binding ligand (L2) – a potential small molecule drug could bind simultaneously to the allosteric site and ATP-binding region.
Figure 7.
Drug design strategies summary and potential binding sites on Bcr-Abl tyrosine kinase. Tyrosine kinases use ATP as a source of phosphate group for the phosphorylation reaction of the phenol groups on the tyrosine residues. A) Substrate (orange) binding to substrate binding site. Huang et al. used peptide inhibitors that binds to the substrate-binding region and were able to inhibit in vitro Bcr-Abl kinase in the micromolar range.Ref However, peptides have several disadvantages to be used as the therapeutics in the clinical settings, and the small molecule drug that binds and significantly inhibits substrate-based region for Bcr-Abl kinase has not yet been reported. B) The binding site for ATP is usually located close to the substrate binding site. Imatinib (shown in red) binds to ATP-binding region. A known T315I mutation, located in ATP-binding site, causes a drug resistance to imatinib, and makes this binding site less attractive target for drug design. C) Allosteric-binding site with asciminib shown in blue. The first discovered allosteric site on Bcr-Abl has been named myristoyl-binding region, since myristoyl acid binds in this binding region and produce inactivation of the kinase. This highly hydrophobic pocket represents a novel target for the site-specific drug design strategy. More drug-like small molecules that could mimic myristate could theoretically bind there and inhibit the kinase. D) A potential, novel dual inhibitor strategy: substrate-binding ligand (L1) is anchored to the ATP-binding ligand (L2) – a potential small molecule drug could bind simultaneously to the allosteric site and ATP-binding region.
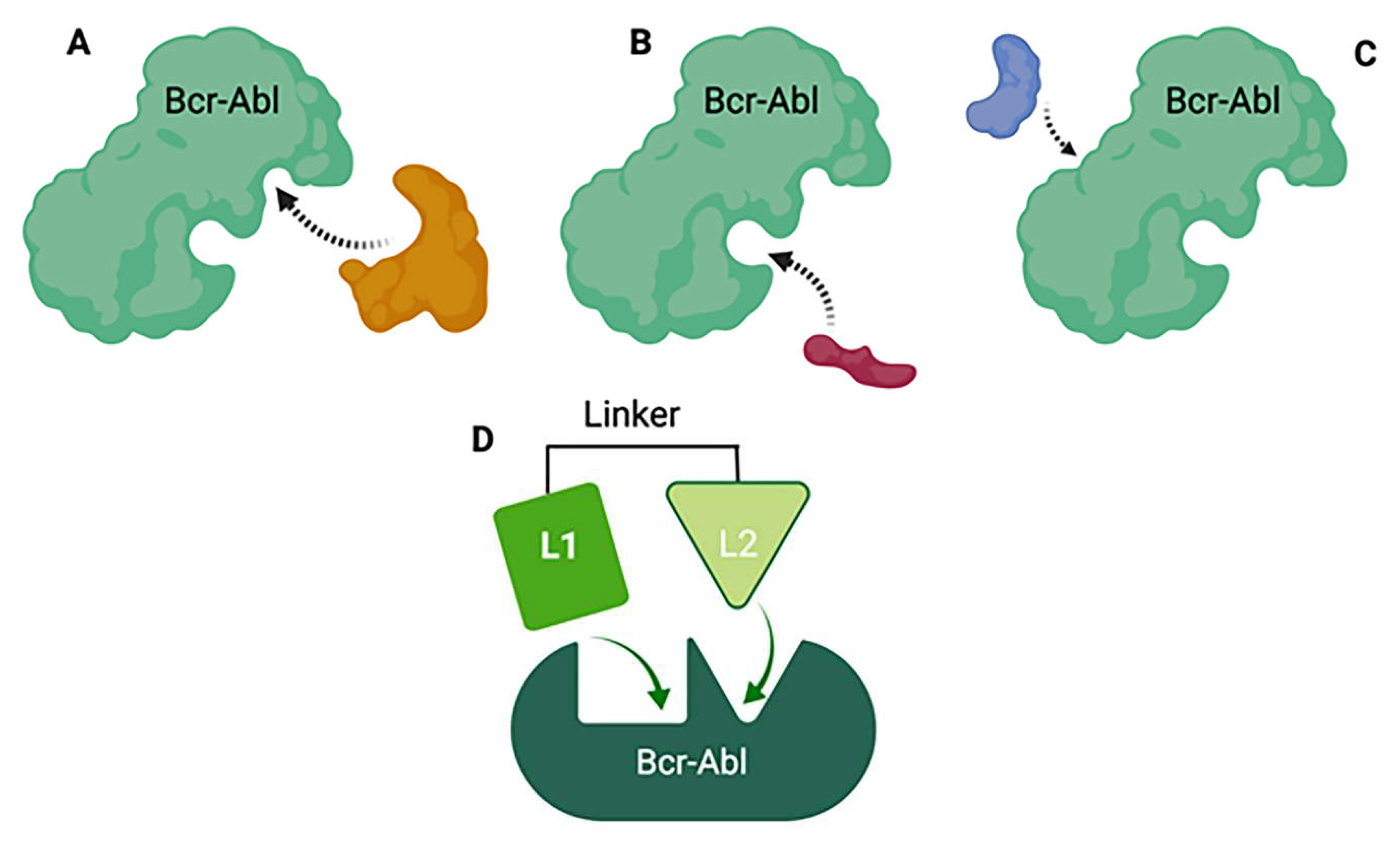
Figure 6.
De novo and Salvage pathways of ceramide synthesis. Enzyme ceramide synthase (CerS) is involved in both pathways and represents a valid target for drug design of ceramide synthesis modulators.
Figure 6.
De novo and Salvage pathways of ceramide synthesis. Enzyme ceramide synthase (CerS) is involved in both pathways and represents a valid target for drug design of ceramide synthesis modulators.
Figure 8.
Polypharmacological approach. Two or more biological targets are selected, and small molecule modulators are identified. Upon determining the pharmacophores, the multi-target designed ligands (MTDLs) are created by linking, fusing or merging pharmacophores. The merged pharmacophores are of the particular interest in the drug discovery due to several pharmacokinetic and pharmacodynamic advantage properties. Once MTDL is designed, synthesized and preliminary structure-activity relationship study is established, the computer-based drug design should follow up in order to create the second generation of lead compounds. Next should follow the crystallographic determination of the lead compound with both targets to better understand the binding interactions of MTDLs with both targets. More specific SAR study that will optimize the drug-target interactions should follow up. .
Figure 8.
Polypharmacological approach. Two or more biological targets are selected, and small molecule modulators are identified. Upon determining the pharmacophores, the multi-target designed ligands (MTDLs) are created by linking, fusing or merging pharmacophores. The merged pharmacophores are of the particular interest in the drug discovery due to several pharmacokinetic and pharmacodynamic advantage properties. Once MTDL is designed, synthesized and preliminary structure-activity relationship study is established, the computer-based drug design should follow up in order to create the second generation of lead compounds. Next should follow the crystallographic determination of the lead compound with both targets to better understand the binding interactions of MTDLs with both targets. More specific SAR study that will optimize the drug-target interactions should follow up. .
Table 1.
Different CerS synthesize ceramides with various acyl chain lengths.
Table 1.
Different CerS synthesize ceramides with various acyl chain lengths.
| CerS |
Synthesized ceramide |
Ref |
| CerS1 |
C18 |
(5) |
| CerS2 |
C20,
C22,
C24,
C26 |
(6) |
| CerS3 |
C22,
C24,
C26 |
(7) |
| CerS4 |
C18,
C20 |
(8) |
| CerS5 |
C16 |
(9) |
| CerS6 |
C14,
C16 |
(10) |
Table 2.
Summary of the role of ceramides and enzymes involved in ceramide metabolism in autophagy regulation in different cancers.
Table 2.
Summary of the role of ceramides and enzymes involved in ceramide metabolism in autophagy regulation in different cancers.
| Ceramide |
Mechanism of Action |
Effect on Autophagy |
Cancer Type |
Ref |
| C2-ceramide |
BNIP3 accumulation, Beclin1:Bcl-2 dissociation |
autophagy induction |
Colon carcinoma cell line (HT-29) |
(151, 213) |
| C18-ceramide |
ceramide and LC3B-II interaction |
lethal autophagy induction |
HNSCC cells (UM-SCC 22A) |
(212) |
| As2O3 induced accumulation of overall ceramide |
upregulation of Beclin-1 |
lethal autophagy induction |
Leukemia cells (HuT-102, C91-Pl, MT-2) |
(215) |
| C2-ceramide |
mitochondrial membrane potential decrease, BNIP3 activation |
lethal autophagy induction |
Glioma cells (U373-MG and T98G) |
(213) |
| C2-ceramide |
c-Jun activation through JNK signaling |
lethal autophagy induction |
Nasopharyngeal carcinoma cells (CNE2), hepatocellular carcinoma cells (Hep3B) |
(146) |
| CAPPs (ceramide activated protein phosphatases) |
Akt-mTOR pathway inhibition |
lethal autophagy induction |
Leukemia cells (HL-60), Chinese hamster ovary cells (CHO) |
(221) |
| S1P |
Akt-mTOR pathway activation |
lethal autophagy inhibition |
Leukemia cells (HL-60), Chinese hamster ovary cells (CHO) |
(221) |
| C6-ceramide |
mitochondrial permeabilization |
autophagy induction |
Breast cancer cells (MCF-7) |
(222) |
| C18-ceramide |
increased the formation of autophagosomes, PI3K/AKT pathway inhibition |
lethal autophagy induction |
brain cancer cells (U251 and A172) |
(218) |
| inhibition of SphK1 |
ATG5 and Beclin-1 |
lethal autophagy induction |
HCT116 human colon cancer cells |
(223) |
| inhibition of SphK2 by ABC294640 |
accumulation of intracellular ceramides |
lethal autophagy induction |
herpes virus-related tumors |
(224) |
Table 3.
Publicly available lipid tools and resources for bioinformatic analysis.
Table 3.
Publicly available lipid tools and resources for bioinformatic analysis.
| Name |
Description |
URL |
| LipidMaps |
The web portal gateway to Lipidomics resources; classification of biological lipids, dividing them into eight general categories |
https://www.lipidmaps.org/ |
| LipidFinder |
An open-source Python workflow which searches several different databases to obtain putative identification of lipids |
https://www.lipidmaps.org/resources/tools/lipidfinder/ |
| LipidSig |
Allows the exploration of the quality and the clustering of samples, correlation between lipids and samples, and the expression and composition of lipids |
http://www.chenglab.cmu.edu.tw/lipidsig/Profiling/ |
| LipidSuite |
Workflow for preprocessing, exploration, differential analysis, and enrichment of untargetted and targeted lipidomics |
https://suite.lipidr.org/ |
| LipidPedia |
Lipid encyclopedia; provides associated biomedical information through text-mining strategy in literatures |
https://lipidpedia.cmdm.tw/ |
| LipidBank |
The official database of Japanese Conference on the Biochemistry of Lipids (JCBL) |
https://lipidbank.jp/ |
| LipidHome |
Database of theoretical lipid structures derived from a seed set of lipid sub classes and some lipid chemical space constraints |
https://apps.lifs.isas.de/lipidhome/ |
| LipidBlast |
In silico tandem mass spectrometry database for lipid identification |
https://fiehnlab.ucdavis.edu/projects/lipidblast |
| LipidMatch |
An automated workflow for rule-based lipid identification using untargeted high-resolution tandem mass spectrometry data |
http://secim.ufl.edu/secim-tools/lipidmatch/ |
| ONION |
Functional approach for integration of lipidomics and transcriptomics data |
https://github.com/wjurkowski/ONION |
| LINT-Web |
Web-based Lipidomic data mining tool using intra-omic integrative correlation strategy |
http://www.lintwebomics.info/ |
| MetaboAnalyst |
Platform dedicated for metabolomics data analysis via user-friendly, web-based interface. |
https://www.metaboanalyst.ca/ |
| SwissTarget Prediction |
Estimate the most probable macromolecular targets of a small molecule, assumed as bioactive |
http://www.swisstargetprediction.ch/predict.php |
| String |
Protein-Protein interaction networks functional enrichment analysis |
https://string-db.org/ |
| Stitch |
Resource to explore known and predicted interactions of chemicals and proteins |
http://stitch.embl.de/ |
| DisGeNET |
Integrative database with human gene-disease associations (GDAs) and variant-disease associations (VDAs) from various repositories including Mendelian, complex and environmental diseases |
https://www.disgenet.org/search |
| VosViewer |
Software tool for constructing and visualizing bibliometric networks |
https://www.vosviewer.com/ |
Table 4.
Studies in adults with ceramide as treatment for cancer.
Table 4.
Studies in adults with ceramide as treatment for cancer.
| # |
Status |
Title |
Conditions |
Drug(s) used |
Phase |
Eligible |
Location |
Reference (NCT) |
| 1 |
Completed |
Topical Ceramide Lipids As Treatment For Cutaneous Breast Cancer |
Breast cancer |
• Drug: ceramide |
2 |
≥ 18, female |
9 states in USA |
NCT00008320 |
| 2 |
Unknown |
C6 Ceramide NanoLiposome in Patients With Advanced Solid Tumors |
Cancer; carcinoma; solid tumors; tumor |
• Drug: Ceramide NanoLiposome |
1 |
≥ 18 |
3 states in USA |
NCT02834611 |
| 3 |
Withdrawn |
Detection of Acid Sphingomyelinase/Ceramide Pathway Activation in Radiotherapy Patients Using Intravoxel Incoherent Motion (IVIM) Diffusion-weighted Magnetic Resonance Imaging and Serum Biomarkers |
Metastatic disease to bone; metastatic disease to soft tissue
|
• Device: MRI with IVIM DW-MRI
• Other: Blood draw
|
N/A |
≥ 18 |
NY, USA |
NCT02465723 |
| 4 |
Not yet recruiting |
C6 Ceramide NanoLiposome (CNL) in Patients With Relapsed/Refractory Acute Myeloid Leukemia (RR-AML) |
Acute Myeloid Leukemia, in Relapse Acute;
Myeloid Leukemia, Refractory
|
• Drug: Ceramide NanoLiposome (Ceraxa) |
1 |
≥ 18 |
3 states in USA |
NCT04716452 |
| 5 |
Completed |
Combination of Lenalidomide (Revlimid, LEN) and Autologous Mature Dendritic Cells Pulsed With α-galactosyl Ceramide (α-GalCer; KRN7000) in Myeloma |
Myeloma |
• Drug: Lenalidomide Biological: Monocyte derived DCs loaded with KRN7000 |
1 |
≥ 18 |
CT, USA |
NCT00698776 |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).