Introduction
Probiotic bacteria, such as Bifidobacteria and Lactobacilli, inhibit the growth and proliferation of potentially pathogenic bacteria in the intestine (1) reports suggest that breast-fed infants are healthy and resistant to infectious diseases. The inclusion of breast-feeding has the objective of maintaining and inoculating beneficial intestinal microbiota. Breast-feeding promotes a healthy intestine by suppressing the growth and proliferation of potentially pathogenic bacteria. The most common species of Bifidobacteria reported in infant faecal material are B. infantis, B. longum, B. bifidum, B. adolescentis, and B. breve. Yet, very rarely is B. catenulatum observed. It is possible to understand the relationship between the healthy infant and the intestinal microbiota by measuring the different groups of bacteria accurately. Only a few studies have evaluated selective agars for the enumeration, isolation, and growth of Bifidobacteria in faecal samples (2).
In the present study, MRS, BHI, LB, and Bifidobacteria agar were assessed for enumeration, isolation, and growth of Bifidobacteria in infant faecal samples (breast-fed and formula-fed infants), and developed an antibiogram. RB (Raffinose Bifidobacterium) was developed for the isolation and enumeration of Bifidobacteria from poultry, caecal samples (6, 7), and different media, such as Beerens agar (3, 4, 5), were suggested for the isolation of Bifidobacteria from gut microbiota. This further indicates that different media are needed for the isolation of Bifidobacteria from different sources. The only medium that contains mupirocin antibiotic is the MW agar, in which Bifidobacteria is resistant and many Lactobacilli are susceptible, therefore it is specific to one species (8). All the media stated and used are complex and cost-effective. Hence, it is very essential to study the growth and isolation of Bifidobacterium spp. in a simpler, commonly used universal medium. One of the primary objectives of the present study lies in understanding, optimizing, isolation, and identification of the Bifidobacterium spp in the common media and, subsequently, antibiogram development.
According to Bifidobacteria population density studies (of gut), Yildrim et al. 1998 state (9) that 25% of adults and 95% of the new-born/infant gut (primarily) consists of Bifidobacterium spp. The members of the various species of B. catenulatum, B. bifidum, B. infantis, B. brevi, and B. adolescentis, etc., along with other probiotic microbes, are used as probiotic supplements in the form of sachets, tablets, in different foods, especially in beverages. The extensive use of antibiotics in recent decades has initiated the emergence of antibiotic resistance in bacterial spp and also led to the creation of antibiotic resistant pathogens by transmitting the resistance factors such as plasmids and insertion sequence elements (Bernardeau and colleagues (2008) (10) (11) (12). Bifidobacteria having antibiotic resistance might transfer the responsible antibiotic resistance factor to harmful and pathogenic bacteria, further leading to the formation of not only antibiotic resistant pathogens, but also resulting in the failure of antibiotic treatment. Based on these relevant incidents and reports, it is critical to investigate and develop Bifidobacterial antibiogram to differentiate between resistant and susceptible varieties, which also aids in understanding the type of resistance, such as intrinsic or acquired. The major side effect of the consumption of antibiotics is antibiotic-associated diarrhoea. It is very well understood that antibiotic therapy not only reduces the viability of indigenous Bifidobacteria (13) but also significantly affects the intestinal microbial flora. Previous reports have concluded that administering B. longum reduced the antibiotic-related diarrhoea and resulted in a reduction in the time required for recovery from rotavirus diarrhoea (14). Very recent reports by Dhanashree et al., 2019 (15) state that Bifidobacteria can also be used for antibiotic and probiotic therapy, specifically for tuberculosis infected. Therefore, the successful prophylactic use of Bifidobacteria against intestinal disorders depends not only on their ability to survive gastrointestinal conditions, compete with, suppress, and eliminate the pathogens, but also circumvent and tolerate antibiotic treatment.
Intestinal environmental conditions such as low pH, the presence of bile salts, and alkaline conditions may all reduce their viability. Also modify their physiological activities and antimicrobial susceptibilities during their passage. Before considering them for prophylactic applications, a thorough investigation of their antibiotic susceptibilities and resistance before and after gastrointestinal passage is required. In the present study, we propose the development of antibiogram in three different test media, especially because there are no suitable and standard protocols available for the susceptibility testing of Bifidobacteria. In addition, we propose to examine changes in susceptibilities to multiple antimicrobial agents between different isolates due to stress caused by alkaline, acidic conditions, and hydrogen peroxide (H2O2).
In this study, Bifidobacteria isolates and strains, mostly of human and dairy origin, were tested against antibiotics of different functional classes. Ten different classes of antibiotics were used, such as β-lactams, macrolides, tetracycline, amino glycosides, and other antibiotics such as amoxyclave, chloramphenicol, kanamycin, pristinomycin, and vancomycin. These strains were tested with E-test methods, all using the standard and specific LAB. The susceptibility test medium (16) (LSM, Klare et al., 2005) specifically designed for antibiotic susceptibility testing of non-enterococcal LAB was followed. Selected Bifidobacteria strains of resistant and susceptible phenotypes characterized by amplification of antibiotic resistance genes were chosen for the study.
Materials and Methods
This study was conducted between December and April of 2010 in the local hospitals. The inclusion criteria were: (i). age under six months upon entering the study, (ii). Either sex is male or female (iii). Cow milk allergy symptoms (iv). No probiotic supplements were used regularly (for more than one week and within six weeks before entering the study). Of the 431 infants referred by local health centres 284 (66%) had parents who wished to participate. Of the 252 infants meeting our inclusion criteria, 230 (aged 3-6 months, mean 6.4) completed the study. One parent of each infant agreed to the consent. Term infants (n = 371) were approached in this study. All infants started breast-feeding. Those who changed to formula-feeding within 4 weeks after birth were randomly assigned to one of the two formula groups. Growth and stool characteristics and side effects that occurred in recruited infants were recorded in a 3-month follow-up period. Nurses in the hospital collected faecal samples from a sub-population and mothers in the homes of infants for analysis of intestinal bacteria (culture technique), acetic acid (gas chromatography) and pH (indicator strip). Of the one hundred samples, only ten were further screened. The ten faecal samples were obtained from the infants of breast-fed and formula-fed infants after 4, 30, and 90 days of delivery and sub-cultured on nonselective and selective media. For each sample, freshly voided faecal material from different infants was/were collected and pooled. All the various bacteria isolated and used in the study are listed in the
Table 1. Most of the faecal samples were obtained from the paediatric departments (hospitals). Out of which, three samples represent (F1 to F3 of the
Table 2) four different paediatric departments, and the remaining samples (F4–F6 of
Table 2) represent formula-fed infants in and around the area.
A total of 5–10 g of faecal samples were processed under sterile conditions, and restricted to aerobic conditions. The total number of anaerobic bacterial cultures was enumerated using anaerobic Wilkins-chalgreen agar (Difco 1805-17-6). The cultures were inoculated and incubated at 37 °C for five days in an anaerobic gas jar. Subsequently, Bifidobacteria were enumerated and isolated through serial dilutions on BHI, LB, MRS, and Bifidobacteria agar. From each selective agar, 25 colonies from the highest dilution of each sample were picked randomly. Subsequently, they were inoculated into reinforced clostridia broth (Merck 5411) with hemin added at a concentration of 0.005 g/ltr. The isolates were stored as glycerol stocks at -70 °C. Subcultures of isolates were grown in 10 mL of Trypticase-peptone-yeast extract broth at 37 °C. The grown cells were harvested by centrifugation (9000 x g for 10 min), and genomic DNA was extracted using a genomic DNA isolation kit (Macherey-Nagel GmbH & Co. KG, Germany) as directed by the manufacturers.
The various Bifidobacteria strains/isolates used in this study are listed in
Table 1. Before subjecting them to antibiotic susceptibility studies, all the listed strains were verified as Bifidobacteria with a species-specific PCR targeting a partial and complete 16S rDNA sequence (650 and 1500 bp) using the method described by Alander
et al. (1999) (17).The Bifidobacteria isolates were grown at 37
°C in an anaerobic gas-jar containing an atmosphere-generating system. In order to allow cultures to adapt to the test media before subjecting them to antibiotic susceptibility tests, they were cultivated on LSM plates for overnight. Following that, isolated and individual colonies were re-suspended in fresh 2.0 mL of sterile 0.9 % saline until an OD
600 nm of 0.16-0.20 was obtained (corresponding to McFarland standard 1.0 and a cell concentration of 3x10
8 cfu/mL).
Growth of Bifidobacteria in Aerobic and Anaerobic Conditions
Bifidobacteria are obligate anaerobes; they are sensitive to oxygen and cannot grow in aerobic conditions. Therefore, they require anaerobic conditions for their growth, essentially in its initial phase. Similar conditions to those for anaerobic cultivation were set and followed for the cultivation of Bifidobacteria in aerobic conditions as well. However, by introducing small changes, firstly the initial growth phase set at low pH, secondly, and more importantly, the partial removal of oxygen from the system before and after autoclaving, and finally by the introduction of 1.0 % of N2 gas into the media.
Antibiotic Susceptibility Determination
The E-test methodology was followed; first, a sterile cotton swab was dipped in the inoculum (approx. 3x10
8 cfu/mL) and spread manually on the MRS/Bifido/TPY/MH agar plates. Subsequently, the plates were allowed to dry for 15–30 min before the E-test strip (Himedia Laboratories Pvt. Ltd., India) was applied. The Minimum Inhibitory Concentration (MIC) values for 10 different antibiotics representing inhibitors of cell wall synthesis (i.e. amoxicillin/amoxyclave, AMX10), protein synthesis (gentamycin GEN10; tetracycline, TET30; chloramphenicol, CHL30; erythromycin, ERY15) and other common antibiotics such as ampicillin, tetracycline, kanamycin, pristomycin, streptomycin, and vancomycin were (obtained from Invitrogen Inc., USA) determined for ten different Bifidobacteria (
Table 1).
The E-test (biodisk, Sweden) method was followed to develop antibiogram on different media such as Brucella agar, LSM agar supplemented with cysteine, TPY and MH agar. Susceptibility or resistance testing were performed according to the NCCLS (2004) (18). In the E-test, the concentrations followed were 0.016-256 µg/mL for all antibiotics. The E-test agar plates finally incubated (anaerobic condition) in a gas jar at 37 ºC for 48 h (Hi-media laboratories, portable anaerobic workstation, India). After incubation, the MICs were determined and made for all the strains in triplicate.
Influence of Oxgall or Bile Salt, Hydrogen Peroxide and Low pH Tolerance on MIC
Gagnon et al., 2004 method was followed (19) in Brief; sterile flat-bottom-96-well micro-titer plates (BD-biosciences, Lincoln Park, NJ) were seeded with various Bifidobacteria. Each Bifidobacteria was treated with bile salts, hydrogen peroxide and low pH. The results presented as MIC of bile salts, hydrogen peroxide and pH that completely inhibit the growth of the organism.
Effect of Bile salts, hydrogen peroxide and low pH challenge on antibiogram
Antibiogram was developed for isolates that were subjected to different concentrations of bile salts (0.3 % physiological concentration), hydrogen peroxide (1.0-2.0 %), and low pH (pH 3.0). MIC for these bacteria was determined on MRS media only, considered as a standard media based on the above studies. 5-10 mL of mid log phase cultures were harvested at 4.0 ºC, the resulted pellet was re-suspended in an equal volumes of MRS broth (fresh) adjusted to pH 3.0 using concentrated Hcl. A second portion of the isolates were re-suspended in MRS broth containing 0.3 % (wt/vol.) bile salts and pH 6.5, the remaining pellet was re-suspended in H2O2 at its MIC. Acid challenge experiments were performed by incubating the cells anaerobically at 37 ºC for 60 min, and 90 min incubation for bile salts and H2O2. Subsequently, antibiogram was developed for all the above samples by diluting in peptone water (0.1 %, wt /vol) plating onto MRS agar plates, and incubated at 37 ºC for 12 - 16 h.
Fructose-6-phosphate Phophoketolase F6PPK assay
The Orban (2000) (20) method was followed, method in brief, the isolated pure Bifidobacterium cultures were grown in MRS broth, (Hi-Media, Mumbai, India) under anaerobic conditions at 37 °C to obtain a stationary phase culture. Subsequently, harvested by centrifugation at 10000 rpm, 4.0 °C for 10-12 min (Remi centrifuge, Mumbai, India) and was washed twice with phosphate buffer. Later, they were treated with CTAB (180 µg) for efficient lysis, and then incubated for 10–12 min at room temperature. The second step involved in the addition of a solution containing sodium fluoride (NaF), fructose-6-phosphate (F6P) and potassium acetate (KoAc) (750 µg, 20 µg, and 1.25 µg, respectively), and then incubated for 5–10 min. At the end, samples were treated with H3NO: Hcl, (195 mg), and incubated at room temperature for 10 min. The contents of the reaction along with the cell lysis components precipitated by the addition of TCA (150 mg), 4.0 N Hcl (1.0 mL) followed by a color development by the addition of ferric chloride (FeCl2 50 mg). Later, incubated the reaction for 10 min at room temperature. The appearance of a reddish violet color indicates the presence of Bifidobacterium, and yellow color indicate the absence.
PCR amplification
To identify the potential ampicillin, erythromycin, streptomycin, and tetracycline resistance genes the PCR amplifications were carried-out. Further, the alkaline lysis method for plasmid DNA isolation was followed to understand the factors responsible for antibiotic resistance are plasmid or chromosomal origin. In addition, genes coding for very common antibiotic resistance as explained above such as chloramphenicol, kanamycin, tetracycline and vancomycin considered, as they are very common antibiotics and prescribed by physicians for bacterial infections. The antibiotic resistance genes represented by the oligonucleotide /primers are given below:
Amino glycoside resistance genes: aac (3)-1a; aac (3)-1b; aac (3)11a.
β - Lactam resistance genes: bla (ACC-01-03).
Chloramphenicol resistance genes: cat 1, cat 2, cat 3 etc.
Macrolides linosamide and streptogramin resistance genes: ere (a), ere (B), mef (A), msr (B), sat (A), vat (A).
Sulfonamide resistance genes: sul1; and sul2
Tetracycline resistance genes: tet(30);tet(31)
Trimethoprim resistance genes: drfA; A1
Vancomycin resistance genes: vanA, vanB
To understand the origin of genes responsible for antibiotic resistance such as intrinsic or plasmid origin were followed by PCR amplification of respective genes with standard primers and PCR reactions. Purified genomic DNA from different Bifidobacteria were used as a template. The PCR was followed with the corresponding primers for different antibiotics by using pfu DNA polymerase and at the following parameters; denaturation 95 ºC for 45 sec’s, annealing 52 ºC for 30 sec’s, and extension 72 ºC for 60 sec’s, and final extension 72 ºC for 4.0 min. The amplified product was subjected to Agarose gel electrophoresis for analysis, followed by DNA sequencing.
Plasmid DNA isolation
Alkaline lysis method of plasmid DNA isolation was followed for only those isolates, which showed antibiotic resistance. In brief, overnight grown Bifidobacteria harvested, lysed with a lysozyme solution, followed by the sequential addition of solution 1, solution 2 and solution 3, finally centrifuged for 15-20 min at room temperature. The resulting lysate obtained was subjected to spin column, after efficient washing of the column, further it was eluted by using TE buffer (Tris, EDTA). The resulting elute was analyzed by Agarose gel electrophoresis for the presence or absence of plasmid DNA.
Results and Discussion
All the bacterial isolates and type cultures used in the study are shown in the
Table 1.
Table 2 shows the colony forming units/bacterial counts (cfu) obtained in the present study. The counts from LB, MRS, Bifido, and BHI agar media were lower than that of total cultivable bacteria. The lowest counts were observed in MRS agar, and in one sample, no colonies appeared on the MRS agar above a detection limit of 1.9 x 10
5 cfu/g (
Table 2). We also observed the various shape and sizes of the colonies on each agar media.
The growth curve studies were conducted for
B. catenulatum (Novel isolate) in aerobic and anaerobic condition. The culmination of the growth curve took around 66 h. The study began with the initial OD of 0.01, as the doubling time is around 24-30 h, therefore, first 40 h it was growing very slowly (may be a lag time). After 40 h the growth was picked-up, and started with the log period, which remained for 20 h. After 60 h of the growth curve, the lag phase began. This phase prolonged for and the samples were isolated. There is a very slight variation of the growth of
B. catenulatum in aerobic and anaerobic condition. The log phase in anaerobic condition was steep, compared with aerobically growing cultures. Overall, the growth rates were slightly different, especially at the log phase stage of the aerobic and anaerobic bacteria (
Figure 1A). In all these studies
B. adolescentis was considered as a control. This further confirms that Bifidobacteria may grow in microaerophilic condition with minor changes such as by removing the existing oxygen in the media, and injection of external N
2. This procedure does not involve skilled manpower high cost equipment/infrastructure facilities such as anaerobic gas chambers.
It was observed that the “bifid” shunt is the phosphoketolase reaction by which D-fructose-6-phosphate is converted to erythrose-4-phosphate and acetyl-1-phosphate and is adopted to test
Bifidobacterium spp. Hence, the same cultures, the positive and negative controls were subjected to the standard (f6ppk) assay. The intensity of reddish violet color formed by different
Bifidobacterium strains were spectrophotometric quantified (absorbance at 505
nm). Test tubes without cells and with cells plus all reagents except for f6ppk were used as a negative control (
Figure 1B, the control yellow color and test reddish brown color showing the isolated cultures). The results further confirm that they are Bifidobacteria isolates.
The Bifidobacteria isolates and the type strains were subjected to PCR amplification of xfp using genus-specific primers (
Figure 1C) (Supplementary Table S1), full-length xfp, conserved regions of xfp (590 bp) (
Figure 1C, and Lane 1, 2, 3 and 1D Lane 5, 8, 9), and full length 16S rRNA (
Figure 1C Lane 4, 5). As expected, the amplifications were observed with an expected DNA. The amplification of various specific and conserved regions of 16S rRNA, and f6ppk with different primers gave an indication that the isolates are Bifidobacteria spp. f6ppk is the marker gene/enzyme found in Bifidobacteria spp. The presence of the gene further confirms that they are Bifidobacteria. Therefore, we conclude biochemically through f6ppk assay, and genetically through PCR amplification of the concerned gene that the isolates are Bifidobacteria.
Farther, the amplified conserved regions of f6ppk were subjected to RFLP.
Figure 1D Lane 1, 2, 4, 6, 7 visualizes the RFLP pattern differing from one to the other.
Figure 1 D, lane 1-4, and 6, 7 are different this concludes they are different Bifidobacteria spp. A low recovery of Bifidobacteria from the selective agar media detected, particularly with the MRS and LB agar, showing that the counts are biased towards too-high bacterial numbers. Bifidobacteria are present in humans normally in numbers between (log) 9.0 and 10.5 per gram-wet weight that is deduced from the data of 65 studies. In the present work, we examined the selectivity of different media for the quantification of total anaerobes, Bifidobacteria and
Lactobacilli. Media were chose on the basis of selectivity, (from the literature data) and use in different studies. All tested media were with fresh human feces of breast-fed and formula-fed infants. No, or very rarely Bifidobacteria were found in the feces of formula-fed babies, even with a detection limit of (approximately.) 4.0 cfu/gram; Bifidobacteria were detected (2.5x10
9 cfu/g) in the majority of fecal samples from breast-fed infants.
Similarly, in a comprehensive study of intestinal bacterial communities analyzed from a library of 401 bacterial 16S rRNA gene sequences cloned from the gut content of 20-25 formula-fed infants, no Bifidobacteria were detected and similar results were observed with our samples (data not shown). These results indicate that the population of Bifidobacteria is numerically low in the gastrointestinal tract of the formula-fed infants, are in good agreement with results from the present study (
Table 2).
In the present study, distinct results of RFLP analysis of the 16S rRNA (data not shown) and xfp gene (
Figure 1C) divided the Bifidobacteria isolates into two different groups. The group 1 isolates showed the close relationship to
B. adolescentis, which was followed for the remaining isolates. Based on the RFLP pattern and 16S rRNA sequence alignment the
B. longum, B. longum longum and
brevi kept in the second group. Based on the previous reports
B. infantis and
B. longum are primarily of human origin. Here, all the isolated strains identified are,
B. catenulatum, noticing, first time that they are also human origin.
The isolated novel
B. catenulatum reported first time in breast-fed infants, and they have the ability to ferment almost all the common sugars except glucose amine (
Table 3). They are unique to ferment lactose, galactose, L-arabinose, sodium gluconate, salicin, and α-methyl-D-glucoside (
Table 3). Mikkelsen et al. 2003 (21) concluded that fermentation patterns of Bifidobacteria isolated from pigs and the type strains do not have the ability to ferment above stated sugars. Hence, we consider the
Bifidobacterium spp isolated from breast-fed infants are unique in many aspects. The fermentation of the lactose by these isolates may be associated with a high dietary intake of lactose from the maternal milk by the infants.
The low selectivity of Bifidobacteria displayed by the MRS and LB agar in the present study showed that these media are not adequate for the enumeration of Bifidobacteria in the breast-fed infant fecal material. Bifidobacteria appear on LB and MRS agar as distinct very small and milky, but similar characteristic colonies shown by
Lactobacilli. Hence, we found it difficult to recognize these characteristics when performing isolations from LB and MRS agar in the present study and these criteria were not accounted. This may partly explain the extremely low number of Bifidobacteria detected in both the media. Bifidobacteria and BHI media showed the highest number of Bifidobacteria and were the only medium that enabled recovery (
Table 3). Non-Bifidobacteria isolates were identified from both the samples, i.e. breast-fed and formula-fed infant fecal material, and their distribution on LB, BHI, MRS and Bifidobacteria agar is given in
Table 4. These results may contribute to the future formulation of improved selective agar for the enumeration of Bifidobacteria.
It is noteworthy that the isolate from formula-fed infant identified as consisting of
Actinomyces spp. In addition, growing especially well on LB, MRS agar showed the characteristic bifid shaped cellular morphology thus, the use of morphology for identifying Bifidobacteria on the selective agar media is not sufficient. The results enclosed in the
Table 6 conclude that the isolates are unique in biochemical tests such as nitrate reductase as well as Voges proskaeuers. The f6ppk assay and xfp gene amplification for the identification of Bifidobacteria essentially followed in the conclusive evaluation of the presence of Bifidobacteria on the selective agar media. The 16S rRNA specific primers and genus-specific primers have highly specific target regions within the 16S rRNA gene of Bifidobacteria, as previously validated by genus-specific
in situ hybridization (22), and DGGE (23). In conclusion, the present investigation showed that LB, MRS agar media are not adequately selective for Bifidobacteria when applied to infant fecal samples, although the Bifidobacteria agar and BHI agar exhibits superiority with respect to both selectivity and sensitivity for Bifidobacteria. The results also indicate that the population of Bifidobacteria in feces of formula-fed infants are numerically low. The isolated Bifidobacteria strains from breast-fed infants conclude that they are predominantly of
B. catenulatum. In addition, a possible new
Bifidobacterium spp was isolated which we will attempt to characterize further in our future studies.
All the strains of
Bifidobacterium showed good growth on Bifidobacterial, MH (Mueller Hinton Hi-Veg media, Hi-media laboratories, India) and most strains on TPY in aerobic as well as anaerobic conditions (
Figure 1 growth curve in aerobic and anaerobic conditions). The growth of some of the Bifidobacteria strains was inadequate for susceptibility testing on BRU (brucella agar). Among them, some showed moderate growth on BRU. Based on species-specific PCR results, all the strains of the study confirmed to be
Bifidobacterium spp (
Table 1). These bacteria in the presence of antibiotics such as ampicillin, kanamycin and erythromycin pinpoint colonies occasionally observed with the E-test method. During the process, it was observed the formation of pinpoint Bifidobacteria after treatment with amoxyclave antibiotic. These pinpoint colonies were observed irrespective of the media used for antibiogram studies. There is a possibility that the formation of pinpoint colonies indicates their susceptibility to the antibiotic exposed. This means there is a close relation between the formation of micro colonies and antibiotic exposure. In these cases the MIC’s were determined a fourth time successfully. A control strain obtained from DSM culture collection center along with a few more multi-drug resistant strains were subjected and successfully developed an antibiogram. The formation of pinpoint colonies and the phenomenon may be a pseudo-resistant, meaning no element is responsible for the resistance. Possibly the presence of tough cell wall restrict the antibiotic diffusion.
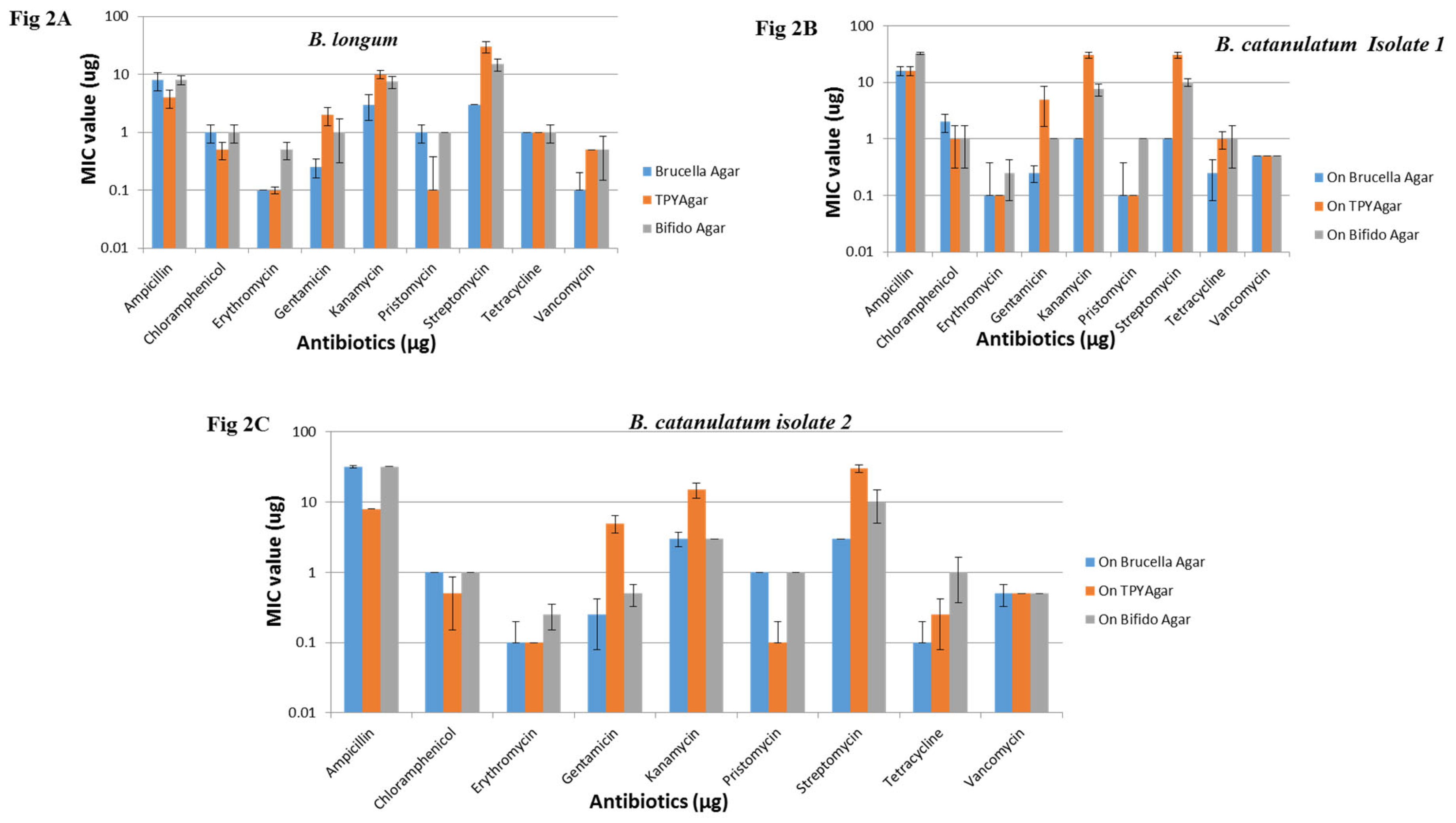
Based on the earlier reports of antibiotic resistance threshold, and according to the microbiological breakpoint regulations prescribed by the panel on additives and products or substances used in animal feed (FEEDAP) (2008) (24) all the Bifidobacteria strains were resistant to amoxyclave, two strains were resistant to ampicillin, and two were resistant to streptomycin determined with E-test only. Examples of outcome of the E-test results on different supplemental agar media are shown in
Figure 2A, B, C,
Table 4. The results obtained were compared and analyzed with the previous Bifidobacteria antibiogram reports such as appelbaum
et al., 1978 (25) and Lim et al., 1993 (26).
The PCR amplification results were altogether contradictory to the phenotypic patterns of the tested species. Known amoxyclave, ampicillin and streptomycin resistance factors failed to detect with PCR. Although they were phenotypically resistant to these antibiotics and the type-strains were also behaving similar to the test strains. Some strains showed unusual characters by showing resistance to ampicillin in Bifido and BRU media, but the identical strains observed to be sensitive in TPY media (
Figure 2A, B, C). Similarly, a few strains were showing sensitivity to kanamycin on Bifidobacteria media, seen resistant on TPY (
Table 3,
Table 4). Similar the case with streptomycin studies in three different media. The results pertaining to MIC values for different antibiotics such as amoxyclave, chloramphenicol, erythromycin, gentamycin, pristomycin, tetracycline and vancomycin on different media (Bifido, TPY and BRU) was identical (
Table 4,
Figure 2 A, B, C). MIC values of ten different Bifidobacteria strains on MH agar were not only consistent but also in the range (
Table 4). The MIC values for amoxyclave, ampicillin and vancomycin showed good correlation between the test media. The MIC values followed for ten different Bifidobacteria strains on MH agar considered as consistent, except with amoxyclave (
Table 4).
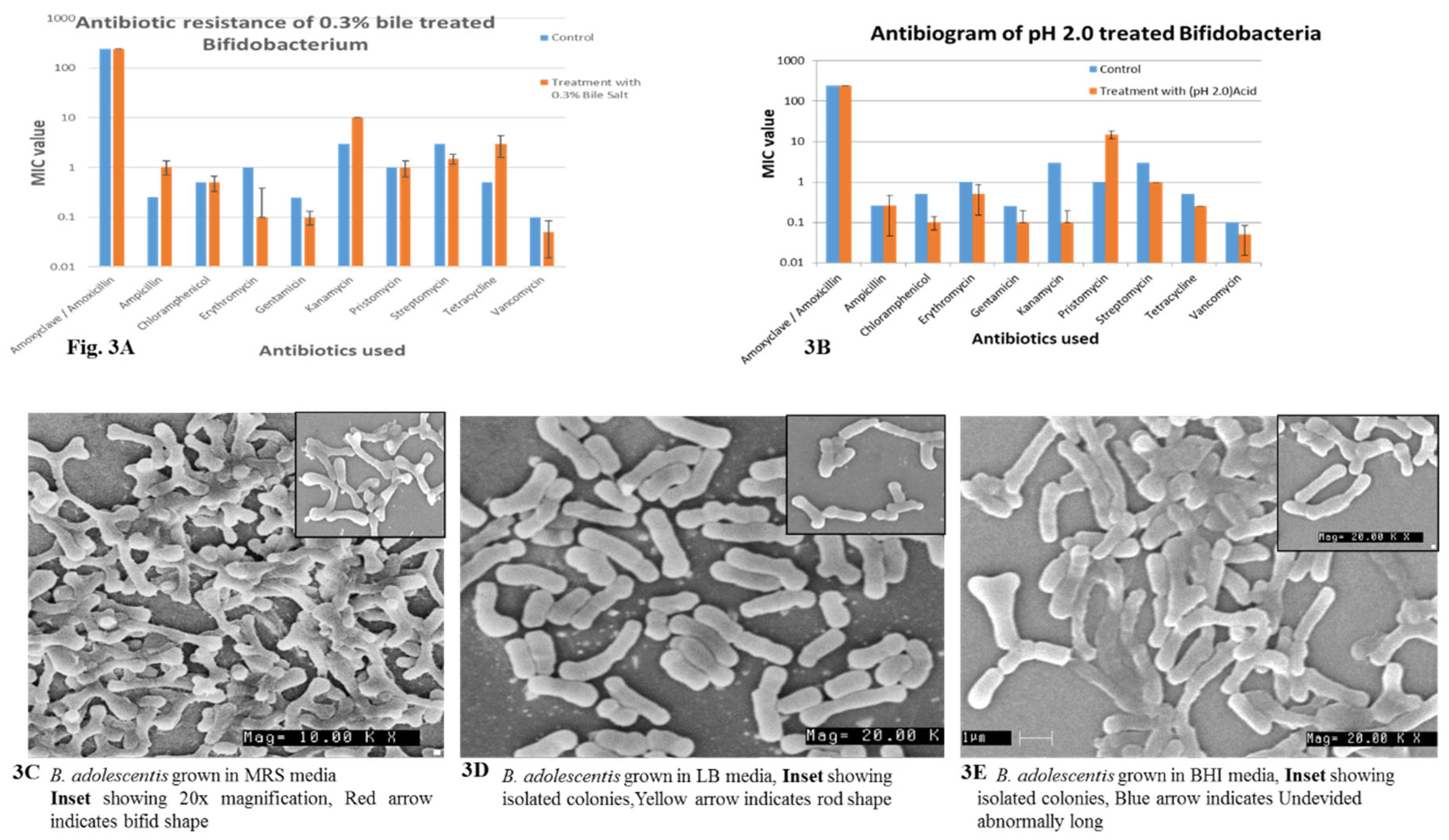
Figure 3A, B Bifidobacteria were treated with 0.3 % bile and subjected to pH 2.0 subsequently they were exposed to various antibiotics to understand their resistance before and after treatment. We observe here that the MIC values does not change with reference to amoxicillin, chloramphenicol, streptomycin and pristinomycin. The values were almost identical before and after the treatment. pH 2.0 exposed Bifidobacteria MIC values does not change with Ampicillin, Amoxicillin, and a slight variations were observed where MIC levels have decreased this is the right phenomenon, where after exposure to low pH, and Bile the cell membrane is compromised therefore the intake will be rapid and the MIC should decrease. The same phenomenon was observed in most. If they have developed intrinsic resistance the phenomenon may be reversible (
Figure 3A, B).
The Bifidobacteria grown on various media such as MRS, LB and BHI media were subjected to SEM and found that only MRS grown Bifidobacteria were in bifid shape (
Figure 3C, D, E). The bacteria grown in LB media are rod shaped, and no bifid shaped were observed (
Figure 3C, D, E). Similarly, the bacteria grown in BHI were elongated rod shaped no bifid shapes were observed (
Figure 3C, D, E). This further confirms that MRS is suitable media for its growth that does not influence cellular morphology, the other media do influence. This is in accordance with the previous reports of the influence of media on bacterial cellular morphology.
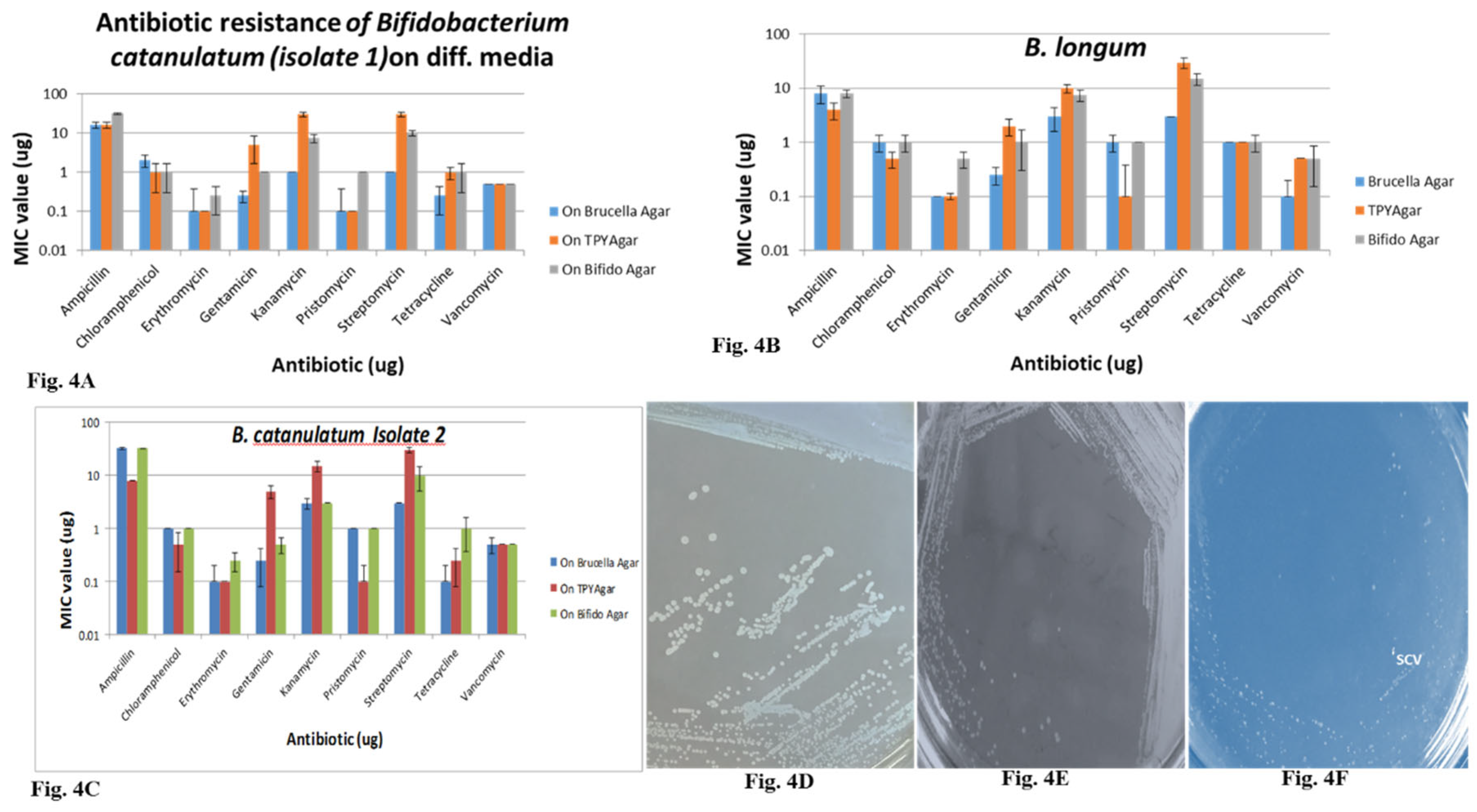
MIC values in different media containing different antibiotics differ from strain to strain, as well as media to media. MIC values of MH media are much less in comparison with the other media. The MIC values for
B. longum and our isolates in different media bifido agar, BRU, TPY and MH agar were differing each other. MIC values were higher in bifido agar in comparison with other media (
Figure 3 A, B, C;
Table 4). The MIC values were much lower in MH agar in comparison with the other. The values differed very drastically in case of ampicillin antibiotic, similar with streptomycin and kanamycin. The MIC values for remaining antibiotics were in the narrow range. The MIC values for three different strains of MH media are in the range.
B. adolescentis is one of the most sensitive microorganism showing sensitivity to all the antibiotics (
Figure 3 A, B,
Table 4), but it is intrinsically resistant to anti-tubercular drugs (30).
Cell morphological analysis: It was observed that the colonies appeared (with a naked Eye) on MRS agar were very small (looking like tip of a pin), delayed and milky white. However, colonies on BHI agar plates were big and milky white (
Figure 4D). These observations confirm that the media components influence the bacterial colony size and texture. The colonies on LB agar are much smaller, no big colonies were observed (
Figure 4 D, E, F). Bifidobacteria grown in different media show different growth pattern, SCV and BCV were observed only in the MRS grown. Very slow growing pattern was observed in LB grown.
Conclusions
Except for the ability of
B. brevi to grow in aerobic conditions such as at pH 6.0 of the media, introduction of 0.2 % glucose, oxygen depleted and N
2 containing media it is difficult to differentiate the closely related species of
Bifidobacterium. Therefore, most of the Bifidobacteria grew at the newly formulated aerobic condition (
Figure 1). Modern biology methods such as species-specific PCR-DGGE, TGGE or PFGE are helpful tools in species or strain determinations (27, 28). All the strains used in this study are shown in
Table 1, were validated confirmed as authenticated. Previous studies by Teresa et al., 2002, reported the use of trans-aldolase for identification, detection, and enumeration of Bifidobacteria (29). In this study the authors have used infant fecal samples. Most of the existing procedures for isolation, identification and enumeration of Bifidobacteria depend on sophisticated, skilled Manpower dependent, laborious and cost effective methodologies. We seek an attention by focusing on very simple and cheaper technologies through which one can isolate, identify and enumerate Bifidobacteria spp from various sources.
All the media such as Bifido, MH, TPY and MRS agar supported the growth of the Bifidobacterium. In addition, the MIC end-point obtained with the E-test on MH agar were easy to interpret. Finally, this study indicates that the MIC values obtained applying different media can vary. This, further, concludes that microbiological breakpoints differentiating resistant and susceptible strains influenced by the test conditions as well. Some of the results of the PCR amplifications as well as lack of plasmid DNA were contradictory to the tested species phenotypic patterns. Known ampicillin, amoxyclave and streptomycin resistance factors were not detected with simple PCR amplification and plasmid presence. Although the strains were phenotypically resistant to these antibiotics and controls demonstrated the expected results. Subsequent, southern hybridization experiments and their analysis revealed that three phenotypically susceptible strains demonstrated a hybridization signal with an oligonucleotide that represents bla (ACC-01), bla (ACC-02), and bla (ACC-03). One of our isolate used in the present study also seemed to possess the phosphotransferase gene (aphE), codes for streptomycin resistance. However, validation of these results failed, since no control strains were available for these specific genes, therefore, these findings should probably viewed as false positives.
To understand the antibiotic resistance of 0.3 % bile treated and pH 2.0 exposed Bifidobacteria. The results
Figure 3 A, B and
Figure 4 A, B, C shows that MIC levels drastically affected by 0.3 % bile treated samples, still shows resistance of many antibiotics. The MIC values or resistance goes down when subjected to low pH (
Figure 3,
Figure 4). This phenomenon anticipated with compromised cell wall at low pH. Therefore, they will be susceptible/sensitive for the antibiotics. The phenomenon of bile treated samples is unexplainable, as no reports exists how bile salts involved in compromising the cell wall.
We were puzzled to see various sizes of bacteria when they grew on MRS agar. They were very small and milky white. However, they were big and milky white colonies on BHI agar. A report by Proctor 2006 clearly envisaged that, small colony variant (SCV) phenotypes are the survival strategies of bacteria during stress conditions here it is antibiotic exposure (30). To date, this phenomenon reported only in the intracellular pathogens such as
Salmonella Typhi (
St),
Mycobacterium tuberculosis (
Mtb), and
Listeria monocytogenes (LM). The identical phenomenon observed in case of Bifidobacteria
, which is non-pathogenic, and a probiotic. Upon 16S rRNA comparison of SCVs and wild-type established that there is no variance. Therefore, it is essential to develop tools to differentiate them. Very less and small isolated colonies were observed on LB agar, and no BCVs were observed (
Figure 4D, E, F).
Table legends
Table 1: The bacterial strains used in this study
Table 2: A. Bacterial counts of total culturable anaerobic bacteria, B. Bacterial counts obtained for the selective agar media, and C number of Bifidobacteria isolates. Recovered from F1 to F3 Breast fed infant faecal samples, F4 to F6 Formula fed infant faecal samples. Total sample were grown in respective media as stated in materials and methods in order to find out total colony forming units (cfu) counts and specific cfu counts of Bifidobacteria under similar conditions.
Table 3: Corresponds to fermentation patterns of different bacteria in the same conditions followed for control. Where, the isolates were grown at 37 ˚C in anaerobic conditions for 2-3 days. The 50 μl of culture was subjected to fermentation study patterns by using Hi-media Hi-carbohydrate kit. The change of color indicates considered for its results.
Table 4: MICs of antibiotics for Bifidobacteria
Table 5: MIC Values of Bifidobacterium spp on different Media
Table 6: Biochemical test patterns of Bifidobacteria isolated in this study and DSM type strains.
The nucleotide sequences deposited in NCBI
The nucleotide data reported in this manuscript submitted to the genebank nucleotide database under the accession numbers FN600543, FN600542 and FN600541.
Institutional Review Board Statement’ of the article”
All subjects gave their informed consent for inclusion before they participated in the study. The study was conducted in accordance with the Declaration of Helsinki, and the protocol was approved by the Ethics Committee of 2010/Probiotic/id.
Acknowledgments
We acknowledge CSIR-CFTRI, and CSIR-IMTECH for facilities and the funding under RC project (of IMTECH, Chandigarh).
Conflicts of Interest
Authors state that there is no conflict of interest.
References
- Crittenden, RG (1999) Prebiotics, In G.W. Tannock (ed.), Probiotics: a critical review. Horizon scientific press, p.141-156, Wymondham, United Kingdom.
- Hartemink, R., Rombouts, F.M (1999) Comparison of media for the detection of Bifidobacteria, Lactobacilli and total anaerobes from faecal samples. J of Microbiol Meth. 36,181-192. [CrossRef]
- Djouzi, Z., Andieux, C (1997) Compared effects of three oligosaccharides on metabolism of intestinal microflora in rats inoculated with human faecal flora. Br. J. Nutr. 78: 313-324. [CrossRef]
- Gavini, F., Pourcher, A.M., Neut, C., Monget, D., Romond, C., Oger, C., Izard, D (1991) Phenotypic differentiation of Bifidobacteria of human and animal origin. Int. J. Syst. Bacteriol. 41: 548-557. [CrossRef]
- Nemcova, R., Bomba, A., Gancarcikova, S., Heich, R., Guba, P (1999) Study of the effect of Lactobacillus paracasei and fructo-oligosaccharides on the faecal microflora in weaning piglets. Berl. Munch. Tierazil. Wochenchr.112; 225-228.
- Hartemink, R., Kok, B.J. Weenk, G.H., Rombouts, FM (1996) Raffinose –Bifidobacterium (RB) agar, a new selective medium for Bifidobacteria. J. Microbiol. Meth. 27: 33-43. [CrossRef]
- Ruda, V., Sirotek, K., Petr, J (1999) Evaluation of selective media for Bifidobacteria in poultry and rabbit cecal samples. J. Vet. Med. Ser. B 46: 369-373. [CrossRef]
- Rada, V (1997) Detection of Bifidobacterium species by enzymatic methods and antimicrobial susceptibility testing. Biotechnol. Tech. 11; 909-912.
- Yildrim, Z, Johnson, MG (1998) Characterization and antimicrobial spectrum of bifidocin B, a bacteriocin produced by Bifidobacterium bifidum NCFB 1454. J. Food Protect, 61:47-51.
- Bernardeau, M., Vernoux, J.P. Henri-Dubernet, S. and Gueguen, M (2007) Safety assessment of dairy microorganisms: the lactic acid bacteria: Int j Food Microbiol, 126: 278-285. [CrossRef]
- Davison, J (1999). Genetic exchange between bacteria in the environment. Plasmid, 42, 73–91. [CrossRef]
- Mathur, S., and Singh, R (2005) Antibiotic resistance in food lactic acid bacteria—A review. Int J Food Microbiol, 105, 281–295. [CrossRef]
- Alesting, K., H. Carlberg, C.E. Nord, and B. Trollfors (1983) Effect of cefoperazone on faecal flora. J. Antimicrob. Agents Chemother. 12:163-167. [CrossRef]
- Black., K. Einarsson, A. Lidbeck, K. Orrhage, and C. E. Nord (1991) Effect of lactic acid producing bacteria on the human intestinal micro flora during ampicillin treatment. Scand J Infect Dis. 23:247-254. [CrossRef]
- Dhanshree, Raman Parkesh, and Kammara. Rajagopal (2018) Bifidobacterium adolescentis is intrinsically resistant to anti-tubercular drugs. Sci rep, 8; 11897. [CrossRef]
- Klare, I., Konstable, G., Huys, G., Vankerchkhoven, V., Kahlmeter, g., Hildebrandt, B., Muller-Bertling, S., and Witte, W (2005) Evaluation of new broth media for micro dilution antibiotic susceptibility testing of lactobacilli, pediococci, lactococci, and bifidobacteria. Appl and Environ Microbiol, 71: 8982-8986. [CrossRef]
- Alander, M., Satokari, R., Korpela, R., Saxelin, M., Vilponnen-Salmella, T., Mattila-Sandholm, T., and Von Wright, A (1999) Persistence of colonization of human colonic mucosa by a probiotic strain, Lactobacillus rhamnosus GG, after oral consumption. Appl and Environ Microbiol, 65: 351-354. [CrossRef]
- NCCLS (2004) Methods for antimicrobial susceptibility testing of anaerobic bacteria; approved standard –sixth edition.M11-A6.
- Gagnon, E., Griffiths, D., Ruffin, Barett, H., Rossman, J., Ogra, PL (1993) Effectiveness of Bifidobacterium bifidum in experimentally induced MRV infection: dietary implications in formulas for new-borns. Endocrine Regulations, 27:223-229.
- Orban, J I., Patterson, JA (2000) Modification of the phosphoketolase assay for rapid identification of Bifidobacteria. J Microbiol Meth, 40(3):221-4. [CrossRef]
- Kathleen Sim, Michael J. Cox, Harm Wopereis, Rocio Martin, Jan Knol, Ming-Shi Li, William O. C. M. Cookson, Miriam F. Moffatt, J. Simon Kroll (2012) Improved Detection of Bifidobacteria with Optimized 16S rRNA-Gene Based Pyrosequencing. PLoS ONE 7(3): e32543. [CrossRef]
- Mikkelsen, N.E., Johansson, K., Karlsson, A., Knecht, W., Andersen, G., Piskur, J., Munch-Petersen, B., Eklund, H (2003) Structural basis for feedback inhibition of the de-oxyribonucleoside salvage pathway: studies of the Drosophila de-oxyribonucleoside kinase. Biochemistry 42(19): 5706--5712. [CrossRef]
- E. Kheadr, N. Dabour, C. Le Lay, C. Lacroix, and I. Fliss (2007) Antibiotic Susceptibility Profile of Bifidobacteria as affected by Oxgall, Acid, and Hydrogen Peroxide Stress. J. Antimicrob agents Chemother. 169-174. [CrossRef]
- Scientific Opinion. Guidance on the assessment of bacterial susceptibility to antimicrobials of human and veterinary importance. EFSA Panel on Additives and Products or Substances used in Animal Feed (FEEDAP) European Food Safety Authority (EFSA), Parma, Italy Endorsed for public consultation on 1 February (2012).
- Appelbaum, P.C., and S.A. Chatterton (1978) Susceptibility of anaerobic bacteria to ten antimicrobial agents. J. Antimicrob Agents Chemother.14:371-376. [CrossRef]
- Lim, K.S., C.S. Huh, and Back Y.J. (1993) Antimicrobial susceptibility of Bifidobacteria. J Dairy Sci. 76; 2168-2174. [CrossRef]
- Proctor RA, von Eiff C, Kahl BC, Becker K, McNamara P, Herrmann M, Peters G (2006) Small colony variants: a pathogenic form of bacteria that facilitates persistent and recurrent infections. Nat Rev Microbiol. 4(4):295-305. [CrossRef]
- Mitsuoka, T., Terada, A., Morishita, Y (1973) Die Dram Flora von Mensch und Tier. Goldschmidt informer 2, 23-41.
- Teresa Requena, Jeremy Burton, Takahiro Matsuki, Karen Munro, Mary Alice Simon, Ryuichiro Tanaka, Koichi Watanabe, and Gerald W. Tannock (2002) Identification, Detection, and Enumeration of Human Bifidobacterium spp by PCR Targeting the Transaldolase Gene. Appl Environ Microbiol.68(5): 2420–2427. [CrossRef]
- Bifid Shape Is Intrinsic to Bifidobacterium adolescentis. Dhanashree, Sharika Rajashekharan, Balamurugan Krishnaswamy and Rajagopal Kammara (2017) Front. Microbiol. [CrossRef]
Figure 1.
Growth curve of Bifidobacteria in Aerobic and anaerobic conditions. Fig. 1B: Phosphoketolase assay was followed according to Orban et al., method. In brief, harvested cells were washed with phosphate buffer, re-suspended in 1.0 ml of phosphate buffer, and were subjected to CTAB. To the lysed cells 250 µl each of sodium fluoride, fructose 6 phosphate substrates added, mixed and vortexed. Further, they were incubated at 37 ˚C for 30 min. following incubation with 1.5 ml of hydroxylamine hydrochloride. Finally, they were treated with TCA and ferric chloride, and formation of color was recorded qualitatively. Fig. 1C: Genomic DNA was purified from respective Bifidobacteria, subjected to 16S rRNA amplification (conserved regions as well as whole gene). The same DNA was further, used for amplification of xfp gene. The amplified products were separated on 2.0 % Agarose gel along with a marker. Sequence of loading, Lane 1. Molecular weight marker of 500 bp, 750, 1000, 2000, 2500, 3000, 4000, 5000, 6000, 8000 and 10000bp, Lane 2. Xfp gene product, Lane 3 and 4. 16S rRNA conserved amplified regions, Lane 5 and 6. Amplified 16S rRNA. Fig. 1D: RFLP pattern of DNA fragments corresponding to conserved regions of xfp. Amplified conserved region such as 590 bp PCR product of different Bifidobacterial spp were single band purified from Agarose gel, and subjected to Xho1 restriction enzyme digestion as directed by manufacturer. The reaction mixture separated on Agarose gel. Sequence of loading is as follows, Lane M.100 bp molecular weight marker, Lane 1. B. adolescentis conserved region digested with Xho1 restriction enzyme. Lane 2. B. longum longum, Lane 3. B. longum infantis, Lane 4. B. breve, Lane 5. 590 bp product as a control, Lane 6 and 7. B. Isolates, Lane 8. PCR product control incubated without enzyme. Lane 9. PCR product amplified.
Figure 1.
Growth curve of Bifidobacteria in Aerobic and anaerobic conditions. Fig. 1B: Phosphoketolase assay was followed according to Orban et al., method. In brief, harvested cells were washed with phosphate buffer, re-suspended in 1.0 ml of phosphate buffer, and were subjected to CTAB. To the lysed cells 250 µl each of sodium fluoride, fructose 6 phosphate substrates added, mixed and vortexed. Further, they were incubated at 37 ˚C for 30 min. following incubation with 1.5 ml of hydroxylamine hydrochloride. Finally, they were treated with TCA and ferric chloride, and formation of color was recorded qualitatively. Fig. 1C: Genomic DNA was purified from respective Bifidobacteria, subjected to 16S rRNA amplification (conserved regions as well as whole gene). The same DNA was further, used for amplification of xfp gene. The amplified products were separated on 2.0 % Agarose gel along with a marker. Sequence of loading, Lane 1. Molecular weight marker of 500 bp, 750, 1000, 2000, 2500, 3000, 4000, 5000, 6000, 8000 and 10000bp, Lane 2. Xfp gene product, Lane 3 and 4. 16S rRNA conserved amplified regions, Lane 5 and 6. Amplified 16S rRNA. Fig. 1D: RFLP pattern of DNA fragments corresponding to conserved regions of xfp. Amplified conserved region such as 590 bp PCR product of different Bifidobacterial spp were single band purified from Agarose gel, and subjected to Xho1 restriction enzyme digestion as directed by manufacturer. The reaction mixture separated on Agarose gel. Sequence of loading is as follows, Lane M.100 bp molecular weight marker, Lane 1. B. adolescentis conserved region digested with Xho1 restriction enzyme. Lane 2. B. longum longum, Lane 3. B. longum infantis, Lane 4. B. breve, Lane 5. 590 bp product as a control, Lane 6 and 7. B. Isolates, Lane 8. PCR product control incubated without enzyme. Lane 9. PCR product amplified.
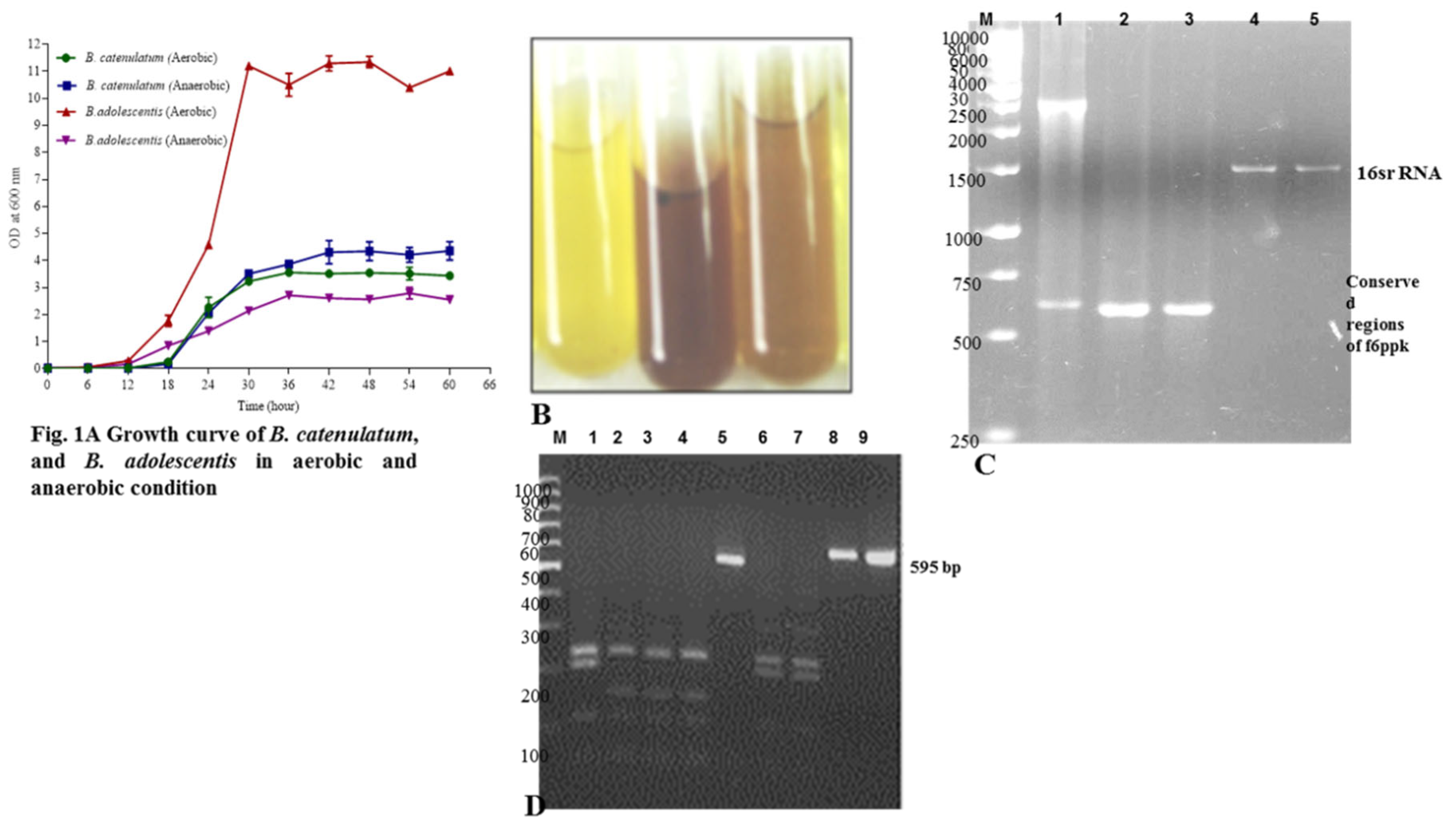
Figure 2.
Fig. 2A. MIC values of B. longum grown on different media ranging from Brucella agar, TPY agar and Bifido agar in presence of various antibiotics. Fig. 2B. MIC values of B. catenulatum (Isolate 1) grown on different media ranging from Brucella agar, TPY agar and Bifido agar in presence of various antibiotics. Fig. 2C. MIC values of B. catenulatum (Isolate 2) grown on different media ranging from Brucella agar, TPY agar and Bifido agar in presence of various antibiotics.
Figure 2.
Fig. 2A. MIC values of B. longum grown on different media ranging from Brucella agar, TPY agar and Bifido agar in presence of various antibiotics. Fig. 2B. MIC values of B. catenulatum (Isolate 1) grown on different media ranging from Brucella agar, TPY agar and Bifido agar in presence of various antibiotics. Fig. 2C. MIC values of B. catenulatum (Isolate 2) grown on different media ranging from Brucella agar, TPY agar and Bifido agar in presence of various antibiotics.
Figure 3.
Fig.3A. Antibiotic resistance of 0.3 % bile treated B. catenulatum Fig.3B. Antibiogram of pH 2.0 treated B. catenulatum (isolate 1). Fig. 3C. B. adolescentis grown in MRS media, Inset showing 20x magnification. Red arrow indicates bifid shape. Dark coloured arrow indicate for big colony variants (BCV). Fig. 3D. B. adolescentis grown in LB media, Inset showing 20x magnification. Yellow arrow indicates rod shape. Fig. 3E. B. adolescentis grown in BHI media, Inset showing 20x magnification. Blue arrow indicates un-devided, abnormally long Bifidobacteria. Light green arrow indicate small colony variants (SCV).
Figure 3.
Fig.3A. Antibiotic resistance of 0.3 % bile treated B. catenulatum Fig.3B. Antibiogram of pH 2.0 treated B. catenulatum (isolate 1). Fig. 3C. B. adolescentis grown in MRS media, Inset showing 20x magnification. Red arrow indicates bifid shape. Dark coloured arrow indicate for big colony variants (BCV). Fig. 3D. B. adolescentis grown in LB media, Inset showing 20x magnification. Yellow arrow indicates rod shape. Fig. 3E. B. adolescentis grown in BHI media, Inset showing 20x magnification. Blue arrow indicates un-devided, abnormally long Bifidobacteria. Light green arrow indicate small colony variants (SCV).
Figure 4.
Fig.4A. Antibiotic resistance of B. catenulatum (Isolate1) on different bacterial media such as Brucella agar, TPY and Bifido agar. Fig.4B. Antibiotic resistance of B. longum on different bacterial media such as Brucella agar, TPY and Bifido agar. Fig.4C. Antibiotic resistance of B. catenulatum (Isolate 2) on different bacterial media such as Brucella agar, TPY and Bifido agar. Fig.4D. Naked eye observation of MRS media grown Bifidobacteria. Fig.4E. Naked eye observation of Bifidobacteria grown in BHI media. Fig.4F. Naked eye observation of Bifidobacteria grown in LB media.
Figure 4.
Fig.4A. Antibiotic resistance of B. catenulatum (Isolate1) on different bacterial media such as Brucella agar, TPY and Bifido agar. Fig.4B. Antibiotic resistance of B. longum on different bacterial media such as Brucella agar, TPY and Bifido agar. Fig.4C. Antibiotic resistance of B. catenulatum (Isolate 2) on different bacterial media such as Brucella agar, TPY and Bifido agar. Fig.4D. Naked eye observation of MRS media grown Bifidobacteria. Fig.4E. Naked eye observation of Bifidobacteria grown in BHI media. Fig.4F. Naked eye observation of Bifidobacteria grown in LB media.
Table 1.
Strains used in this study.
Table 1.
Strains used in this study.
| Sample |
Bacterial Counts of total |
LB Agar |
BHI Agar |
MRS Agar |
Bifido Agar |
| Culturable bacteria (CFU/g) |
A |
B |
C |
A |
B |
C |
A |
B |
C |
A |
B |
C |
| F1 |
8 × 10 9
|
3.5 × 10 9
|
25 |
1 |
9 × 10 9
|
25 |
8 |
3×105
|
25 |
1 |
8.5 × 109
|
22 |
10 |
| F2 |
7 × 108
|
4 × 10 8
|
22 |
0 |
6 × 10 8
|
25 |
2 |
2×107
|
22 |
0 |
9 × 107
|
22 |
9 |
| F3 |
9 × 109
|
8 × 10 7
|
25 |
1 |
7 × 10 9
|
23 |
5 |
3×107
|
23 |
0 |
9 × 109
|
25 |
8 |
| F4 |
6 × 107
|
3 × 105
|
25 |
0 |
3 × 10 8
|
25 |
1 |
2×105
|
25 |
0 |
4 x 109
|
25 |
0 |
| F5 |
5 × 107
|
3 × 10 7
|
24 |
0 |
5 × 10 9
|
23 |
0 |
2×106
|
22 |
0 |
5 × 108
|
25 |
1 |
| F6 |
6 × 109
|
3 × 10 4
|
24 |
0 |
7 × 10 9
|
23 |
0 |
2×105
|
20 |
0 |
8 × 10 9
|
22 |
0 |
Table 2.
A. Bacterial counts of total culturable anaerobic bacteria, B. Bacterial counts obtained for the selective agar media, and C. Number of Bifidobacteria isolates. Recovered from F1 to F3 Breast fed infant fecal samples, F4 to F6 Formula fed infant fecal samples.
Table 2.
A. Bacterial counts of total culturable anaerobic bacteria, B. Bacterial counts obtained for the selective agar media, and C. Number of Bifidobacteria isolates. Recovered from F1 to F3 Breast fed infant fecal samples, F4 to F6 Formula fed infant fecal samples.
Table 3.
Fermentation patterns of Bifidobacteria isolated in this study and Type strain of DSM.
Table 3.
Fermentation patterns of Bifidobacteria isolated in this study and Type strain of DSM.
| |
Bifidobacterial isolates |
|
| Substrate |
B.breve (DSM) |
| |
Group I |
Group II |
|
| Lactose |
+ |
+ |
+ |
| Xylose |
+ |
+ |
+ |
| Maltose |
+ |
+ |
+ |
| Fructose |
+ |
+ |
+ |
| Dextrose |
+ |
+ |
+ |
| Galactose |
+ |
+ |
+ |
| Raffinose |
+ |
+ |
+ |
| Trehalose |
+ |
+ |
+ |
| Melibiose |
+ |
+ |
+ |
| Sucrose |
+ |
+ |
+ |
| L-Arabinose |
+ |
+ |
+ |
| Mannose |
+ |
+ |
+ |
| Inulin |
+ |
+ |
+ |
| Sodium Gluconate |
+ |
+ |
+ |
| Glycerol |
+ |
+ |
+ |
| Salicin |
+ |
+ |
+ |
| Glucosamine |
- |
- |
- |
| Dulcitol |
+ |
+ |
+ |
| Inositol |
+ |
+ |
+ |
| Sorbitol |
+ |
+ |
+ |
| Mannitol |
+ |
+ |
+ |
| Adonitol |
+ |
+ |
+ |
| α-Methyl-D-Mannoside |
+ |
+ |
+ |
| Xylitol |
+ |
+ |
+ |
| ONPG |
- |
- |
- |
| Esculin Hydrolysis |
- |
- |
- |
| D-Arabinose |
+ |
+ |
+ |
| Sorbose |
+ |
+ |
+ |
| Citrate utilization |
- |
- |
- |
| Malonate utilization |
- |
- |
- |
Table 4.
MICs of antibiotics for Bifidobacteria
Table 4.
MICs of antibiotics for Bifidobacteria
| |
MIC ((µg/ml) of the following antibiotica for the indicated organism on MH media |
| Organismb
|
Amo |
Amp |
Chl |
Ery |
Gen |
Kan |
Pri |
Str |
Tet |
Van |
| B. adolescentis |
0.5 |
0.016 |
0.1 |
0.25 |
0.01 |
3.0 |
1.0 |
1.0 |
0.01 |
0.5 |
| B. animalis |
>240.0 |
0.512 |
0.1 |
0.1 |
0.25 |
3.0 |
1.0 |
5.0 |
0.1 |
0.05 |
| B. asteroides |
>240.0 |
0.512 |
0.1 |
0.1 |
0.25 |
3.0 |
0.1 |
5.0 |
0.1 |
0.1 |
| B. breve |
>240.0 |
0.512 |
0.1 |
0.5 |
0.25 |
5.0 |
1.0 |
3.0 |
2.0 |
0.1 |
| B. indicum |
0.2 |
0.032 |
0.1 |
0.5 |
0.1 |
1.0 |
1.0 |
1.0 |
0.01 |
0.05 |
| B. infantis |
1.0 |
0.032 |
0.01 |
0.5 |
0.1 |
1.0 |
1.0 |
1.0 |
0.01 |
0.1 |
| B. longum |
>240.0 |
8.0 |
1.0 |
0.25 |
0.5 |
3.0 |
1.0 |
3.0 |
1.0 |
0.1 |
| B. thermoacidophilum |
>240.0 |
0.512 |
0.1 |
0.5 |
0.25 |
5.0 |
1.0 |
7.5 |
1.0 |
0.1 |
| Isolate1 |
>240.0 |
0.512 |
0.5 |
0.25 |
0.25 |
3.0 |
1.0 |
3.0 |
1.0 |
0.1 |
| Isolate2 |
>240.0 |
0.256 |
0.5 |
1.0 |
0.25 |
3.0 |
1.0 |
3.0 |
0.5 |
0.1 |
Table 5.
MIC Values of Bifidobacterium spp on different Media a Novel isolate from seven month old infant fecal material b Control strain.
Table 5.
MIC Values of Bifidobacterium spp on different Media a Novel isolate from seven month old infant fecal material b Control strain.
| |
|
Brucella Agar |
TPYAgar |
Bifido Agar |
| S. |
|
| No |
Antibiotics |
| |
(µg/ml) |
bB. longum |
aB. |
aB. |
aB.longum
|
aB. |
aB. |
bB. longum |
aB. |
aB. |
| |
|
catenulatum |
catenulatum |
catenulatum |
catenulatum |
catenulatum |
catenulatum |
| 1. |
Amoxyclave / Amoxicillin |
>240 |
>240 |
>240 |
>240 |
>240 |
>240 |
>240 |
>240 |
>240 |
| 2. |
Ampicillin |
8 |
16 |
32 |
4 |
16 |
8 |
8 |
32 |
32 |
| 3. |
Chloramphenicol |
1 |
2 |
1 |
0.5 |
1 |
0.5 |
1 |
1 |
1 |
| 4. |
Erythromycin |
0.1 |
0.1 |
0.1 |
0.1 |
0.1 |
0.1 |
0.5 |
0.25 |
0.25 |
| 5. |
Gentamicin |
0.25 |
0.25 |
0.25 |
2 |
5 |
5 |
1 |
1 |
0.5 |
| 6. |
Kanamycin |
3 |
1 |
3 |
10 |
30 |
15 |
7.5 |
7.5 |
3.0 |
| 7. |
Pristomycin |
1 |
0.1 |
1 |
0.1 |
0.1 |
0.1 |
1 |
1 |
1 |
| 8. |
Streptomycin |
3 |
1 |
3 |
30 |
30 |
30 |
15 |
10 |
10 |
| 9. |
Tetracycline |
1 |
0.25 |
0.1 |
1 |
1 |
0.25 |
1 |
1 |
1 |
| 10. |
Vancomycin |
0.1 |
0.5 |
0.5 |
0.5 |
0.5 |
0.5 |
0.5 |
0.5 |
0.5 |
Table 6.
Biochemical test patterns of Bifidobacteria isolated in this study and type strain from DSM.
Table 6.
Biochemical test patterns of Bifidobacteria isolated in this study and type strain from DSM.
| Name of the test |
Bifidobacterial isolates |
B.breve (DSM) |
| Group I |
Group II |
| 1.ONPG |
- |
- |
- |
| 2.Lysine Utilization |
- |
- |
- |
| 3.Ornithine Utilization |
- |
- |
- |
| 4.Urease |
- |
- |
- |
| 5.Phenylalanine Deamination |
- |
- |
- |
| 6.Nitrate reductase |
+ |
- |
- |
| 7.HS production |
- |
- |
- |
| 8.Citrate utilization |
- |
- |
- |
| 9.Voges proskaeuers |
- |
+ |
- |
| 10.Methyl red |
+ |
+ |
+ |
| 11.Indole |
- |
- |
- |
| 12.Malonate utilization |
- |
- |
- |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).