1. Introduction
Since March 2020, the world is facing a biological threat caused by the emergence of a new virus: The Severe Acute Respiratory Syndrome-Coronavirus 2 (SARS-CoV-2) [
1]. Named by the International Committee on Taxonomy of Viruses (CITV), SARS-CoV-2 is a 30 kb enveloped virus with a helical capsid whose genome consists of single-stranded, non-segmented, positive polarity ribonucleic acid (RNA) [
2]. It has four essential structural proteins: spike surface protein (S), envelope protein (E), membrane protein (M), and nucleocapsid protein (N) [
3]. To usurp the human organism, its spike protein (S) binds by affinity and avidity forces to cellular angiotensin-converting enzyme 2 (ACE 2) receptors mainly expressed by respiratory epithelial cells from the nasal mucosa and secondarily by type 2 pneumocytes hence its tropism for the respiratory tract and thus the preferential pulmonary involvement where SARS-CoV-2 causes emerging and potentially lethal atypical pneumonitis [
4].
Since its appearance almost four waves of SARS-CoV-2 have been experienced so far [
5]. As of 27 November 2022, the world has recorded 637 million confirmed cases and 6.6 million deaths globally [
5]. In general, the United States of America (USA) represents so far the most affected country in the world with above 98,972,375 cases [
6]. In Africa, COVID has affected all 47 African Region countries with 8.887.814 cumulative cases, which represented around two percent of the infections around the world [
7]. South Africa is the most drastically affected country, with more than 3.6 million infections, followed by Cameroon with 123 993 cases of COVID-19, of which 1965 died and above 7774 were cured [
7].
Many aspects of COVID-19 remains unknown as it is asymptomatic in about 50% of cases where the subject recovers spontaneously (acute or moderate form) [
8]. Never-theless, in these acute forms, symptoms such as cough, moderate fever, asthenia, headache, and loss of taste and/or smell may be noted [
9]. In addition, in the absence of a cure, the contamination of the subject evolves into an infection characterized by the appearance of symptoms. These symptoms appear progressively and correlates with the severity of the SARS-CoV-2 infection, which is the severe form [
9]. This severe form would be the result of a particular exaggerated inflammatory reaction charac-terized by a cytokine storm [
3]. Subjects of younger age are described as less likely to develop severe COVID-19 forms than adults.
Transmission of SARS-CoV-2 in the young depends on the local transmission rates; the types of variants circulating; the epidemiology of COVID-19 among children, adoles-cents, and adults; vaccine coverage for those eligible; and mitigation measures in place to prevent transmission [
10]. At the heart of the COVID-19 pandemic, population re-striction measures, including school closures and the introduction of barrier measures were practiced worldwide to curb the spread of the pandemic [
11]. Some evidence indicates that SARS-CoV-2 might spread more easily within high school settings than in elementary school settings [
12,
13,
14,
15] suggesting that SARS-CoV-2 transmission among children and adolescents is relatively rare, particularly when prevention strategies were in place[
16]. However, close contact with persons with COVID-19, attending gatherings, and having visitors at home can increase its trans-mission rate [
17]. In Cameroon, epidemiological data of COVID-19 infection in elementary as well as high School are rare. To limit the spread of the pandemic, easy access to testing is important. However, the available diagnostic tools have not been in many settings. This study sought to determine the infection rate (PCR-positivity) and spread (IgM/IgG) of SARS-CoV-2 in settlements around 3 Cameroonian universities as well as suggest a suitable diagnostic algorithm to SARS-CoV -2 there-in.
2. Materials and Methods
A. Study design and population
A multicenter cross-sectional study was conducted from September 28, 2020 to September 06, 2021, among 291 students in three privates universities from two regions of Cameroon: Université des Montagnes (UdM-Bangangte) located in Bangangté, in the west region; the Higher Institute of Biological and Applied Sciences (ISSBA-Yaounde) and the Estuary Academy and Strategic Institute (IUEs / INSAM-Yaounde), located both in Yaounde, in the central region. Several reasons guided the choice of these two towns (regions): i) Both are the most populated and highly heterogeneous cities in their respective regions; ii) Yaounde, the capital city of Cameroon is bigger than Bangangte and is equipped with an international airport of entry of foreigners; iii) Yaounde was one of the starting point of the COVID-19 and thus, was among the first site endorsed by the Ministry of Public Health for molecular diagnostic and management of COVID–19 cases; iv) Unlike Bangangte, Yaounde was basically among the two first sites of multifunctional reference laboratories for SARS–CoV–2 molecular detection in Cameroon before extension to others cities across country thereafter.
After obtaining the required administrative authorizations, we included in the study all students aged>18 years old from those three private universities (symptomatic and/or not) who signed the inform consent. Conversely, patients who declined invitation and refused to sign informed consent form were excluded. Additionally, those admitted at intensive care units, and individuals who declined either blood stick or nasopharyngeal swab were also excluded. From each participant, blood was collected by a prick on the middle finger and nasopharyngeal swab respectively for SARS-CoV-2 IgM/IgG antibodies and RNA detection. The protocol of sample collection was performed WHO guidelines for COVID-19 samples collection. And samples were transferred using a cooler at +4˚C to the Center for Research on emerging and reemerging Diseases (CREMER).
An individual was considered symptomatic when he/she had at least three COVID-19-related symptoms such as taste disorders, loss of smell, fever, dry cough, breathing/shortness of breath, chest pain/pressure, aches and pains, sore throat, diarrhea, conjunctivitis,headache(
https://www.who.int/health-topics/coronavirus#tab=tab_3). The study aim was well explained to all the participants, those who agreed to participate were recruited consecutively and filled the structured questionnaire with a member of the research team. Information on participant’s sex, age, university, history of COVID-19 symptoms, treatment, and comorbidity were collected. B. Ethics Approval and Consent
B. Ethics Approval and Consent
The research proposal was evaluated, and the ethical clearance was obtained from the Institutional Review Board (IRB) of the National Ethics Committee for Human Health Research (N°: 2020/05/1218/CE/CRERSHC/SP from 06th May 2020). Additionally, we obtained authorization from the directors of the three Universities selected. An information note was given to all the eligible participants, who then provided their written informed consent before enrollment into the study. The confidentiality of study participants was secured by the use of identification codes.
C. Determination of Minimum Sample Size
The minimum sample size was obtained using the standard formula:
n= z2.p (1-p)/m2, with “z” = the standard deviation of 1.96 (95% confidence interval); “p”= seroprevalence of SARS-CoV-2 antibodies found in Cameroon (IgM: 20% and IgG: 24%) [
9], “m”=degree of precision (0.05) and “n”= minimum sample size.
D. Sample Collection and Conservation
Eligible participants who gave their approval were subjected to the collection and blood and a nasopharyngeal fluid.
- 1.
Blood Sampling
Blood was collected according to the aseptic and barrier measures of COVID-19. One drop of whole blood was required for the “on-site” testing according to the SOP presented to us by the " Abbott PanbioTMCOVID-19 " rapid diagnostic test kit (Panbio COVID-19 IgG/IgM Rapid Test | Point-of-Care – Abbott (globalpointofcare.abbott))
- 2.
Nasopharyngeal sampling
Safety procedures were performed according to procedural references for safe nasopharyngeal swab collection previously described [
18,
19]. Samples collected on nasopharyngeal swabs were moved to collection tubes containing a viral transport medium by breaking the swab at the groove. Sealed state and labeling was checked, and surface disinfection was performed [
20].
- 3.
Samples Transportation
Antibodies testing was performed on-site, whereas nasopharyngeal swabs were transported to CREMER, Yaounde for diagnosis. All Viral transport mediums containing nasopharyngeal swabs were temporarily stored between 2-8°C immediately after collection. Nasopharyngeal swabs collected at ISSBA-Yaounde and IUEs/INSAM in Yaounde were directly transferred to CREMER, and analyzed within 48 hours. On the other sites, samples collected in Banganté were firstly frozen (-20˚C) in the Laboratory of UdM-Bangangte, then transferred once per week between 2-8˚C to the virology laboratory CREMER located in Yaounde(~ 400 km away) where they were analyzed upon arrival.
- 4.
COVID-19 Testing
SARS-CoV-2 IgG/IgM antibodies were detected in whole blood using Panbio™ COVID-19 IgG/IgM Rapid Test Device, REF: ICO-T40203, LOT: COV0052132, Expiration: 2021-04-30) according to the manufacturer’s instructions. The Panbio™ COVID-19 IgG/IgM Rapid Test is an immunochromatographic lateral flow test kit used for the qualitative detection of IgG and IgM antibodies to SARSCoV-2 in human serum, plasma, venous, and capillary whole blood. In this study, all Panbio™ COVID-19 IgG/IgM Rapid Tests were performed on-site using capillary whole blood. That technique is reported to have a specificity of 99.4% and a sensitivity of 98.2% [
21].
Molecular diagnosis of SARS-CoV-2 was performed using realtime RT-qPCR [
22]. In all, viral genome was detected by the analysis of the RdRp, E, et N genes using a DaAnGene® kit (DaAn Gene Co., Ltd, Guangzhou, Guangdong, China) after RNA extraction. Amplification of the SARS–CoV–2 genes were performed on a QuantStudioTM 7 real time thermocycler (Applied Biosystems, Massachusetts, USA). Cycling conditions were as follows: reverse transcription (45 °C/15 seconds) followed by initial denaturation (95 °C/2 minutes), and 45 cycles of [denaturation (95 °C/15 seconds), annealing (60 °C/30 seconds), and extension (72 °C/60 seconds)]. Internal control was included in each amplification. Cycle threshold (Ct) value was used to determine viremia and classified patients (negative or positive) as reported previously [
22]. The prevalence of SARS–CoV–2 infection was defined as the proportion of individuals with positive RT–qPCR.
E. Data analysis
The analyses were done using the software package Stat view 5.1 for Windows (SAS Institute Inc, USA). The continuous variables are presented in terms of mean ± Standard deviation (Std) and categorical variables in absolute number (proportion in %). The associations between the SARS-CoV-2 positivity or serological profile and demographic and clinical characteristics were investigated by Chi-Square (Pearson or for trend) test, Mann Witney or Kruskal-Wallis test as appropriate. The univariate and multivariate regression analyses were conducted to identify factors independently associated with the risk of SARS-CoV-2 infection and serological profile. Only participants with all informations were included during analysis. For all the analyses, the significance level was set at p<0.05.
3. Results
3.1. Determination of baseline characteristics of the study population
A total of 291 participants in three universities was were surveyed between September 28, 2020, to September 06, 2021, in of Yaoundé and Bangangté. Males and females had similar representation in the study with sex ratio of 1:1. The mean age of the study participant was 22.59years old [min 18, max 27] and the age range [21-24[years was most represented (41.24 %). The majority of these students were from the city of Bangangté 68.04% while 31.96% were from Yaoundé. Based on clinical presentation, almost all our most of the participants were asymptomatic (80.41%vs 19.59%symptomatic) with less comorbidity (4.13%). It appears that the majority of the population have not haven’t yet been in closed contact with an infected person (80.76%). In addition, most were sedentary
2 (93.13% vs. 6.87% nomadic
1) and had not yet taken any drugs against COVID-19 (98.63%) (see
Table 1).
3.2. Prevalence of SARS-CoV-2 in students and associated risk factors
The overall prevalence of SARS-CoV-2 (positive PCR)among students was 21.31%. However, the PCR positivity rates differed across universities as follow; UdM-Bangante: 25.25% (50/198), IUEs/INSAM-Yaounde: 5% (3/60), and ISSBA-Yaounde: 27.27% (9/33). The identification of risk factors (sociodemographic and clinical characteristics) by binary logistic regression showed that females were more affected by SARS CoV-2 infection than male by two-fold (28.76% vs 13.04%; OR=2.21, 95%CI=1.5-4.5; P=0.0007). Similarly, students in the city of Banganté were almost two times more affected by COVID-19 than students from Yaounde (25.25%vs 12.9%; OR=1.95, 95%CI=1.2-3.8; P=0.01) (See
Table 2).
3.3. IgM/IgG serological profile and their associated factors
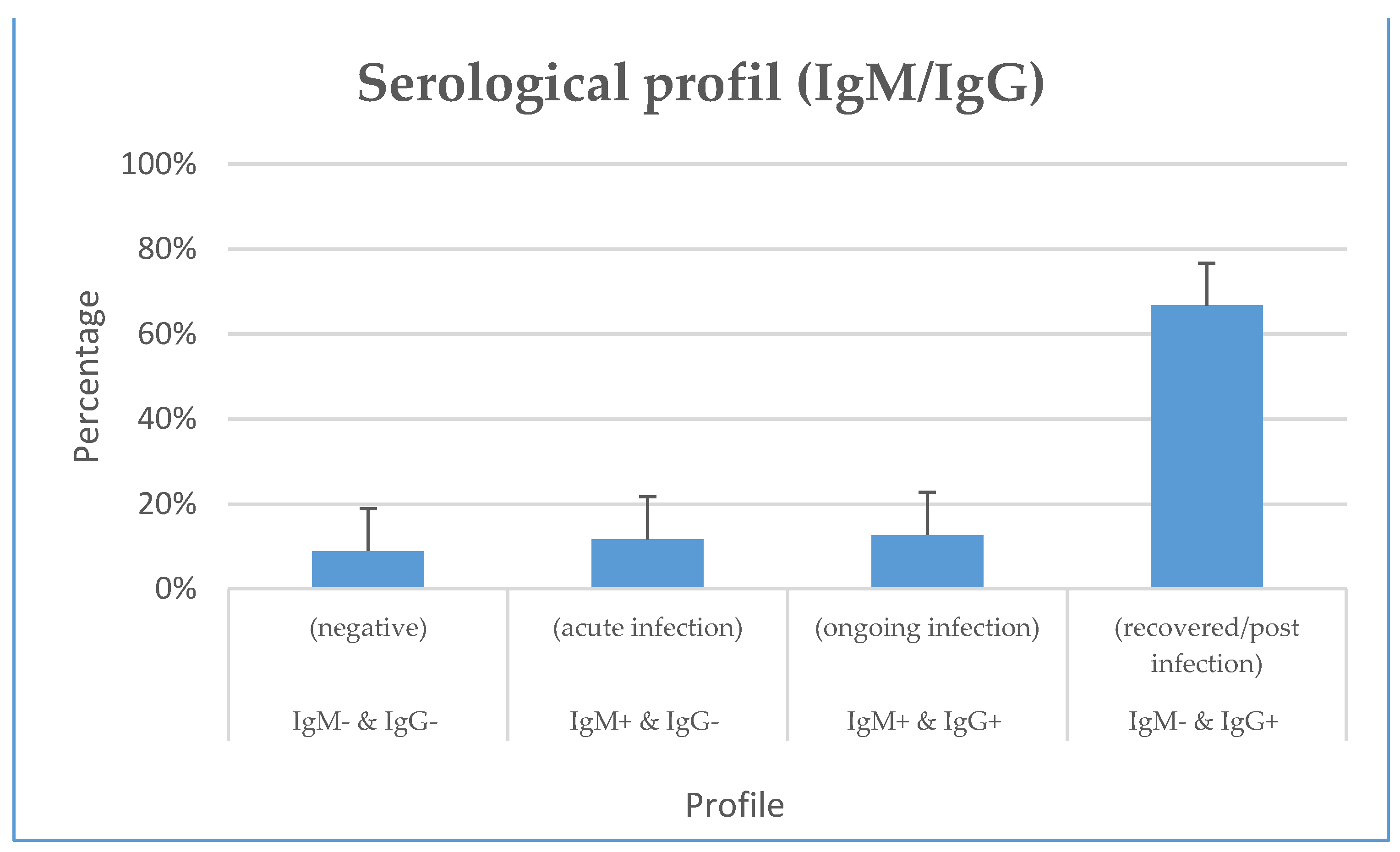
From the rapid serological testing (IgM/IgG), we noticed an overall IgM (IgM+/IgG- and IgM+/IgG+) and IgG (IgM-/IgG+ and IgM+/IgG+) seropositivity of 20.62 % (60/291) and 24.4% (71/291) respectively. Also, 11.68% (34/291) and 8.93% (26/291) of our participants had typical progressive (IgM+/IgG+) and acute (IgM+/IgG-) infection profiles. In addition, more than half of our participants did not have previous exposure to SARS-COV-2 (IgM- & IgG-) 66.67% (194/291) (
Figure 1). The identification of factors (socio-demographic and clinical predictors) associated with the serological profile showed that two time more females had the progressive (IgM+/IgG+) profile compared to males (15.69% vs 7.25%; OR=2, 95%CI=; p=0.02).
Students from the city of Bangangté in our sample had the majority of active infection (IgM+&IgG+) compared to those from Yaoundé (14.14% vs. 4.45%; OR=3.5, 95%CI=; p=0.05). Students reported as "case-contact" were the least affected by active infection (IgM+&IgG+) (7.14%, p=0.05); on the other hand, nomads were more affected by active infection (20%, p=0.01) than sedentary participants (
Table 4).
3.4. IgM/IgG serological profile and SARS-CoV-2 RNA detection
Comparing serological and molecular profiles we found that a proportion of 26.92% (7/26) presenting COVID-19 IgM+/IgG- had negative PCR compared to 73.08 % (19/26) for those with positive PCR (p<0.0001). Furthermore, 17.65% (6/34) negative PCR versus 82.35% (28/34) positive PCR were found in students with COVID-19 IgM+/IgG+, (p<0.0001). Lastly, 7.22% (14/194) with IgM-/IgG- had a positive PCR (see
Table 5).
4. Discussion
During this study conducted in three private universities in Cameroon (UdM-Bangangte, IUEs/INSAM-Yaounde, and ISSBA-yaounde), 291students were enrolled. The mean age was 22.59±10.43 years old [min 18, max 27], females were predominant compared to males (52.58% vs. 47.42%) and the majority of the population resided in the city of Bangangté (68.04% vs. 31.96% in Yaounde). Approximately 3/4 of the overall study population was asymptomatic (80.41%). This high number of asymptomatic participants must be related to the young age of the study population (mean=22 years). In fact, studies conducted worldwide presented COVID-19 as an asymptomatic disease in more than 85% of young subjects [
23].
The overall prevalence of COVID-19 by RT-qPCR was 21.31% (62/291), with 25.25% at UdM-Bangangte, 27.27% at ISSBA-Yaounde, and 5% at IUEs/INSAM-Yaounde private Universities. Similarly, students of Banganté had twice as many cases of COVID-19 than those in Yaoundé (25.25% vs. 12.90%, p =0.01). These disproportionate prevalences between private High Schools in Yaounde and Bangante could be explained by the fact that Yaoundé was among the cities considered as the starting point of the COVID-19 epidemic in Cameroon in early 2020 and thus, the initial point of the anti-COVID response [
24]. The spread of the pandemic through smaller cities like Bangangte was amplified by their lower response capacity. The detection of SARS-CoV-2 in these two cities of Cameroon informs us of the importance of the continuous application of barrier measures to prevent the spread of SARS-COV-2. Our findings portraying women to be twice as affected as men (28.76% vs. 13.04%, p<0.0007) and was differed from several studies [
25,
26,
27,
28]. Studies reported that even when men and women have the same prevalence, men with COVID-19 are more at risk for worse outcomes and death, independent of age [
29]. Furthermore, most of the COVID-19 cases confirmed by RT-qPCR were asymptomatic (22.65% vs. 15.79%symptomatic). No severe case was reported in this study. This could be due to the relative young age of the study population. However, there is evidence that asymptomatic SARS-CoV-2 carriers can transmit the virus [
30].
We noted a predominance of IgG (24.40 %) compared to IgM (20.62 %). Similar trends have already been reported with higher IgG level in the general population in USA [
31], Cameroon [
7,
32,
33,
34], Congo [
35], India [
36] and Brazil [
37] as well as in students in Spain [
38]. In fact, experience earned from the kinetics of antibody response from other viral infections taught us that unlike IgM that appears in the acute phase of infection, IgG is the marker of the chronic infection and should appear later on in greater amount. Only 12.71% of our participant presented the IgM-/IgG+ profile. This serological profile in this study should reflect recovering or post infectional immunity as a result of infection with SARS-CoV-2 since none of our participant were vaccinated.
Concerning symptomatic subjects, Only 9 over 57 (15.78%) were PCR positive whereas 31.57% (18/57) were presenting at least one COVID-19 IgM and or IgG antibody [31.57% (18/57)]. SARS-CoV-2 Prevalence [
39,
40,
41,
42] as well as IgM/IgG antibodies detection [
43,
44,
45] vary according to identified symptoms. In fact, COVID-19 share similar flu-like symptoms make it difficult to differentiate from other respiratory diseases [
46,
47,
48]. Thus, the need of standard method looking for SARS-COV—2 RNA such RT-PCR for its diagnosis in clinical practice [
49]. Furthermore, as compared to PCR, antibodies tests look for antibodies in the blood that fight the virus that causes COVID-19 [
50]. The presence of antibodies such IgM/IgG does not always signs infection as they can also be detected in the blood of people who have recovered from COVID-19 or people who have been vaccinated against COVID-19[
50]. That is why anti-SARS-CoV-2 IgM and IgG antibodies have been expected to be useful as complementary tests, in addition to RT-PCR, for the diagnosis of COVID-19[
43].
Four serological profiles were identified: 12.71% IgM-/IgG+, 11.68% IgM+/IgG+, 8.93% of IgM+/IgG-, 66.67 % of IgM-/IgG-. Similar results have been reported in Congo [
35], in the USA [
31], and in Cameroon [
32,
33] with a sample size of 684, 368, 291 and 971 respectively. No association has been found between serological profiles and all the socio-demographic characteristics. Our results diverge from those of Congo where hospitalizations due to COVID-19 were correlates to immune response [
35]. However, our findings are similar to other studies in Cameroon reporting this association [
32,
33]. The differences seen between Cameroon and Congo may be accounted by the difference in sample size and study context. In fact, the study in Congo was done on 684 travelers [
51] and those in Cameroun by Nguwoh [
32] and Nwosu [
33] were conducted on 368 and 971 health care workers. Moreover, more than half of our study participants did not have previous exposure to COVID (IgM- & IgG-) 66.67% (194/291). This large proportion of non-exposure could reflect the success of the nationwide measures implemented by the Cameroonian government to stall COVID-19 transmission [
24].
In this study, about 1/4th (26.92%) of the patients with serological markers of acute COVID-19 infection (IgM+IgG-) and 1/5th(17.65%) with serological markers of ongoing COVID-19 infection (IgM+IgG+)had a negative PCR result (p<0.0001). The difference observed between antibodies and PCR is logical and supported by previous findings [
16,
33,
52,
54,
55]. The observed disparity between serologic (IgG/IgM) and molecular (PCR) profiles could be accounted for the acute nature of such an infection with an incubation time of 2-14days, which differs from the peak production of IgM and IgG (~10-14days) [
52,
55]. This further supports the use of PCR for diagnosis and Antibodies for sero-surveys.
Only 2.7% of the participants with serological markers of postinfectional immunity (IgM-IgG+) still had positive PCR results. In fact, the IgM-IgG+ is the conventional serological profile of past exposure to infections, or chronic infection, an indicator of cure, or natural immunization [
56]. Finally, 7.22% (14/194) of the participants negative by IgM- and IgG- had a positive PCR result. These may be false-negative or evidence that these are early infections with levels of antibodies that cannot be detected by the serological tests we used [
35]. Moreover, it could be related to seroconversion, a period the onset of antibody production which will the rise to detectable [
57,
58]. In addition, numerous studies have already reported a low sensitivity of the COVID IgM/IgG serological tests compared to techniques such as ELISA or LUMINEX [
59,
60,
61,
62,
63] suggesting a continuous readaptation of prefabricated RDTs in the emergency anti-COVID response is needed, especially as circulating strains arise and cause new waves.
5. Conclusions
During the COVID-19 pandemic, there was an upsurge (above 20%) of COVID-19 confirmed cases within the University settings of Cameroon, calling for a increased preparedness and rapid response strategies to implement in higher institutes in case of any new pathogen with pandemic or epidemic potentials arises. The observed disparity between IgG/IgM and viral profile supports prioritizing assays targeting the virus (nucleic acid or antigen) for diagnosis and antibody screening for sero-surveys.
Author Contributions
“Conceptualization, R.K.W, C.I.D., M.S.N, TA, and E.M.M.; methodology, R.K.W, L.A.K.S., P.D.D.D., M.T.; software, P.D.D.D. and M.T; validation, R.K.W., L.A.K.S., L.F.E. and J.F.; formal analysis, M.T., C.I.D.; investigation, R.K.W., M.T; resources, MSN and E.M.M.; data curation, P.D.D.D.; writing—original draft preparation, L.A.K.S., R.K.W., L.F.E. and J.F. writing—review and editing, R.K.W., L.F.E. and J.F.; supervision, T.A. and E.M.M.; project administration, T.A. and E.M.M.; All authors have read and agreed to the published version of the manuscript.”.
Funding
This research received no external funding.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved) by the Institutional Review Board (IRB) of the National Ethics Committee for Human Health Research in Cameroon (N°: 2020/05/1218/CE/CRERSHC/SP from 06th May 2020). Additionally, we obtained authorization from the directors of the three Universities selected.
Informed Consent Statement
An information note was given to all the eligible participants, who then provided their written informed consent before enrollment into the study. The confidentiality of study participants was secured by the use of identification codes.
Acknowledgments
we will like to acknowledge all the participants (Students), directors of each private universities and administrative authorities, the American Society of Microbiology (ASM)-ASM Cameroon and the staff of the laboratory of UdM-Bangante and CREMER-Yaounde.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Van Caeseele, P.; Bailey, D.; Forgie, S. E.; Dingle, T. C. et Krajden, M. SARS-CoV-2 (COVID-19) serology: implications for clinical practice, laboratory medicine and public health, Can. Med. Assoc. J., vol. 192, no 34, p. E973, août 2020. [CrossRef]
- Zhou, F.; Yu, T.; Du, R.; Fan, G.; Liu, Y.; Liu, Z.; Xiang, J.; Wang, Y.; Song, B.; Gu, X.; Guan, L.; Wei, Y. et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study, The Lancet, vol. 395, no 10229, p. 1054-1062, mars 2020. [CrossRef]
- Zhao, J.; Yuan, Q.; Wang, H.; Liu, W.; Liao, X.; Su, Y.; Wang, X.; Yuan, J.; Li, T.; Li, J.; Qian, S.; Hong, C. et al. Antibody Responses to SARS-CoV-2 in Patients With Novel Coronavirus Disease 2019 , Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am., vol. 71, no 16, p. 2027-2034, nov. 2020. [CrossRef]
- Oran, D. P.; and Topol, E. J. Prevalence of asymptomatic SARS-CoV-2 infection: a narrative review. Ann. Intern. Med 2020, 173, 362–367. [Google Scholar] [CrossRef] [PubMed]
- WHO. Weekly epidemiological update on COVID-19 - 30 November 2022, Edition 120, 30 November 2022, Emergency Situational Updates. (https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19---30-november-2022, consulted on 16/01/2023 at 19h30).
- Johns Hopkins University of Medicine, Mortality Analyses, Coronavirus Resource Center. (https://coronavirus.jhu.edu/data/mortality, consulted on 16/01/2023 at 20h00).
- WHO Africa, The future of WHO Covid-19 response operations in Africa in 2022. (https://www.afro.who.int/publications/future-who-covid-19-response-operations-africa-2022, consulted on 16/01/2023 at 20h10).
- Rabi, F. A.; Al Zoubi, M. S.; Kasasbeh, G. A.; Salameh, D. M.; et Al-Nasser, A. D. SARS-CoV-2 and Coronavirus Disease 2019: What We Know So Far », Pathog. Basel Switz., vol. 9, no 3, p. E231, mars 2020. [CrossRef]
- Bonny, V.; Maillard, A.; Mousseaux, C.; Plaçais, L.; et Richier, Q. COVID-19 : physiopathologie d’une maladie à plusieurs visages, Rev. Médecine Interne, vol. 41, no 6, p. 375-389, juin 2020. [CrossRef]
- CDC COVID-19 Science Briefs, Science Brief: Transmission of SARS-CoV-2 in K-12 Schools and Early Care and Education Programs – Updated, December 17, 2021. (https://www.ncbi.nlm.nih.gov/books/NBK570438/, 17/01/2023, 15h).
- Ravens-Sieberer, U.; Kaman, A.; Erhart, M.; Devine, J.; Schlack, R.; Otto, C. Impact of the COVID-19 pandemic on quality of life and mental health in children and adolescents in Germany. Eur Child Adolesc Psychiatry. Published online January 25, 2021. [CrossRef]
- Goldstein, E:; Lipsitch, M.; Cevik, M. On the Effect of Age on the Transmission of SARS-CoV-2 in Households, Schools, and the Community. J Infect Dis. 2021, 223, 362–369. [Google Scholar] [CrossRef] [PubMed]
- National Centre for Immunisation Research and Surveillance. COVID-19 in schools and early childhood education and care services – the Term 3 experience in NSW. Report from National Centre for Immunisation Research and Surveillance. October 9, 2020. Accessed June 30, 2021. https://www.ncirs.org.au/sites/default/files/2020-10/COVID-19%20Transmission%20in%20educational%20settings%20in%20NSW%20Term%203%20report_0.pdf.
- Larosa E.; Djuric, O.; Cassinadri, M.; et al. Secondary transmission of COVID-19 in preschool and school settings in northern Italy after their reopening in September 2020: a population-based study. Euro Surveill. 2020, 25. [Google Scholar]
- Murillo-Llorente, M.T.; Perez-Bermejo, M. COVID-19: Social Irresponsibility of Teenagers Towards the Second Wave in Spain. J Epidemiol. 2020, 30, 483. [CrossRef] [PubMed]
- Zhu, Y.; Bloxham, C.J.; Hulme, KD.; et al. A Meta-analysis on the Role of Children in Severe Acute Respiratory Syndrome Coronavirus 2 in Household Transmission Clusters. Clin Infect Dis. 2021, 72, e1146–e1153. [Google Scholar] [CrossRef]
- Hobbs, C.V.; Drobeniuc, J.; Kittle, T.; et al. Estimated SARS-CoV-2 Seroprevalence Among Persons Aged <18 Years – Mississippi, May-September 2020. MMWR Morb Mortal Wkly Rep. 2021, 70, 312–315. [Google Scholar]
- Kim, D. H.; Kim, D. ; Moon, J. W.; Chae, S.-W.; et Rhyu, I. J. Complications of Nasopharyngeal Swabs and Safe Procedures for COVID-19 Testing Based on Anatomical Knowledge , J. Korean Med. Sci., vol. 37, no 11, p. e88, mars 2022. [CrossRef]
- Marty, F. M.; Chen, K.; et Verrill, K. A. How to Obtain a Nasopharyngeal Swab Specimen. Reply », N. Engl. J. Med., vol. 383, no 3, p. e14, juill. 2020. [CrossRef]
- Di Maio, P.; Iocca, O.; Cavallero, A. et Giudice, M. Performing the nasopharyngeal and oropharyngeal swab for 2019-novel coronavirus (SARS-CoV-2) safely: How to dress, undress, and technical notes », Head Neck, vol. 42, no 7, p. 1548-1551, juill. 2020. [Google Scholar] [CrossRef]
- Batra, R.; Olivieri, L.G.; Rubin, D.; Vallari, A.; Pearce, S.; Olivo, A.; Prostko, J.; Nebbia, G.; Douthwaite, S.; Rodgers, M.; Cloherty, G. A comparative evaluation between the Abbott PanbioTM COVID-19 IgG/IgM rapid test device and Abbott ArchitectTM SARS CoV-2 IgG assay, J. Clin. Virol. Off. Publ. Pan Am. Soc. Clin. Virol., vol. 132, p. 104645, nov. 2020. [Google Scholar] [CrossRef]
- Yamada, S.; Fukushi, S.; Kinoshita, H.; Ohnishi, M.; Suzuki, T.; Fujimoto, T.; Saijo, M.; Maeda, K. ; Virus Diagnosis Group (NIID Toyama. Assessment of SARS-CoV-2 infectivity of upper respiratory specimens from COVID-19 patients by virus isolation using VeroE6/TMPRSS2 cells, BMJ Open Respir. Res., vol. 8, no 1, p. e000830, févr. 2021. [Google Scholar] [CrossRef]
- Pratha, S.; Meagan, C. F.; Zimmer, C. F.; Abdollahi, E.; Juden-Kelly, L.; Moghadas, S. M. ; Singer, Burton H.; et Galvani, A. P. Asymptomatic SARS-CoV-2 infection: A systematic review and meta-analysis », Proc. Natl. Acad. Sci. U. S. A., vol. 118, no 34, p. e2109229118, août 2021. [CrossRef]
- Esso, L.; Epée, E.; Bilounga, C.; Abah, A.; Hamadou, A.; Dibongue, E.; Kamga, Y.; Belinga, S.; Eyangoh, S.; Okomo, M.C.; Mounagué, C. et al. Cameroon’s bold response to the COVID-19 pandemic during the first and second waves , Lancet Infect. Dis., vol. 21, no 8, p. 1064-1065, août 2021. [CrossRef]
- Abate, B.B.; Kassie, A.M.; Kassaw, M.W.; et al. Sex difference in coronavirus disease (COVID-19): a systematic review and meta-analysis, BMJ Open 2020;10:e040129. [CrossRef]
- Doerre, A.; Doblhammer, G. The influence of gender on COVID-19 infections and mortality in Germany: Insights from age- and gender-specific modeling of contact rates, infections, and deaths in the early phase of the pandemic. PLoS ONE, 2022, 17(5): e0268119. [CrossRef]
- Griffith, D.M.; Sharma, G.; Holliday, C.S.; Enyia, O.K.; Valliere, M.; Semlow, A.R.; et al. Men and COVID-19: A Biopsychosocial Approach to Understanding Sex Differences in Mortality and Recommendations for Practice and Policy Interventions. Prev Chronic Dis 2020, 17:200247. [CrossRef]
- Perlis, R.H.; Santillana, M.; Ognyanova, K.; et al. Prevalence and Correlates of Long COVID Symptoms Among US Adults. JAMA Netw Open. 2022;5(10):e2238804. [CrossRef]
- Jin, J-M.; Bai, P.; He, W.; Wu, F.; Liu, X-F.; Han, D-M.; Liu, S.; and Yang, J-K. Gender Differences in Patients With COVID-19: Focus on Severity and Mortality. Front. Public Health, 2020, 8:152. [CrossRef]
- Muller, C. P. Do asymptomatic carriers of SARS-COV-2 transmit the virus? , Lancet Reg. Health – Eur., vol. 4, mai 2021. [CrossRef]
- Chang, S. E.; Feng, A.; Meng, W.; Apostolidis, A. S.; Mack, E.; Artandi, M.; Barman, L.; Bennett, K.; Chakraborty, S.; Chang, I.; Cheung, P. et al. New-onset IgG autoantibodies in hospitalized patients with COVID-19 », Nat. Commun., vol. 12, no 1, Art. no 1, sept. 2021. [Google Scholar] [CrossRef]
- Nguwoh, P. S.; Mboringong, A. B.; Fokam, J.; Ngounouh Taheu, C.; Halilou, I.; Djieudeu Nouwe, S. H.; Moussa, N. I.; Al-Mayé Bit Younouss, A.; Likeng, J. L. N.; Tchoffo, D.; Essomba, G. R.; Kamga, H. L. et Okomo, M. C. A. Seroprevalence of SARS-CoV-2 (COVID-19) among Health Care Workers in Three Health Facilities of Yaounde, Center Region of Cameroon », Eur. J. Med. Health Sci., vol. 3, no 6, p. 89-94, déc. 2021. [Google Scholar] [CrossRef]
- Nwosu, K.; Fokam, J.; Wanda, F.; Mama, L.; Orel, E.; Ray, N.; Meke, J.; Tassegning, A.; Takou, D.; Mimbe, E.; Stoll, B.; et al. SARS-CoV-2 antibody seroprevalence and associated risk factors in an urban district in Cameroon. Nat Commun, 2021 Oct 6;12(1):5851. PMID: 34615863; PMCID: PMC8494753. [CrossRef]
- Ndongo, F.A.; Guichet, E.; Mimbé, E.D.; Ndié, J.; Pelloquin, R.; Varloteaux, M.; Esemu, L.; Mpoudi-Etame, M.; Lamare, N.; Edoul, G.; Wouambo R.K. et al. Rapid Increase of Community SARS-CoV-2 Seroprevalence during Second Wave of COVID-19, Yaoundé, Cameroon, Emerg. Infect. Dis., vol. 28, no 6, p. 1233-1236, juin 2022. [CrossRef]
- Lobaloba Ingoba, L.; Djontu, J.C.; Mfoutou Mapangui, C. C.; Mouziga, F.; Diafouka, K. S.; Vouvoungui, C.; Kuisma, E.; Nguimbi, E. et Ntoumi, F. Seroprevalence of anti-SARS-CoV-2 antibodies in a population living in Bomassa village, Republic of Congo », IJID Reg. Online, vol. 2, p. 130-136, mars 2022. [CrossRef]
- Lone, K.S.; Khan, S.M.S.; Qurieshi, M.A.; Majid, S.; Pandit, M.I.; Haq, I.; Ahmad, J.; Bhat, A.A.; Bashir, K.; Bilquees, S.; Fazili, A.B.; Hassan, M.; Jan, Y., Kaul, R-uR.; et al. Seroprevalence of SARS-CoV-2-specific anti-spike IgM, IgG, and anti-nucleocapsid IgG antibodies during the second wave of the pandemic: A population-based cross-sectional survey across Kashmir, India. Front. Public Health, 2022, 10:967447. [CrossRef]
- Borges, L.P.; Martins, A.F.; de Melo, M.S.; de Oliveira, M. G. B.; Neto, J.M.R.; Dósea, M.B.; Cabral, B. C. M.; Menezes, R. F.; Santos, A. A.; Matos, I. L. S.; Borges, P. C.; Dos Santos, K. A.; Ribeiro, A. A. et al. Seroprevalence of SARS-CoV-2 IgM and IgG antibodies in an asymptomatic population in Sergipe, Brazil. Rev Panam Salud Publica, 2020 Oct 6, 44:e108. PMID: 33042199; PMCID: PMC7541966. [CrossRef]
- Pérez-Tanoira, R.; Lledó García, L.; Torralba González de Suso, M.; Rodríguez Zapata, M.; Arroyo Serrano, T.; Giménez Pardo, C.; Rodríguez Pedrosa, M. I.; Romero Badía, M. N.; Pérez-García, F.; González López, P.; Villaescusa García, C.; Cuadros González, J. High Seroprevalence Against SARS-CoV-2 Among Faculty of Medicine and Health Sciences Personnel and Students of the University of Alcalá, Spain: Contributing Factors. Int J Gen Med, 2021 Oct 21, 14:7017-7024. PMID: 34707393; PMCID: PMC8544125. [CrossRef]
- Guan, W-j.; Ni, Z-y.; Hu, Y.; et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med 2020, 382, 1708–20. [Google Scholar] [CrossRef]
- Chen, N.; Zhou, M.; Dong, X.; et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. The Lancet 2020, 395, 507–13. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Han, X.; Jiang, N.; et al. Radiological findings from 81 patients with COVID-19 pneumonia in Wuhan, China: a descriptive study. Lancet Infect Dis 2020, 20, 425–34. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Wang, Y.; Li, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan. China. The Lancet, 2020; 395, 497–506. [Google Scholar]
- Nakano, Y.; Kurano, M.; Morita, Y.; et al. Time course of the sensitivity and specificity of anti-SARS-CoV-2 IgM and IgG antibodies for symptomatic COVID-19 in Japan. Sci Rep 11, 2776,2021. [CrossRef]
- Zhao, J.; et al. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. Clin. Infect. Dis 2020. [Google Scholar] [CrossRef]
- Qu, J.; et al. Profile of IgG and IgM antibodies against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Clin. Infect Dis 2020. [Google Scholar] [CrossRef]
- Wu, D.; Wu, T.; Liu, Q.; Yang, Z. The SARS-CoV-2 outbreak: what we know. Int J Infect Dis 2020, 94, 44–8. [Google Scholar] [CrossRef] [PubMed]
- Li, L.Q.; Huang, T.; Wang, Y.Q.; et al. 2019 novel coronavirus patients' clinical characteristics, discharge rate and fatality rate of meta-analysis. J Med Virol 2020, 92, 577–83. [Google Scholar]
- Sun, P.; Lu, X.; Xu, C.; Sun, W.; Pan B. Understanding of COVID-19 based on current evidence. J Med Virol 2020, 92, 548–51. [Google Scholar] [CrossRef] [PubMed]
- Pascarella, G. et al. COVID-19 diagnosis and management: a comprehensive review. J. Intern. Med, 2020. [CrossRef]
- CDC, Interim Guidelines for COVID-19 Antibody Testing in Clinical and Public Health Settings, Updated Dec. 16, 2022. https://www.cdc.gov/coronavirus/2019-ncov/hcp/testing/antibody-tests-guidelines.html.
- Katchunga, P.B.; Murhula, A.; Akilimali, P.; Zaluka, J.C.; Karhikalembu, R.; Makombo, M.; Bisimwa, J.; et Mubalama E. Seroprevalence of SARS-CoV-2 antibodies among travellers and workers screened at the Saint Luc Clinic in Bukavu, a city in eastern Democratic Republic of the Congo, from May to August 2020, Pan Afr. Med. J., vol. 38, p. 93, 2021. [CrossRef]
- De Donno, A.; Lobreglio, G.; Panico, A.; Grassi, T.; Bagordo, F.; Bozzetti, M.P.; Massari, S.; Siculella, L.; Damiano, F.; Guerra, F. et al. IgM and IgG Profiles Reveal Peculiar Features of Humoral Immunity Response to SARS-CoV-2 Infection. Int J Environ Res Public Health. 2021 Feb 1;18(3):1318. [CrossRef]
- Munitz, A. , Edry-Botzer, L., Itan, M.; Tur-Kaspa, R.; Dicker, D.; Marcoviciu, D.; Goren, M. G.; Mor, M.; Lev, S. ; Gottesman, T. et al. Rapid seroconversion and persistent functional IgG antibodies in severe COVID-19 patients correlates with an IL-12p70 and IL-33 signature. Sci Rep 11, 3461 (2021). [CrossRef]
- Meireles, P.; Costa, J. P.; Novais, M.J.; Miranda, D.; Lopes, M. M.; Severo, M.; Barros, H. The SARS-CoV-2 Infection Among Students in the University of Porto: A Cross-Sectional Study. Int J Public Health. 2022 Oct 20;67:1604548. [CrossRef]
- Oran, D.P.; Topol, E.J. Prevalence of Asymptomatic SARS-CoV-2 Infection : A Narrative Review. Ann Intern Med. 2020 Sep 1;173(5):362-367. [CrossRef]
- Ozturk, T.; Howell, C.; Benameur, K.; Ramonell, R.P.; Cashman, K.; Pirmohammed, S.; Bassit, L.; Roback, J.; Marconi, V.C.; Schinazi, R.F.; Wharton, W.; Lee, F.E.; Hu, W.T. Cross-sectional IgM and IgG profiles in SARS-CoV-2 infection , medRxiv, p. 2020.05.10.20097535, mai 2020. [CrossRef]
- Fernández-Rojas, M.A.; Luna-Ruiz Esparza, M.A.; Campos-Romero, A.; Calva-Espinosa, D.Y.; Moreno-Camacho, J.L.; Mendlovic, F.; Plett-Torres, T.; Alcántar-Fernández, J. Seroconversion dynamic and SARS-CoV-2 seropositivity in unvaccinated population during the first and second outbreaks in Mexico, Sci. Rep., vol. 12, no 1, Art. no 1, mars 2022. [CrossRef]
- Yadav, A.K.; Ghosh, S.; Kotwal, A.; Kaushik, S.K.;, Bobdey, S.; Sahu, R.; Kapoor, S.; Faujdar, D.S., Teli, P.T.; Anand, V. Seroconversion among COVID-19 patients admitted in a dedicated COVID hospital: A longitudinal prospective study of 1000 patients », Med. J. Armed Forces India, vol. 77, p. S379-S384, juill. 2021. [CrossRef]
- Mohit, E.; Rostami, Z.; et Vahidi, H. A comparative review of immunoassays for COVID-19 detection », Expert Rev. Clin. Immunol., vol. 17, no 6, p. 573-599, juin 2021. [CrossRef]
- Gambino, C.M.; Lo Sasso, B.; Colomba, C.; Giglio, R.V.; Agnello, L.; Bivona, G.; Ciaccio, M. Comparison of a rapid immunochromatographic test with a chemiluminescence immunoassay for detection of anti-SARS-CoV-2 IgM and IgG, Biochem. Medica, vol. 30, no 3, p. 030901, oct. 2020. [CrossRef]
- Chansaenroj, J.; Yorsaeng, R.; Posuwan, N.; Puenpa, J.; Sudhinaraset, N.; Chirathaworn, C.; Poovorawan, Y. Detection of SARS-CoV-2-specific antibodies via rapid diagnostic immunoassays in COVID-19 patients », Virol. J., vol. 18, no 1, p. 52, mars 2021. [CrossRef]
- Gong, F.; Wei, H.-X. ; Li, Q. ; Liu, L. ; et Li, B. Evaluation and Comparison of Serological Methods for COVID-19 Diagnosis », Front. Mol. Biosci., vol. 8, p. 682405, 2021. [CrossRef]
- Andrey, D.O.; Cohen, P.; Meyer, B.; Torriani, G.; Yerly, S.; Mazza, L.; Calame, A.; Arm-Vernez, I.; Guessous, I.; Stringhini, S.; et al. Head-to-Head Accuracy Comparison of Three Commercial COVID-19 IgM/IgG Serology Rapid Tests », J. Clin. Med., vol. 9, no 8, p. E2369, juill. 2020. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).