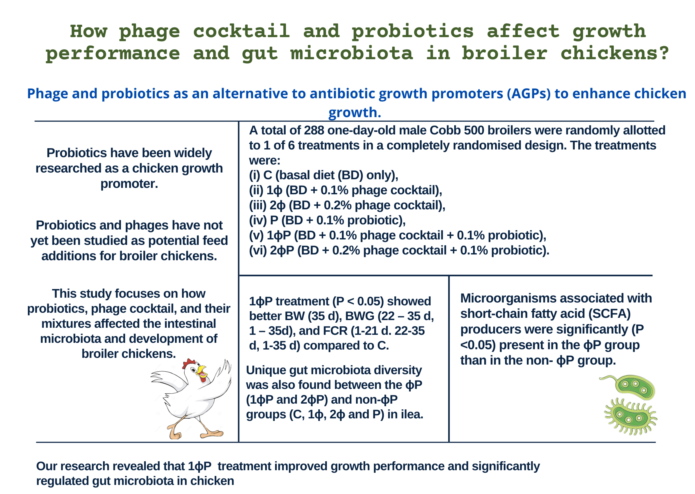
Introduction
Poultry is the most important agricultural sector providing the cheapest protein source to the global population. The use of antibiotic growth promoters (AGPs) in poultry production may be the important factor that reduces production costs. AGPs are supplemented in feed at a subliminal amount that can improve growth performance and reduce diseases in chickens (Dibner and Richards 2005, Leeson 2008, Mehdi et al. 2018). This practice, however, has led to the occurrence of antibiotic-resistant bacteria at an alarming rate, resulting in serious health problems for humans due to the inefficacy of antibiotics to treat infections (Dunshea et al. 2014, Nhung et al. 2017). Hence, AGPs use in livestock has been banned in certain parts of the world, such as European Union countries and South Korea (Flynn 2011, Maron et al. 2013, McEwen et al. 2017). The abolition of AGPs has affected the performance and health of broilers, necessitating the development of an effective substitute to provide the same level of sustainable chicken production as when AGPs are utilised (M'Sadeq et al. 2015, Karavolias et al. 2018). Phage usage has then begun to attract attention as a possible AGP replacement.
Phages are bacteria-infecting viruses that have recently attracted attention as an alternative to antibiotics. They have various advantages over antibiotics, including a narrow killing spectrum for targeting certain bacterial species, the ability to rapidly proliferate in the presence of a host and eventually kill the host, and is easily isolated from the environment (Keen & Adhya, 2015; Rose et al., 2014; Ryan et al., 2011). Phages show similar mechanisms of action as AGPs, but the phage target is specific. Its application can reduce selected target bacterial species, thereby reducing other normal gut microflora competition for adhesion sites and nutrient utilisation. Phages are commonly employed for therapeutic purposes, but recently, they have been explored for gut modulation. Pathogens such as Escherichia coli, Salmonella spp., Clostridium spp., and coliforms were significantly reduced, and beneficial bacteria such as Bifidobacterium spp. and Lactobacillus spp. increased in pigs and chickens when phages were used to modulate gut microbiota (Yan et al. 2012, Zhao et al. 2012, Wang et al. 2013, Kim et al. 2014, Kim et al. 2017, Ngu et al. 2022). These studies indicated that phages could be utilised to modulate the gut microbiome to increase beneficial bacteria and decrease harmful bacteria. Probiotics, which are extensively studied as potential for AGPs replacement, have never been used in conjunction with phage treatment in broiler chickens. Probiotics are live microorganisms that may benefit the host when administered sufficiently (De Vuyst et al. 2004, Kabir 2009). Previous studies have shown that probiotics improve the growth and health of broiler chickens (Mountzouris et al. 2010, Blajman et al. 2014, Ye et al. 2021, Yu et al. 2022).
In this study, phage that targets non-pathogenic E. coli does not require to be thoroughly purified using Caesium Chloride (CsCl) and ultracentrifugation as the phage that targets pathogens for therapeutic purposes (Steele et al. 2020, Liu et al. 2021). In the case of phages that target non-pathogens, the bacterial debris present in the unpurified lysate may not provoke an immune response and kill the host. E. coli was also selected as the target bacterial host for the gut modification investigation because of its continuous presence in the ilea and caeca of broiler chickens at different ages (Mohd Shaufi et al. 2015, Shaufi et al. 2017). Both E. coli host and phages were isolated previously from ilea and caeca of healthy broiler chickens and characterised. Only non-pathogenic E. coli was selected for phage isolation which was then used for further study. There is a paucity of knowledge about the impact of phages targeting non-pathogenic E. coli, probiotics, and their combinations in chickens. Therefore, this study aimed to assess the effects of phage cocktail that targets non-pathogenic E. coli, commercial probiotics, and their combinations on the growth performance and gut microbiota of broiler chickens.
Materials and Methods
Probiotics and phage preparation
Escherichia coli P1, P2, P3 and P4 phages (prepared as freeze-dried phage cocktail) at titre of 1010 PFU/g each were prepared. Briefly, the carrier materials of skim milk (Oxoid, UK) for protecting phage from high temperature, industrial maltodextrin as cryoprotectants, and calcium carbonate (CaCO3) (Merck, Germany) as antacids, were pre-dissolved with each diluted phage lysates at the ratio of 2:2:1:5, respectively. The phage lysate was diluted in sterile SM buffer (50 mM Tris-HCl [pH 7.5], 0.10 M NaCl, 8 mM MgSO4.7H2O) to a final titre of ~1-5 x 1010 PFU/g. The mixture was then freeze dried in Labconco freeze dryer (Labconco, USA) by first pressing vacuum when the temperature was at -40oC or colder. Samples were then added when the vacuum was 1.33 x 10-3 mBar or less. The titre of freeze-dried powder of each phage was quantified weekly until the week of chicken trial. This was to ensure their survivability and concentration maintained at ~1010 PFU/g. The powder form of each phage (C1, C2, C3 and C4 phages) were then mixed at the ratio of 1:1:1:1 to obtain a final phage cocktail form that will be supplemented into the chicken feeds. Commercial PrimaLac® probiotic (Starlabs, USA) used in this study consisted of Lactobacillus acidophilus, Lactobacillus casei, Bifidobacterium termophilum, Enterococcus faecium and Aspergillus oryzae at a concentration of 109 CFU/g each. The phage cocktail and commercial probiotic were mixed with basal feed fresh daily before being fed to the chickens.
Chicken management
A total of 288 one-day-old male Cobb 500 broiler chickens (initial body weight (BW) = 42.95 ± 2.26 g) were obtained from a local commercial hatchery. They were housed in stainless steel three-tiered battery cages with raised wire floors (Dimension: 116 cm width x 89 cm length x 46 cm height) in an open house facility at Animal Research Centre (ARC), Institute of Tropical Agriculture, Universiti Putra Malaysia, Malaysia. The cages were cleaned and disinfected through fumigation beforehand, and strict hygiene and biosecurity measures were practiced throughout the experiment. The feeders and drinkers were cleaned and filled with fresh feed and water daily. The temperature and relative humidity were recorded twice daily in the morning and the afternoon. For the chicken brooding period that lasts for the first 14 days, lighting from 100 W bulb per cage was provided for each replicates cages. The raised wire floors were covered with newspaper and cleaned daily. Procedures pertaining to chicken management, experimental design, procedures and analyses have been approved by Institutional Animal Care and Use Committee (IACUC) Universiti Putra Malaysia (Ref: UPM/IACUC/AUP-R101/2015).
Experimental design, animals and diets
A total of 288 one-day-old male Cobb 500 broilers were randomly allotted to 1 of 6 dietary treatments with 6 replicates cages containing 8 chicks per cage in a completely randomised design. The dietary treatments were: (i) basal diet only (BD)(Control, C), (ii) BD + 1 g/kg phage cocktail (1ɸ), (iii) BD + 2 g/kg phage cocktail (2ɸ), (iv) BD + 1 g/kg probiotic (P), (v) BD + 1 g/kg phage cocktail + 1 g/kg probiotic (1ɸP), (vi) BD + 2 g/kg phage cocktail + 1 g/kg probiotic (2ɸP). Both phage cocktail and probiotic for respective treatments were supplemented at the expense of corn to achieve a final equal percentage for each treatment group. The basal diet formulated for starter (1 to 21 days) (
Table 1) and finisher (22 to 35 days) (
Table 2) periods were antibiotic-free, in mash form, and ensured to meet or exceed the energy and nutrient requirements as recommended by NRC (1994) for each growing phase. Both feeds and water were provided
ad libitum.
Chemical analysis of experimental diets
The formulated starter feed, finisher feed, phage cocktail and probiotic were chemically analysed, based on feed proximate analyses (Malaysian Agricultural Research and Development Institute (MARDI)) on crude protein, crude fat, crude fibre, calcium, phosphorus and sodium (
Table 1 and
Table 2).
Sampling
For every sampling period on day 21 and 35, twelve chickens per treatment (2 chickens per replicate cage) were randomly selected, weighed and euthanised by severing the jugular veins. The mucosal contents of ilea and caeca samples were used for gut microbiota study based on high-throughput next-generation sequencing (HT-NGS) of 16S rRNA gene amplicons. All samples were kept on ice before the respective samples were processed. They were then frozen in -80oC freezer until analysis.
Chicken gut microbiota study
Illumina sequencing of the V3-V4 region of the 16S rRNA gene
V3-V4 hypervariable region of 16S rRNA gene was amplified using forward primer (5’-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGNNNNCCTACGGGNG GCWGCAG-3') and reverse primer (5’-GTCTCGTGGGCTATAAGAGACA GGACTACHVGGGTATCTAATCC-3') (Integrated DNA Technologies (IDT), Singapore) as per described by Klindworth et al. (2013) with some modifications. Four degenerate bases (N) were added to maximise the diversity for unique cluster identification. The purified amplicons were quantified by using Qubit Fluorometer (ThermoFisher Scientific, USA) and normalised to 2 nM and subjected to Illumina Miseq desktop sequencer by using paired 300 bp reads Miseq Reagent Kit v3 (600-cycle) (Illumina, USA) at Monash University Malaysia Genomic Facility.
Nucleotide sequence accession numbers
The V3-V4 region of 16S rRNA gene sequences from this study have been deposited in the NCBI sequence read archive (
https://www.ncbi.nlm.nih.gov/sra) under BioSample Accession numbers of SAMN06027949-SAMN06028092.
Statistical analysis
The experimental data were analysed based on a one-way analysis of variance (ANOVA) and paired t-test using Statistical Package for Social Science (SPSS) Statistics version 22 (IBM, USA). The replicate cage was used as the experimental unit for all parameters unless stated otherwise. Results were stated as means or means ± standard error (SE). Significant means with P < 0.05 were compared within samples by Duncan's multiple range test (Duncan 1955).
Results
Multivariate analysis on gut microbiota
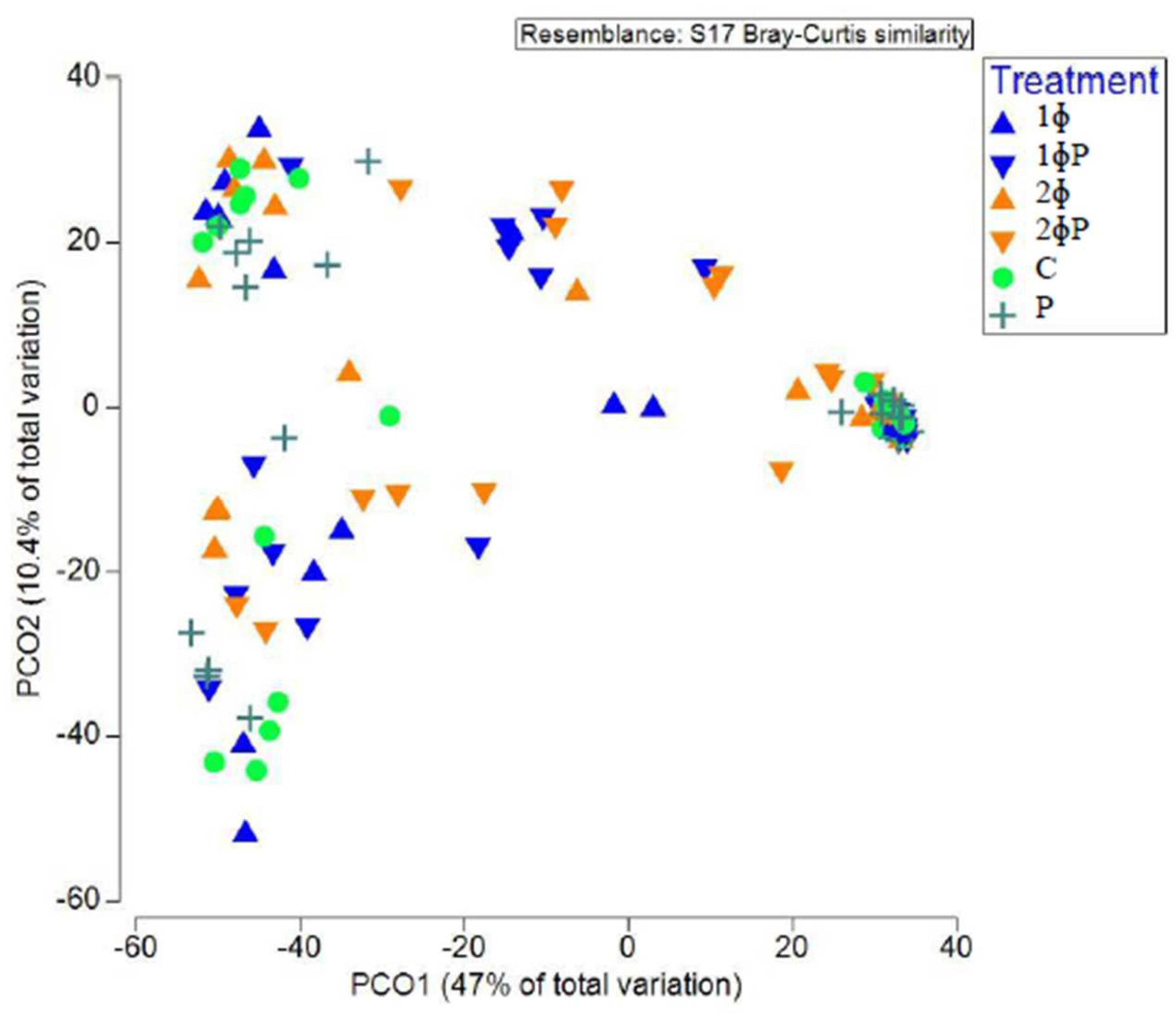
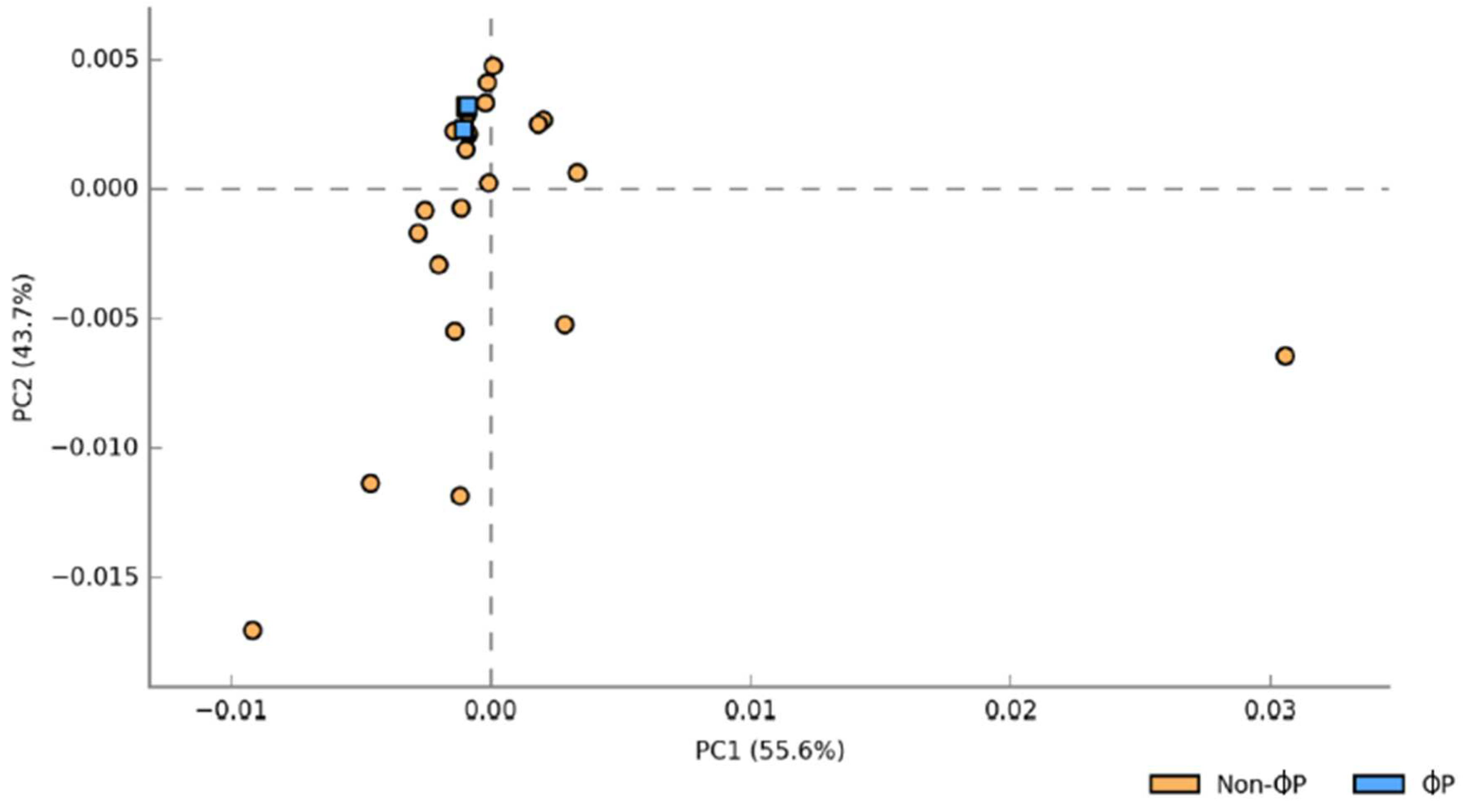
The principal coordinate analysis (PCO) plot of Bray-Curtis similarity index was employed to investigate the structure of gut microbiota in different dietary treatments, age (21 and 35 d) and part of intestine (ilea and caeca). A clear separation of chicken gut microbiota between age (21 and 35 d) (
Figure S2) and part of intestine (ilea and caeca) (
Figure S3) were observed. ɸP (1ɸP and 2ɸP) gut microbiota was distinctively different compared to non-ɸP group (C, 1ɸ, 2ɸ and P) (
Figure 1).
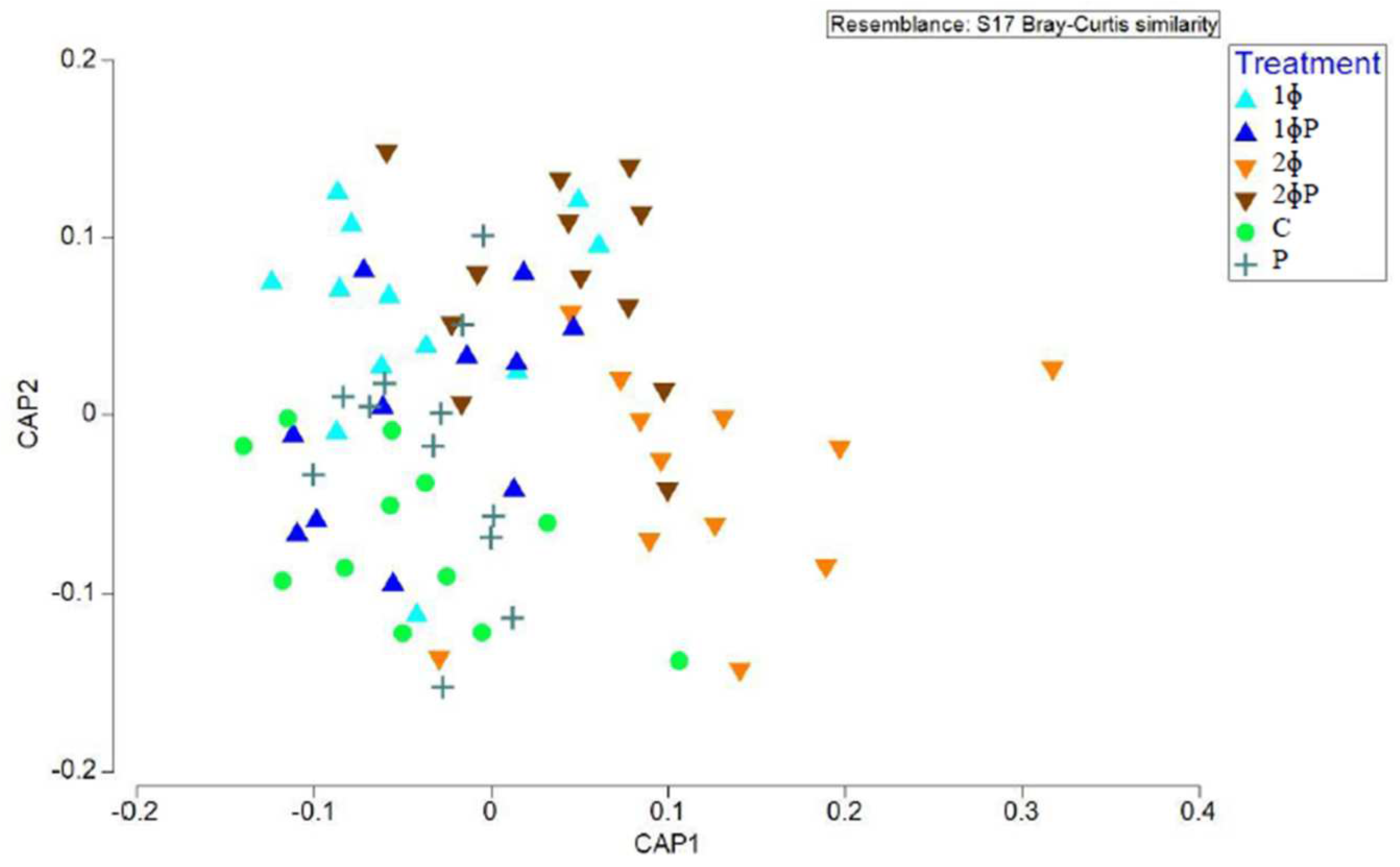
Further analysis based on the canonical analysis of principal coordinates (CAP) plot was performed to study the difference of gut microbiota structure based on dietary treatments. The CAP plot also showed that ɸP gut microbiota was distinctively different compared to non-ɸP group especially in ilea of 21 and 35 d chickens (
Figure 2) and ilea of 35 d chickens (
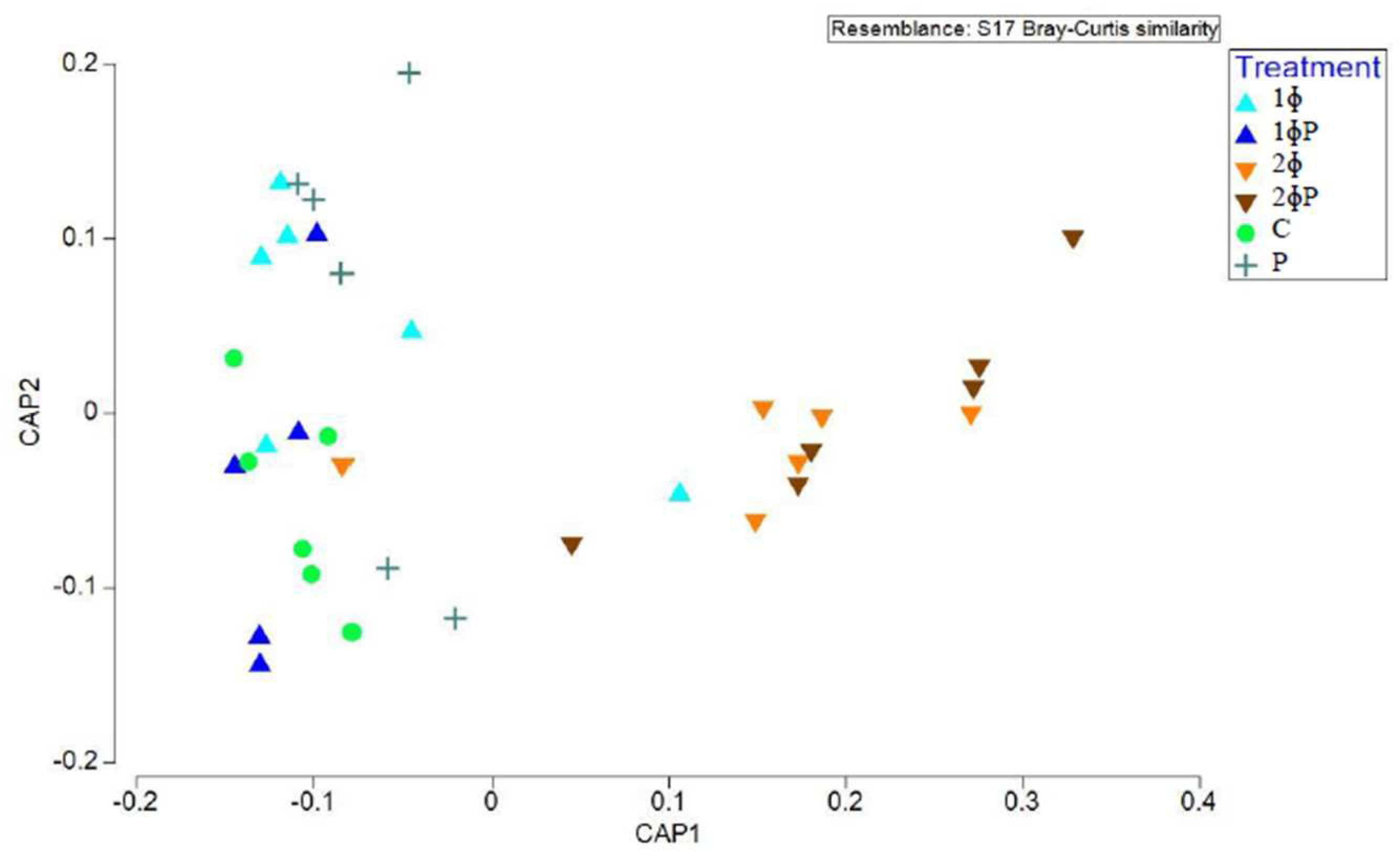
Figure 3).
Further verifications were performed to validate the earlier patterns observed from the PCA plot. The hypothesis test on the difference among gut microbiota between age, part of the intestine, and dietary treatments was verified based on the permutational multivariate analysis of variance (PERMANOVA) marginal test.
Based on PERMANOVA marginal test, the gut microbiota diversity was significantly different between age (p = 0.002) (
Table S2), part of intestine (p = 0.001) (
Table S2), dietary treatments in ilea (p = 0.005) (
Table 4) and dietary treatments in ilea of 35 d chickens (p = 0.001) (Table 8). PERMANOVA pairwise test calculated based on dietary treatments in ilea of 21 and 35 d chickens (
Table 4 (b)) and ilea of 35 d chickens (
Table 5 (b)) further verified the patterns observed earlier in CAP plot where a significant difference in gut microbiota was observed between ɸP and non-ɸP groups.
Significant OTUs present in the ɸP compared to the non-ɸP groups
The OTUs that were significantly expressed in phage cocktail and probiotic combinations groups (ɸP) were identified based on 'fitFeatureModel' in the metagenomeSeq package (
Table 6). The value of logFC is directly related to their abundance in the ɸP groups. Out of the top 50 OTUs selected, the most common bacterial genera or species that were significantly elevated in ɸP groups, compared to non-ɸP groups were
Bacteroides, Odoribacter, Alistipes, Anaerotruncus, Ruminococcaceae, Lachnospiraceae, Ruminococcus, Desulfovibrio, Anaerostipes, Clostridium, Coprobacillus, Butyricimonas, Faecalibacterium prausnitzii and
Oscillopira.
Discussion
This is the first study that investigate the effects of dietary supplementation of freeze-dried Escherichia coli phage cocktail at different dosages (four different phages that lyse four different E. coli strains at 1010 PFU/g each), commercial probiotics (Lactobacillus acidophilus, Lactobacillus casei, Bifidobacterium termophilum, Enterococcus faecium and Aspergillus oryzae at 109 CFU/g each) and their combinations on growth performance and gut microbiota diversity of broiler chickens. The experiment was performed in normal physiological conditions without bacterial (E. coli) challenge. The present study has also demonstrated, for the first time, that phage targeting non-pathogenic E. coli was used. This study showed that chickens fed with combination of phage cocktail and probiotics enhanced growth performance and positively modulate the gut microbiota.
Growth performance parameters are the most important parameters to evaluate the efficacy of feed supplements in broiler production. High BW, BWG, FI, and low FCR showed that the feed supplements enhance growth performance. The results of the present study showed that chickens fed with 1ɸP without bacterial challenge had significantly (P < 0.05) better BW (35 d), BWG (22 - 35 d, 1 – 35d), and FCR (1-21 d, 22-35 d, 1-35 d) compared to control. The 1ɸP also showed better FCR (1-21 d) compared to control than the P group. No study was done on the effects of combination of both phages and probiotics in chicken, which made comparison difficult. However, studies in pigs have shown that supplementation with 1 g/kg and 1.5 g/kg commercial phage cocktail (S. typhimurium, S. enteritidis, S. cholerasuis and S. derby), Staphylococcus aureus, Escherichia coli (k88, k99 and f41) and Clostridium perfringens types A and C at 109 PFU/g each) without bacterial challenge and in combination with 3 g/kgprobiotics (Lactobacillus acidophilus K31, Bacillus subtilis K 42, Saccharomyces cerevisiae K47) at 108, 109 and 104 CFU/g, respectively) did not show any significant improvement on growth performance compared to phage alone (Kim et al., 2016). This contrasts with the findings reported here because pigs and broilers have distinct physiological gut environments.
In the current study, 1ɸ and 2ɸ groups showed significantly (P < 0.05) better FCR (1-21 d. 22-35 d, 1-35 d) than the control agrees with other studies. Previous study demonstrated that the FCR of broilers supplemented with 1 ml of 1010 PFU/ml Salmonella typhimurium phage with the bacterial challenge was significantly better than control (Ngu et al. 2022). This result was in contrast to the findings of Upadhaya et al. (2021), who found that broilers supplemented with 0.5 g/kg and 1 g/kg phage cocktail (Salmonella gallinarum, Salmonella typhimurium, S. enteritidis, E. coli at 1.0 × 108 PFU/g each and C. perfringens at 1.0 × 106 PFU/g) without bacterial challenge only showed slightly (not significant) better BWG, FI and FCR than control. In our study, there was no difference in growth performance between low (1 g/kg of 1010 PFU/g) and high (2 g/kg of 1010 PFU/g) dosages of phages. Our result was in contrast with a study by Wang et al. (2013), who reported that a higher dosage of 0.5 g/kg S. typhimurium phage (108 PFU/g) without bacterial challenge showed significantly better FCR (1-14d) compared to control than the lower dosage of 0.25 g/kg S. typhimurium phage (108 PFU /g). This inconsistency may be due to the concentration of phage used in our study was already very high at 1010 PFU/g compared to others at 108 PFU/g. A note of caution is also due here since previous studies mentioned earlier incorporated phages targeting pathogens such as S. typhimurium, S. gallinarum, S. enteritidis, Clostridium perfringens, and E. coli. It is evidenced from this study that phage supplements that target non-pathogenic E. coli could also result in significant improvement in growth performance.
According to CAP plot and PERMANOVA pairwise test, distinct gut microbiota were found between the ɸP (1ɸP and 2ɸP) and non-ɸP groups (C, 1ɸ, 2ɸ and P) in ilea, particularly in the ilea of 35 d chickens. No prior research has examined the effects of phage, probiotics, and their combinations on the diversity of the gut microbiota based on 16S rRNA HT-NGS made comparisons challenging. Upadhaya et al. (2021), however, demonstrated that chickens supplied with 0.5 g/kg phage cocktail, 1 g/kg phage cocktail, and 0.25 g/kg Avilamix (antibiotic) had distinctively different gut microbiota based on unweighted UniFrac. This demonstrates that the diversity of the gut microbiota was altered by phage supplementation in broiler chickens. On the other note, in accordance with earlier studies, the chicken gut microbiota showed distinct differences based on age (p = 0.002) (21 d and 35 d) and section of the intestine (p = 0.001) (ilea and caeca) (Gong et al. 2002, Lu et al. 2003, Torok et al. 2008, Mohd Shaufi et al. 2015).
In the current study, the OTUs that were significantly (P < 0.05) present in ɸP than the non-ɸP groups were related to short-chain fatty acids (SCFAs) producers (e.g., Bacteroides, Odoribacter, Alistipes, Anaerotruncus, Ruminococcus, Clostridiales, Clostridium, Desulfovibrio, Butyricimonas, Faecalibacterium prausnitzii, Anaerostipes and Phascolarctobacterium). These SCFAs producers have known roles of excreting various enzymes to facilitate the breakdown of non-starch polysaccharides (NSP) to SCFAs such as acetic, succinic, propionic and butyric acid in chickens (Eeckhaut et al. 2011, Kaakoush et al. 2014, Oakley et al. 2014, Polansky et al. 2016). The presence of these SCFAs producers were associated with better digestion and energy production that improved growth performance in chickens (Scheppach and Weiler 2004). The SCFAs produced from the breakdown and fermentation of polysaccharides (e.g., cellulose and hemicellulose) were also recognised as an important source of energy for the host. Furthermore, the SCFAs producers have also been reported to inhibit the growth of pathogens such as Salmonella spp. and C. perfringens based on the reduction of intestinal pH, excretion of mucin, and host antimicrobial peptides (Oakley et al. 2014, Sergeant et al. 2014) (Hu and Guo 2007, Timbermont et al. 2010). The beneficial effects reported on the presence of SCFAs producers could be responsible for the improvement of chickens growth performance in the ɸP groups, especially for the 1ɸP group. However, the roles of phages and probiotic combinations on promoting the SCFAs producers were not clear.
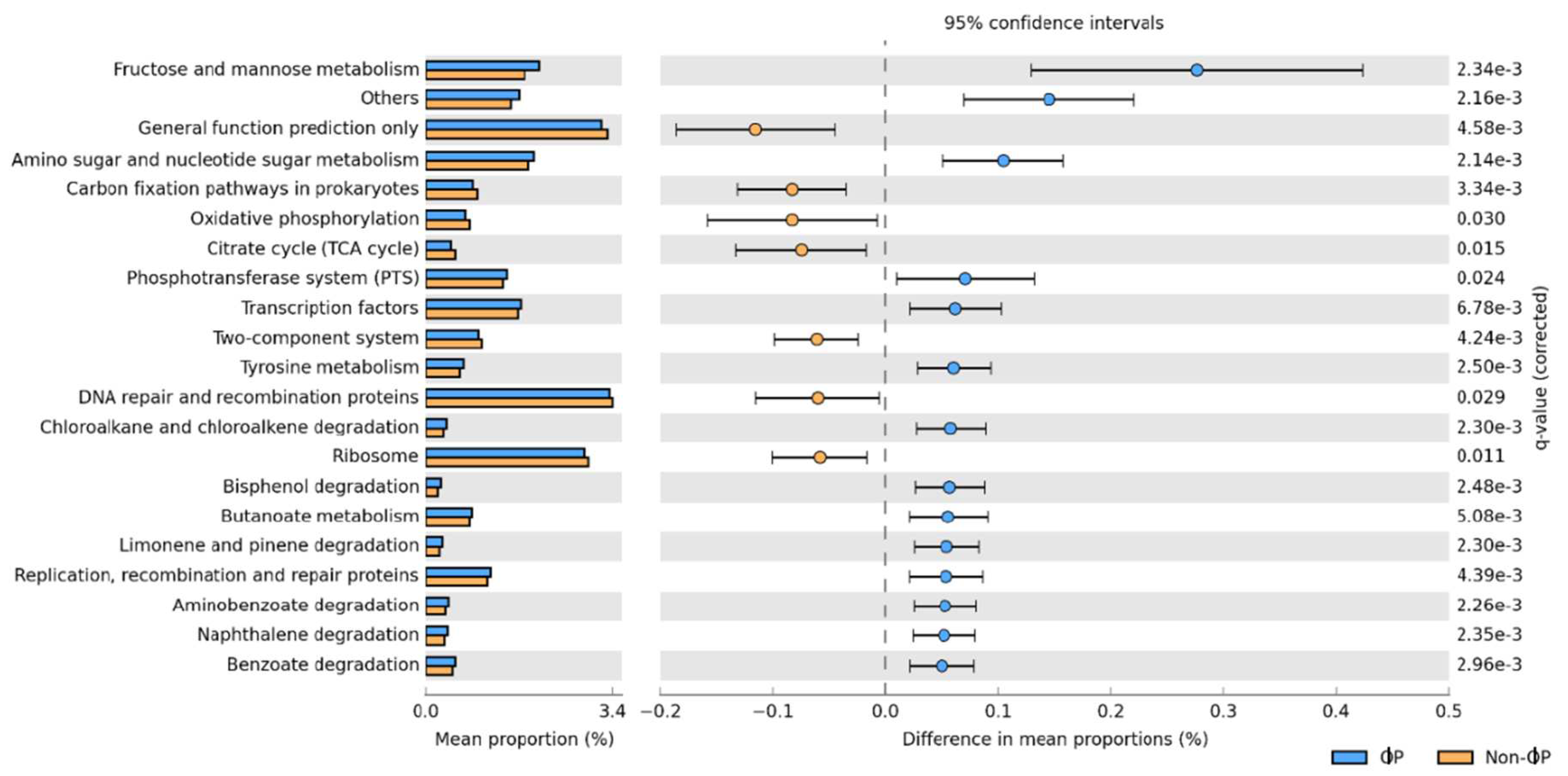
Based on the microbial predicted metagenome study, genes related to nutrient digestion and absorption and energy production, such as carbohydrate metabolism (fructose and mannose metabolism; butanoate metabolism) and amino acid metabolism (amino sugar and nucleotide sugar metabolism) were significantly upregulated in ɸP compared to non-ɸP groups. These findings were consistent with our earlier findings that SCFAs producers that were significantly present in ɸP groups could be capable of hydrolysing carbohydrate such as non-starch polysaccharides (NSPs) (e.g., glucose, fructose, starch and fructooligosaccharide) (Bjerrum et al. 2006). Another predicted function is the phosphotransferase system (PTS). The PTS has been demonstrated to facilitate the nutrient uptake of carbohydrate, glycerol and phosphate in members of Firmicutes bacteria (Polansky et al. 2016). The higher microbial predicted function related to metabolism and nutrient absorption suggests that ɸP groups had modulated the gut microbiota which may result in the improvement of chicken growth performance in 1ɸP supplemented group, especially in the period of 1-21 d. However, these microbial predicted metagenomes data need to be interpreted with caution as they can only be accurately assigned to the sequences present in the database, while there is no assignment available for novel bacteria (Waite and Taylor 2014). In addition, the predicted genes will not necessarily be expressed in the host. Therefore, a further validation step using targeted reverse transcriptase or shotgun metagenomics is required to study the RNA expression of the target genes.
It is unclear exactly how phages and probiotics affected the production of these SCFAs in the gut microbiota to increase growth performance. Further research is required to understand the mechanisms underlying the efficacy of phage and probiotic combinations in enhancing growth performance and gut microbiota diversity. It was proposed that the synergistic effects of the E. coli phage cocktail and probiotic combination may have supplied additional adhesion sites and nutrients and lowered toxins that favour the colonisation of these SCFAs producers. Although the advent of 16S rRNA of HT-NGS provides an unprecedented depth of sequencing gut microbiota, it unable to differentiate between viable and non-viable microorganisms (Ercolini 2013). Knowing whether or not a microorganism is viable is important, particularly in research utilising phages. Future study could focus on treating DNA samples with propidium monoazide (PMA) that able to distinguish the viability of microorganisms (Erkus et al. 2016).
This study was carried out to determine the effects of E. coli phage cocktail, commercial probiotics and their combinations on growth performance and modulation of gut microbiota in ilea and caeca of 21 and 35 d broiler chicken. Specifically, this study identified that supplementing a combination of 1 g/kg phage cocktail and probiotic (1ɸP) to broiler chickens significantly improved chicken growth performance and positively modulated gut microbiota. The 1ɸP group also had significantly better FCR (1-21 d) compared to the control than the probiotic (P) group. The OTUs related to SCFAs producers were dominantly observed in ɸP compared to the non-ɸP groups. They might be responsible for gut microbiota modulation that facilitates carbohydrate and amino acid metabolism and nutrient uptake, which provides energy for chicken growth. The microbial predicted metagenomes study also showed that the genes related to carbohydrate and amino acid metabolism and nutrient uptake were significantly present in ɸP compared to the non-ɸP groups. The results indicated that the 1ɸP supplement could be considered a potential alternative to AGPs for poultry.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org.
References
- Bjerrum, L., R. M. Engberg, T. D. Leser, B. B. Jensen, K. Finster and K. Pedersen (2006). "Microbial community composition of the ileum and cecum of broiler chickens as revealed by molecular and culture-based techniques." Poultry Science 85(7): 1151-1164. [CrossRef]
- Blajman, J. E., L. S. Frizzo, M. V. Zbrun, D. M. Astesana, M. L. Fusari, L. P. Soto, M. R. Rosmini and M. L. Signorini (2014). "Probiotics and broiler growth performance: a meta-analysis of randomised controlled trials." British Poultry Science 55(4): 483-494. [CrossRef]
- Clarke, K. and R. Gorley (2007). PRIMER-6. 6.1, PRIMER-E Ltd, Plymouth.
- De Vuyst, L., L. Avonts and L. Makras (2004). Probiotics, prebiotics and gut health: 416-482.
- Dibner, J. J. and J. D. Richards (2005). "Antibiotic growth promoters in agriculture: history and mode of action." Poultry Science 84(4): 634-643. [CrossRef]
- Duncan, D. B. (1955). "Multiple range and multiple F tests." Biometrics 11(1): 1-42. [CrossRef]
- Dunshea, F. R., D. N. D'Souza, B. B. Jensen and R. M. Engberg (2014). MEAT, ANIMAL, POULTRY AND FISH PRODUCTION AND MANAGEMENT | Antibiotic Growth Promotants. Encyclopedia of Meat Sciences. Oxford, Academic Press: 172-176.
- Eeckhaut, V., F. Van Immerseel, S. Croubels, S. De Baere, F. Haesebrouck, R. Ducatelle, P. Louis and P. Vandamme (2011). "Butyrate production in phylogenetically diverse Firmicutes isolated from the chicken caecum." Microbial Biotechnology 4(4): 503-512. [CrossRef]
- Ercolini, D. (2013). "High-throughput sequencing and metagenomics: moving forward in the culture-independent analysis of food microbial ecology." Applied and Environmental Microbiology 79(10): 3148-3155.
- Erkus, O., V. C. de Jager, R. T. Geene, I. van Alen-Boerrigter, L. Hazelwood, S. A. van Hijum, M. Kleerebezem and E. J. Smid (2016). "Use of propidium monoazide for selective profiling of viable microbial cells during Gouda cheese ripening." International Journal of Food Microbiology 228: 1-9. [CrossRef]
- Flynn, D. (2011). South Korea bans antibiotics in animal feed. Seattle: Food Safety News.
- Gong, J., R. J. Forster, H. Yu, J. R. Chambers, R. Wheatcroft, P. M. Sabour and S. Chen (2002). "Molecular analysis of bacterial populations in the ileum of broiler chickens and comparison with bacteria in the cecum." FEMS Microbiology Ecology 41(3): 171-179.
- Hu, Z. and Y. Guo (2007). "Effects of dietary sodium butyrate supplementation on the intestinal morphological structure, absorptive function and gut flora in chickens." Animal Feed Science and Technology 132(3): 240-249.
- Kaakoush, N., N. Sodhi, J. Chenu, J. Cox, S. Riordan and H. Mitchell (2014). "The interplay between Campylobacter and Helicobacter species and other gastrointestinal microbiota of commercial broiler chickens." Gut Pathogens 6(1): 18. [CrossRef]
- Kabir, S. (2009). "The role of probiotics in the poultry industry." International Journal of Molecular Sciences 10(8): 3531-3546.
- Karavolias, J., M. J. Salois, K. T. Baker and K. Watkins (2018). "Raised without antibiotics: impact on animal welfare and implications for food policy." Translational Animal Science 2(4): 337-348. [CrossRef]
- Kim, J. S., A. Hosseindoust, S. H. Lee, Y. H. Choi, M. J. Kim, J. H. Lee, I. K. Kwon and B. J. Chae (2017). "Bacteriophage cocktail and multi-strain probiotics in the feed for weanling pigs: effects on intestine morphology and targeted intestinal coliforms and Clostridium." Animal 11(1): 45-53. [CrossRef]
- Kim, K. H., S. L. Ingale, J. S. Kim, S. H. Lee, J. H. Lee, I. K. Kwon and B. J. Chae (2014). "Bacteriophage and probiotics both enhance the performance of growing pigs but bacteriophage are more effective." Animal Feed Science and Technology 196: 88-95. [CrossRef]
- Klindworth, A., E. Pruesse, T. Schweer, J. Peplies, C. Quast, M. Horn and F. O. Glockner (2013). "Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies." Nucleic Acids Research 41(1): e1. [CrossRef]
- Langille, M. G. I., J. Zaneveld, J. G. Caporaso, D. McDonald, D. Knights, J. A. Reyes, J. C. Clemente, D. E. Burkepile, R. L. Vega Thurber, R. Knight, R. G. Beiko and C. Huttenhower (2013). "Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences." Nature Biotechnology 31(9): 814-821. [CrossRef]
- Leeson, S. (2008). "Predictions for Commercial Poultry Nutrition." Journal of Applied Poultry Research 17(2): 315-322.
- Liu, D., J. D. Van Belleghem, C. R. de Vries, E. Burgener, Q. Chen, R. Manasherob, J. R. Aronson, D. F. Amanatullah, P. D. Tamma and G. A. Suh (2021). "The safety and toxicity of phage therapy: a review of animal and clinical studies." Viruses 13(7): 1268. [CrossRef]
- Lu, J., U. Idris, B. Harmon, C. Hofacre, J. J. Maurer and M. D. Lee (2003). "Diversity and succession of the intestinal bacterial community of the maturing broiler chicken." Applied and Environmental Microbiology 69(11): 6816-6824.
- M'Sadeq, S. A., S. Wu, R. A. Swick and M. Choct (2015). "Towards the control of necrotic enteritis in broiler chickens with in-feed antibiotics phasing-out worldwide." Animal Nutrition 1(1): 1-11. [CrossRef]
- Maron, D. F., T. J. Smith and K. E. Nachman (2013). "Restrictions on antimicrobial use in food animal production: an international regulatory and economic survey." Globalization and Health 9(1): 1-11.
- McEwen, S. A., F. J. Angulo, P. J. Collignon and J. Conly (2017). Potential unintended consequences associated with restrictions on antimicrobial use in food-producing animals. WHO Guidelines on Use of Medically Important Antimicrobials in Food-Producing Animals, World Health Organization.
- Mehdi, Y., M. P. Letourneau-Montminy, M. L. Gaucher, Y. Chorfi, G. Suresh, T. Rouissi, S. K. Brar, C. Cote, A. A. Ramirez and S. Godbout (2018). "Use of antibiotics in broiler production: Global impacts and alternatives." Animal Nutrition 4(2): 170-178. [CrossRef]
- Mohd Shaufi, M. A., C. C. Sieo, C. W. Chong, H. M. Gan and Y. W. Ho (2015). "Deciphering chicken gut microbial dynamics based on high-throughput 16S rRNA metagenomics analyses." Gut Pathogens 7(1): 4. [CrossRef]
- Mountzouris, K. C., P. Tsitrsikos, I. Palamidi, A. Arvaniti, M. Mohnl, G. Schatzmayr and K. Fegeros (2010). "Effects of probiotic inclusion levels in broiler nutrition on growth performance, nutrient digestibility, plasma immunoglobulins, and cecal microflora composition." Poultry Science 89(1): 58-67.
- Naidoo, V., L. J. McGaw, S. P. Bisschop, N. Duncan and J. N. Eloff (2008). "The value of plant extracts with antioxidant activity in attenuating coccidiosis in broiler chickens." Veterinary Parasitology 153(3-4): 214-219. [CrossRef]
- Ngu, N., L. Phuong, L. Anh, H. Loc, N. Tam, P. Huan, T. Diep and K. Kamei (2022). "The Efficiency of Bacteriophages Against Salmonella Typhimurium Infection in Native Noi Broilers." Brazilian Journal of Poultry Science 24. [CrossRef]
- Nhung, N. T., N. Chansiripornchai and J. J. Carrique-Mas (2017). "Antimicrobial Resistance in Bacterial Poultry Pathogens: A Review." Frontiers in Veterinary Science 4: 126. [CrossRef]
- NRC (1994). Nutrient Requirements of Poultry. Washington, DC, The National Academies Press.
- Oakley, B. B., H. S. Lillehoj, M. H. Kogut, W. K. Kim, J. J. Maurer, A. Pedroso, M. D. Lee, S. R. Collett, T. J. Johnson and N. A. Cox (2014). "The chicken gastrointestinal microbiome." FEMS Microbiology Letter 360(2): 100-112.
- Parks, D. H., G. W. Tyson, P. Hugenholtz and R. G. Beiko (2014). "STAMP: statistical analysis of taxonomic and functional profiles." Bioinformatics 30(21): 3123-3124.
- Paulson, J. N. (2014). "metagenomeSeq: Statistical analysis for sparse high-throughput sequencing." Bioconductor package 1(0).
- Polansky, O., Z. Sekelova, M. Faldynova, A. Sebkova, F. Sisak and I. Rychlik (2016). "Important Metabolic Pathways and Biological Processes Expressed by Chicken Cecal Microbiota." Applied and Environmental Microbiology 82(5): 1569-1576.
- Scheppach, W. and F. Weiler (2004). "The butyrate story: old wine in new bottles?" Current Opinion in Clinical Nutrition & Metabolic Care 7(5): 563-567.
- Schloss, P. D., S. L. Westcott, T. Ryabin, J. R. Hall, M. Hartmann, E. B. Hollister, R. A. Lesniewski, B. B. Oakley, D. H. Parks, C. J. Robinson, J. W. Sahl, B. Stres, G. G. Thallinger, D. J. Van Horn and C. F. Weber (2009). "Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities." Applied and Environmental Microbiology 75(23): 7537-7541.
- Sergeant, M. J., C. Constantinidou, T. A. Cogan, M. R. Bedford, C. W. Penn and M. J. Pallen (2014). "Extensive microbial and functional diversity within the chicken cecal microbiome." PLoS One 9(3): e91941. [CrossRef]
- Shaufi, M. A. M., C. C. Sieo, Y.-K. Cheah, C. W. Chong, A. R. Omar, Y. W. Ho and G. H. Tan (2017). "Discrimination of Escherichia coli isolates recovered from mucosal contents of chicken intestines and different age by repetitive elements sequence-based PCR." Journal of Biochemistry, Microbiology and Biotechnology 5(1): 7-12. [CrossRef]
- Steele, A., H. J. Stacey, S. de Soir and J. D. Jones (2020). "The Safety and Efficacy of Phage Therapy for Superficial Bacterial Infections: A Systematic Review." Antibiotics 9(11): 754. [CrossRef]
- Timbermont, L., A. Lanckriet, J. Dewulf, N. Nollet, K. Schwarzer, F. Haesebrouck, R. Ducatelle and F. Van Immerseel (2010). "Control of Clostridium perfringens-induced necrotic enteritis in broilers by target-released butyric acid, fatty acids and essential oils." Avian Pathology 39(2): 117-121.
- Torok, V. A., K. Ophel-Keller, M. Loo and R. J. Hughes (2008). "Application of methods for identifying broiler chicken gut bacterial species linked with increased energy metabolism." Applied and Environmental Microbiology 74(3): 783-791.
- Upadhaya, S. D., J. M. Ahn, J. H. Cho, J. Y. Kim, D. K. Kang, S. W. Kim, H. B. Kim and I. H. Kim (2021). "Bacteriophage cocktail supplementation improves growth performance, gut microbiome and production traits in broiler chickens." Journal of Animal Science and Biotechnology 12(1): 49.
- Waite, D. W. and M. W. Taylor (2014). "Characterizing the avian gut microbiota: membership, driving influences, and potential function." Frontiers in Microbiology 5: 223.
- Wang, J. P., L. Yan, J. H. Lee and I. H. Kim (2013). "Evaluation of bacteriophage supplementation on growth performance, blood characteristics, relative organ weight, breast muscle characteristics and excreta microbial shedding in broilers." Asian-Australasian Journal of Animal Sciences 26(4): 573-578. [CrossRef]
- Wang, Y.-B. and B.-H. Xu (2008). "Effect of different selenium source (sodium selenite and selenium yeast) on broiler chickens." Animal Feed Science and Technology 144(3): 306-314. [CrossRef]
- Yan, L., S. M. Hong and I. H. Kim (2012). "Effect of bacteriophage supplementation on the growth performance, nutrient digestibility, blood characteristics, and fecal microbial shedding in growing pigs." Asian-Australasian Journal of Animal Sciences 25(10): 1451-1456.
- Ye, Y., Z. Li, P. Wang, B. Zhu, M. Zhao, D. Huang, Y. Ye, Z. Ding, L. Li, G. Wan, Q. Wu, D. Song and Y. Tang (2021). "Effects of probiotic supplements on growth performance and intestinal microbiota of partridge shank broiler chicks." PeerJ 9: e12538. [CrossRef]
- Yu, Y., Q. Li, X. Zeng, Y. Xu, K. Jin, J. Liu and G. Cao (2022). "Effects of Probiotics on the Growth Performance, Antioxidant Functions, Immune Responses, and Caecal Microbiota of Broilers Challenged by Lipopolysaccharide." Frontiers in Veterinary Science 9: 846649. [CrossRef]
- Zhao, P. Y., H. Y. Baek and I. H. Kim (2012). "Effects of bacteriophage supplementation on egg performance, egg quality, excreta microflora, and moisture content in laying hens." Asian-Australasian Journal of Animal Sciences 25(7): 1015-1020.
Figure 1.
Structure of gut microbiota supplemented with different dietary treatments in ilea and caeca of 21 and 35 d chickens investigated based on Principal coordinate analysis (PCO) of Bray-Curtis similarity index. For treatment (C = control (basal diet); 1ɸ = BD + 1 g/kg phage cocktail; 2ɸ = BD + 2 g/kg phage cocktail; P = BD + 1 g/kg probiotic; 1ɸP = BD + 1 g/kg phage cocktail + 1 g/kg probiotic; 2ɸP = BD + 2 g/kg phage cocktail + 1 g/kg probiotic).
Figure 1.
Structure of gut microbiota supplemented with different dietary treatments in ilea and caeca of 21 and 35 d chickens investigated based on Principal coordinate analysis (PCO) of Bray-Curtis similarity index. For treatment (C = control (basal diet); 1ɸ = BD + 1 g/kg phage cocktail; 2ɸ = BD + 2 g/kg phage cocktail; P = BD + 1 g/kg probiotic; 1ɸP = BD + 1 g/kg phage cocktail + 1 g/kg probiotic; 2ɸP = BD + 2 g/kg phage cocktail + 1 g/kg probiotic).
Figure 2.
Structure of gut microbiota supplemented with different dietary treatments in ilea of 21 and 35 d chickens investigated based on Canonical analysis of principal coordinates (CAP) of Bray-Curtis similarity index. For treatment (C = control (basal diet); 1ɸ = BD + 1 g/kg phage cocktail; 2ɸ = BD + 2 g/kg phage cocktail; P = BD + 1 g/kg probiotic; 1ɸP = BD + 1 g/kg phage cocktail + 1 g/kg probiotic; 2ɸP = BD + 2 g/kg phage cocktail + 1 g/kg probiotic), age (21 = 21-day-old; 35 = 35-day-old) and part of intestine (I = ilea, C = caeca).
Figure 2.
Structure of gut microbiota supplemented with different dietary treatments in ilea of 21 and 35 d chickens investigated based on Canonical analysis of principal coordinates (CAP) of Bray-Curtis similarity index. For treatment (C = control (basal diet); 1ɸ = BD + 1 g/kg phage cocktail; 2ɸ = BD + 2 g/kg phage cocktail; P = BD + 1 g/kg probiotic; 1ɸP = BD + 1 g/kg phage cocktail + 1 g/kg probiotic; 2ɸP = BD + 2 g/kg phage cocktail + 1 g/kg probiotic), age (21 = 21-day-old; 35 = 35-day-old) and part of intestine (I = ilea, C = caeca).
Figure 3.
Structure of gut microbiota supplemented with different dietary treatments in ilea of 35 d chickens investigated based on Canonical analysis of principal coordinates (CAP) of Bray-Curtis similarity index. For treatment (C = control (basal diet); 1ɸ = BD + 1 g/kg phage cocktail; 2ɸ = BD + 2 g/kg phage cocktail; P = BD + 1 g/kg probiotic; 1ɸP = BD + 1 g/kg phage cocktail + 1 g/kg probiotic; 2ɸP = BD + 2 g/kg phage cocktail + 1 g/kg probiotic), age (21 = 21-day-old; 35 = 35-day-old) and part of intestine (I = ilea, C = caeca).
Figure 3.
Structure of gut microbiota supplemented with different dietary treatments in ilea of 35 d chickens investigated based on Canonical analysis of principal coordinates (CAP) of Bray-Curtis similarity index. For treatment (C = control (basal diet); 1ɸ = BD + 1 g/kg phage cocktail; 2ɸ = BD + 2 g/kg phage cocktail; P = BD + 1 g/kg probiotic; 1ɸP = BD + 1 g/kg phage cocktail + 1 g/kg probiotic; 2ɸP = BD + 2 g/kg phage cocktail + 1 g/kg probiotic), age (21 = 21-day-old; 35 = 35-day-old) and part of intestine (I = ilea, C = caeca).
Figure 4.
Principal component analysis (PCA) of predicted functional metagenomes based on ɸP versus non-ɸP groups. For phage cocktail and probiotic combinations groups (ɸP; 1ɸP and 2ɸP) and other groups (Non-ɸP; C, 1ɸ, 2ɸ and P).
Figure 4.
Principal component analysis (PCA) of predicted functional metagenomes based on ɸP versus non-ɸP groups. For phage cocktail and probiotic combinations groups (ɸP; 1ɸP and 2ɸP) and other groups (Non-ɸP; C, 1ɸ, 2ɸ and P).
Figure 5.
Pairwise comparison of the predicted functional metagenomes using Storey's FDR multiple test correction methods based on ɸP versus non-ɸP groups. For phage cocktail and probiotic combinations groups (ɸP; 1ɸP and 2ɸP) and other groups (Non-ɸP; C, 1ɸ, 2ɸ and P).
Figure 5.
Pairwise comparison of the predicted functional metagenomes using Storey's FDR multiple test correction methods based on ɸP versus non-ɸP groups. For phage cocktail and probiotic combinations groups (ɸP; 1ɸP and 2ɸP) and other groups (Non-ɸP; C, 1ɸ, 2ɸ and P).
Table 1.
Feed composition of dietary treatments for starter phase (1 to 21 days).
Table 1.
Feed composition of dietary treatments for starter phase (1 to 21 days).
| |
Basal diet (C) |
BD + 1 g/kg Phage cocktail
(1ɸ) |
BD + 2 g/kg Phage cocktail
(2ɸ) |
BD + 1 g/kg Probiotic
(P) |
BD + 1 g/kg Phage cocktail + 1 g/kg Probiotic
(1ɸP) |
BD + 2 g/kg Phage cocktail + 1 g/kg Probiotic
(2ɸP) |
| Ingredient (g/kg) |
|
|
|
|
|
|
| Corn |
538.60 |
537.60 |
536.60 |
537.60 |
536.60 |
535.60 |
| Soybean Meal, 48% Cp |
361.90 |
361.90 |
361.90 |
361.90 |
361.90 |
361.90 |
| Fish meal |
30.00 |
30.00 |
30.00 |
30.00 |
30.00 |
30.00 |
| Palm Oil |
37.40 |
37.40 |
37.40 |
37.40 |
37.40 |
37.40 |
| 60% choline chloride |
2.50 |
2.50 |
2.50 |
2.50 |
2.50 |
2.50 |
| Vitamin premix† |
0.30 |
0.30 |
0.30 |
0.30 |
0.30 |
0.30 |
| Mineral premix‡ |
1.00 |
1.00 |
1.00 |
1.00 |
1.00 |
1.00 |
| Salt (NaCl) |
2.00 |
2.00 |
2.00 |
2.00 |
2.00 |
2.00 |
| DL-Methionine |
1.80 |
1.80 |
1.80 |
1.80 |
1.80 |
1.80 |
| Limestone |
13.00 |
13.00 |
13.00 |
13.00 |
13.00 |
13.00 |
| Dicalcium phosphate |
11.50 |
11.50 |
11.50 |
11.50 |
11.50 |
11.50 |
| Phage cocktail |
0.00 |
1.00 |
2.00 |
0.00 |
1.00 |
2.00 |
| Probiotic |
0.00 |
0.00 |
0.00 |
1.00 |
1.00 |
1.00 |
| Total |
1000.00 |
1000.00 |
1000.00 |
1000.00 |
1000.00 |
1000.00 |
| Calculated analysis |
|
|
|
|
|
|
| Metabolisable energy (MJ/kg) |
13.06 |
13.06 |
13.06 |
13.06 |
13.06 |
13.06 |
| Crude protein |
220.00 |
220.00 |
220.00 |
220.00 |
220.00 |
220.00 |
| Crude fat |
63.10 |
63.10 |
63.10 |
63.10 |
63.10 |
63.10 |
| Crude fibre |
38.00 |
38.00 |
38.00 |
38.00 |
38.00 |
38.00 |
| Methionine + Cysteine |
9.50 |
9.50 |
9.50 |
9.50 |
9.50 |
9.50 |
| Lysine |
13.70 |
13.70 |
13.70 |
13.70 |
13.70 |
13.70 |
| Calcium |
10.20 |
10.20 |
10.20 |
10.20 |
10.20 |
10.20 |
| Phosphorus |
4.50 |
4.50 |
4.50 |
4.50 |
4.50 |
4.50 |
| Chemical analysis |
|
|
|
|
|
|
| Crude protein |
217.70 |
217.70 |
217.70 |
217.70 |
217.70 |
217.70 |
| Crude fat |
54.80 |
54.80 |
54.80 |
54.80 |
54.80 |
54.80 |
| Crude fibre |
38.80 |
38.80 |
38.80 |
39.01 |
39.02 |
39.02 |
| Calcium |
6.50 |
6.55 |
6.61 |
6.66 |
6.71 |
6.77 |
| Phosphorus |
5.16 |
5.16 |
5.16 |
5.16 |
5.16 |
5.16 |
| Sodium |
6.70 |
6.71 |
6.72 |
6.70 |
6.71 |
6.72 |
Table 2.
Feed composition of dietary treatments for finisher phase (22 to 35 days).
Table 2.
Feed composition of dietary treatments for finisher phase (22 to 35 days).
| |
Basal diet (C) |
BD + 1 g/kg Phage cocktail
(1ɸ) |
BD + 2 g/kg Phage cocktail
(2ɸ) |
BD + 1 g/kg Probiotic
(P) |
BD + 1 g/kg Phage cocktail + 1 g/kg Probiotic
(1ɸP) |
BD + 2 g/kg Phage cocktail + 1 g/kg Probiotic
(2ɸP) |
| Ingredient (g/kg) |
|
|
|
|
|
|
| Corn |
602.70 |
601.70 |
600.70 |
601.70 |
600.70 |
599.70 |
| Soybean Meal, 48% Cp |
318.60 |
318.60 |
318.60 |
318.60 |
318.60 |
318.60 |
| Fish meal |
30.00 |
30.00 |
30.00 |
30.00 |
30.00 |
30.00 |
| Palm Oil |
24.50 |
24.50 |
24.50 |
24.50 |
24.50 |
24.50 |
| 60% choline chloride |
2.00 |
2.00 |
2.00 |
2.00 |
2.00 |
2.00 |
| Vitamin premix† |
0.30 |
0.30 |
0.30 |
0.30 |
0.30 |
0.30 |
| Mineral premix‡ |
1.00 |
1.00 |
1.00 |
1.00 |
1.00 |
1.00 |
| Salt (NaCl) |
1.00 |
1.00 |
1.00 |
1.00 |
1.00 |
1.00 |
| DL-Methionine |
0.40 |
0.40 |
0.40 |
0.40 |
0.40 |
0.40 |
| Limestone |
13.00 |
13.00 |
13.00 |
13.00 |
13.00 |
13.00 |
| Dicalcium phosphate |
6.50 |
6.50 |
6.50 |
6.50 |
6.50 |
6.50 |
| Phage cocktail |
0.00 |
1.00 |
2.00 |
0.00 |
1.00 |
2.00 |
| Probiotic |
0.00 |
0.00 |
0.00 |
1.00 |
1.00 |
1.00 |
| Total |
1000.00 |
1000.00 |
1000.00 |
1000.00 |
1000.00 |
1000.00 |
| Calculated analysis |
|
|
|
|
|
|
| Metabolisable energy (MJ/kg) |
13.06 |
13.06 |
13.06 |
13.06 |
13.06 |
13.06 |
| Crude protein |
199.90 |
199.90 |
199.90 |
199.90 |
199.90 |
199.90 |
| Crude fat |
52.20 |
52.20 |
52.20 |
52.20 |
52.20 |
52.20 |
| Crude fibre |
36.50 |
36.50 |
36.50 |
36.50 |
36.50 |
36.50 |
| Methionine + Cysteine |
8.50 |
8.50 |
8.50 |
8.50 |
8.50 |
8.50 |
| Lysine |
12.00 |
12.00 |
12.00 |
12.00 |
12.00 |
12.00 |
| Calcium |
9.00 |
9.00 |
9.00 |
9.00 |
9.00 |
9.00 |
| Phosphorus |
3.50 |
3.50 |
3.50 |
3.50 |
3.50 |
3.50 |
| Chemical analysis |
|
|
|
|
|
|
| Crude protein |
212.05 |
212.50 |
212.50 |
212.50 |
212.50 |
212.50 |
| Crude fat |
45.00 |
45.00 |
45.00 |
45.00 |
45.00 |
45.00 |
| Crude fibre |
40.50 |
40.50 |
40.50 |
40.71 |
40.71 |
40.71 |
| Calcium |
6.80 |
6.85 |
6.91 |
6.96 |
7.01 |
7.07 |
| Phosphorus |
4.50 |
4.50 |
4.50 |
4.50 |
4.50 |
4.50 |
| Sodium |
1.90 |
1.91 |
1.92 |
1.90 |
1.91 |
1.92 |
Table 3.
Effects of different dietary treatments on growth performances of chickens.
Table 3.
Effects of different dietary treatments on growth performances of chickens.
| Item† |
Age |
C |
1ɸ |
2ɸ |
P |
1ɸP |
2ɸP |
| BW (g) |
1 d |
43.00 ± 1.10 |
42.78 ± 1.09 |
42.98 ± 0.78 |
42.82 ± 0.96 |
42.78 ± 1.05 |
43.32 ± 0.38 |
| |
21 d |
859.58 ± 14.20 |
874.07 ± 8.85 |
864.67 ± 11.05 |
871.78 ± 6.09 |
874.02 ± 12.31 |
855.10 ± 13.74 |
| |
35 d |
1605.30 ± 39.45 a
|
1734.17 ± 47.55 ab
|
1741.27 ± 56.58 ab
|
1690.43 ± 40.05 ab
|
1785.75 ± 31.93 b
|
1733.63 ± 56.00 ab
|
| BWG (g/bird/d) |
1-21 d |
816.63 ± 13.97 |
861.32 ± 9.47 |
821.70 ± 11.09 |
828.98 ± 6.04 |
831.25 ± 11.91 |
811.80 ± 12.95 |
| |
22-35 d |
745.72 ± 40.37 a
|
860.10 ± 50.69 ab
|
876.60 ± 46.20 ab
|
818.65 ± 41.87 ab
|
911.73 ± 32.82 b
|
878.53 ± 47.67 ab
|
| |
1-35 d |
1562.35 ± 40.42 a
|
1691.42 ± 47.67 ab
|
1698.30 ± 56.55 ab
|
1647.63 ± 39.82 ab
|
1742.98 ± 32.76 b
|
1690.33 ± 55.75 ab
|
| FI (g/bird/d) |
1-21 d |
1169.88 ± 18.03 |
1153.10 ± 10.46 |
1111.35 ± 30.52 |
1104.87 ± 37.66 |
1160.58 ± 20.14 |
1131.58 ± 10.33 |
| |
22-35 d |
1748.38 ± 24.66 |
1761.45 ± 51.10 |
1650.32 ± 104.62 |
1653.68 ± 40.06 |
1740.27 ± 63.75 |
1760.85 ± 44.20 |
| |
1-35 d |
2918.27 ± 38.40 |
2914.55 ± 55.02 |
2922.88 ± 158.00 |
2758.53 ± 51.98 |
2900.85 ± 70.40 |
2892.42 ± 53.92 |
| FCR (feed/gain) |
1-21 d |
1.43 ± 0.17 c
|
1.35 ± 0.17 a
|
1.37 ± 0.15 ab
|
1.40 ± 0.10 bc
|
1.37 ± 0.11 ab
|
1.34 ± 0.15 a
|
| |
22-35 d |
2.53 ± 0.12 b
|
2.10 ± 0.08 a
|
2.06 ± 0.06 a
|
1.97 ± 0.06 a
|
1.89 ± 0.04 a
|
2.01 ± 0.07 a
|
| |
1-35 d |
1.87 ± 0.04 b
|
1.68 ± 0.03 a
|
1.65 ± 0.03 a
|
1.61 ± 0.03 a
|
1.60 ± 0.03 a
|
1.61 ± 0.03 a
|
Table 4.
Structure of gut microbiota supplemented with different dietary treatments in ilea of 21 and 35 d old chickens based on PERMANOVA (a) marginal and (b) pairwise test of Bray-Curtis similarities. The test includes degrees of freedom (Df), sum of squares (SS), mean square (MS) and P value under Monte-Carlo correction (PMC).
Table 4.
Structure of gut microbiota supplemented with different dietary treatments in ilea of 21 and 35 d old chickens based on PERMANOVA (a) marginal and (b) pairwise test of Bray-Curtis similarities. The test includes degrees of freedom (Df), sum of squares (SS), mean square (MS) and P value under Monte-Carlo correction (PMC).
|
| Source |
Df |
SS |
MS |
Pseudo-F |
PMC
|
| Treatment |
5 |
23528 |
4705.6 |
1.855 |
0.005 |
| Residual |
65 |
1.6488E+05 |
2536.7 |
|
|
| Total |
70 |
1.8841E+05 |
|
|
|
- b.
Pairwise test |
| Groups† |
t |
Unique perms |
PMC
|
| 1ɸ, 1ɸP |
1.0853 |
998 |
0.299 |
| 1ɸ, 2ɸ |
0.73964 |
998 |
0.81 |
| 1ɸ, 2ɸP |
1.5589 |
998 |
0.043 |
| 1ɸ, C |
0.96796 |
999 |
0.457 |
| 1ɸ, P |
0.66535 |
997 |
0.867 |
| 1ɸP, 2ɸ |
1.2802 |
998 |
0.137 |
| 1ɸP, 2ɸP |
1.1864 |
999 |
0.179 |
| 1ɸP, C |
1.6169 |
996 |
0.026 |
| 1ɸP, P |
1.4116 |
998 |
0.051 |
| 2ɸ, 2ɸP |
1.9504 |
998 |
0.004 |
| 2ɸ, C |
1.0898 |
999 |
0.325 |
| 2ɸ, P |
0.86084 |
998 |
0.595 |
| 2ɸP, C |
2.2436 |
999 |
0.001 |
| 2ɸP, P |
2.0671 |
999 |
0.002 |
| C, P |
0.73884 |
999 |
0.804 |
Table 5.
Structure of gut microbiota supplemented with different dietary treatments in ilea of 35 d old chickens based on PERMANOVA (a) marginal and (b) pairwise test of Bray-Curtis similarities The test includes degrees of freedom (Df), sum of squares (SS), mean square (MS) and P value under Monte-Carlo correction (PMC).
Table 5.
Structure of gut microbiota supplemented with different dietary treatments in ilea of 35 d old chickens based on PERMANOVA (a) marginal and (b) pairwise test of Bray-Curtis similarities The test includes degrees of freedom (Df), sum of squares (SS), mean square (MS) and P value under Monte-Carlo correction (PMC).
|
| Source |
Df |
SS |
MS |
Pseudo-F |
PMC
|
| Treatment |
5 |
28910 |
5782.1 |
4.0189 |
0.001 |
| Residual |
29 |
41723 |
1438.7 |
|
|
| Total |
34 |
70634 |
|
|
|
- b.
Pairwise test |
| Groups† |
t |
Unique perms |
PMC
|
| 1ɸ, 1ɸP |
2.2179 |
403 |
0.011 |
| 1ɸ, 2ɸ |
0.99346 |
405 |
0.419 |
| 1ɸ, 2ɸP |
2.7819 |
405 |
0.002 |
| 1ɸ, C |
0.97549 |
408 |
0.437 |
| 1ɸ, P |
0.78783 |
413 |
0.672 |
| 1ɸP, 2O |
2.6116 |
01 |
0.002 |
| 1ɸP, 2OP |
1.179 |
418 |
0.234 |
| 1ɸP, C |
2.4102 |
403 |
0.006 |
| 1ɸP, P |
2.2141 |
410 |
0.005 |
| 2ɸ, 2OP |
3.2675 |
401 |
0.002 |
| 2ɸ, C |
0.80525 |
408 |
0.651 |
| 2ɸ, P |
0.98786 |
397 |
0.393 |
| 2ɸP, C |
3.167 |
399 |
0.001 |
| 2ɸP, P |
2.7201 |
400 |
0.002 |
| C, P |
1.0209 |
394 |
.3850 |
Table 6.
List of OTUs that significantly higher in ɸP than the non- ɸP groups.
Table 6.
List of OTUs that significantly higher in ɸP than the non- ɸP groups.
| OTUs |
Taxonomy |
LogFC |
Standard error (SE) |
P-values |
Adjusted P-values |
| Otu000006 |
Bacteroides uniformis |
3.238460356 |
0.513486 |
2.85E-10 |
1.64E-08 |
| Otu000001 |
Bacteroides |
3.082429046 |
0.511617 |
1.69E-09 |
6.49E-08 |
| Otu000585 |
Odoribacter |
2.884646375 |
1.034084 |
0.005278 |
0.02529 |
| Otu000009 |
Alistipes |
2.764961394 |
0.432556 |
1.64E-10 |
1.64E-08 |
| Otu000139 |
Alistipes finegoldii |
2.723944494 |
0.680663 |
6.28E-05 |
0.000723 |
| Otu000269 |
Ruminococcaceae_unclassified |
2.720698332 |
1.02679 |
0.008056 |
0.034313 |
| Otu000070 |
Alistipes |
2.647180277 |
0.526343 |
4.92E-07 |
1.41E-05 |
| Otu000624 |
Ruminococcaceae UCG-014 |
2.618442967 |
1.056871 |
0.013229 |
0.04612 |
| Otu000111 |
Anaerotruncus |
2.539148885 |
0.862369 |
0.003236 |
0.018607 |
| Otu000014 |
Ruminococcus |
2.495897314 |
0.525048 |
2.00E-06 |
4.59E-05 |
| Otu000031 |
Lachnospiraceae_unclassified |
2.470502349 |
0.558793 |
9.82E-06 |
0.000188 |
| Otu000117 |
Ruminococcaceae UCG-005 |
2.445622307 |
0.891623 |
0.00609 |
0.028015 |
| Otu000048 |
Bacillaceae_unclassified |
2.308772376 |
0.862055 |
0.007401 |
0.032737 |
| Otu000170 |
Bacteroides |
2.245170744 |
0.57669 |
9.89E-05 |
0.001034 |
| Otu000319 |
Rhodospirillaceae |
2.162299849 |
0.88805 |
0.014897 |
0.047586 |
| Otu000005 |
Alistipes onderdonkii |
2.121163741 |
0.521993 |
4.83E-05 |
0.000695 |
| Otu000210 |
Anaerotruncus |
2.025946607 |
0.945538 |
0.032142 |
0.080355 |
| Otu000015 |
Clostridium X1Vb |
2.019893556 |
0.617525 |
0.001072 |
0.007704 |
| Otu000337 |
Desulfovibrio |
2.014867537 |
0.841755 |
0.016682 |
0.049954 |
| Otu000675 |
Anaerostipes |
2.013275931 |
1.087486 |
0.064125 |
0.122906 |
| Otu000032 |
Clostridium X1Va |
1.982675782 |
0.536143 |
0.000217 |
0.002082 |
| Otu000643 |
Vampirovibrio |
1.967914419 |
1.072608 |
0.06655 |
0.125463 |
| Otu000353 |
Alistipes putredinis |
1.899173634 |
0.770005 |
0.013646 |
0.046157 |
| Otu000204 |
Ruminococcaceae UCG-014 |
1.859164848 |
0.750237 |
0.013208 |
0.04612 |
| Otu000157 |
Ruminococcaceae_unclassified |
1.841791976 |
0.706923 |
0.009178 |
0.036394 |
| Otu000866 |
Clostridium |
1.840566564 |
0.885046 |
0.03756 |
0.086387 |
| Otu000367 |
Coprobacillus |
1.822128533 |
1.039682 |
0.079674 |
0.143164 |
| Otu000694 |
Lachnospiraceae_unclassified |
1.815009851 |
1.076386 |
0.091756 |
0.158407 |
| Otu000066 |
Bacteroides fragilis |
1.803001647 |
0.592086 |
0.002325 |
0.014857 |
| Otu000025 |
Lachnospiraceae_unclassified |
1.784599464 |
0.409758 |
1.33E-05 |
0.000218 |
| Otu000426 |
Ruminococcaceae UCG-014 |
1.782185523 |
0.793533 |
0.024711 |
0.063151 |
| Otu000027 |
Eisenbergiella |
1.766953157 |
0.495082 |
0.000358 |
0.00317 |
| Otu000067 |
Butyricimonas |
1.753087241 |
0.611499 |
0.004146 |
0.02165 |
| Otu000075 |
Lachnospiraceae_unclassified |
1.747367694 |
0.612453 |
0.00433 |
0.02165 |
| Otu000247 |
Bacteria_unclassified |
1.741509274 |
0.770742 |
0.023851 |
0.062338 |
| Otu000022 |
Faecalibacterium prausnitzii |
1.738699859 |
0.433282 |
6.00E-05 |
0.000723 |
| Otu000104 |
Lachnospiraceae_unclassified |
1.715040783 |
0.493786 |
0.000514 |
0.004224 |
| Otu002794 |
Lachnospiraceae_unclassified |
1.702047151 |
1.080272 |
0.115124 |
0.194695 |
| Otu000095 |
Oscillospira |
1.686705907 |
0.725797 |
0.020129 |
0.055115 |
| Otu000338 |
Ruminococcaceae UCG-014 |
1.677289282 |
0.969175 |
0.083517 |
0.14776 |
| Otu000186 |
Anaerotruncus |
1.668325958 |
0.655373 |
0.010909 |
0.041816 |
| Otu000148 |
Oscillospira |
1.665393273 |
0.699204 |
0.017226 |
0.049954 |
| Otu000947 |
Clostridiales |
1.659154189 |
1.306282 |
0.204037 |
0.312856 |
| Otu000802 |
Clostridium IV |
1.654944242 |
0.813506 |
0.041918 |
0.090954 |
| Otu000041 |
Clostridium X1Va |
1.644739429 |
0.517848 |
0.001493 |
0.010098 |
| Otu000069 |
Ruminococcus |
1.601694881 |
0.542143 |
0.003133 |
0.018607 |
| Otu000044 |
Lachnospiraceae_unclassified |
1.575177456 |
0.551342 |
0.004277 |
0.02165 |
| Otu000573 |
Ruminococcus |
1.546199668 |
1.1055 |
0.16192 |
0.258623 |
| Otu000057 |
Ruminococcus |
1.517227727 |
0.458007 |
0.000924 |
0.007085 |
| Otu000007 |
Phascolarctobacterium |
1.505713845 |
0.607781 |
0.013234 |
0.04612 |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).