Submitted:
31 January 2023
Posted:
03 February 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. The discovery of the DGF-1 family
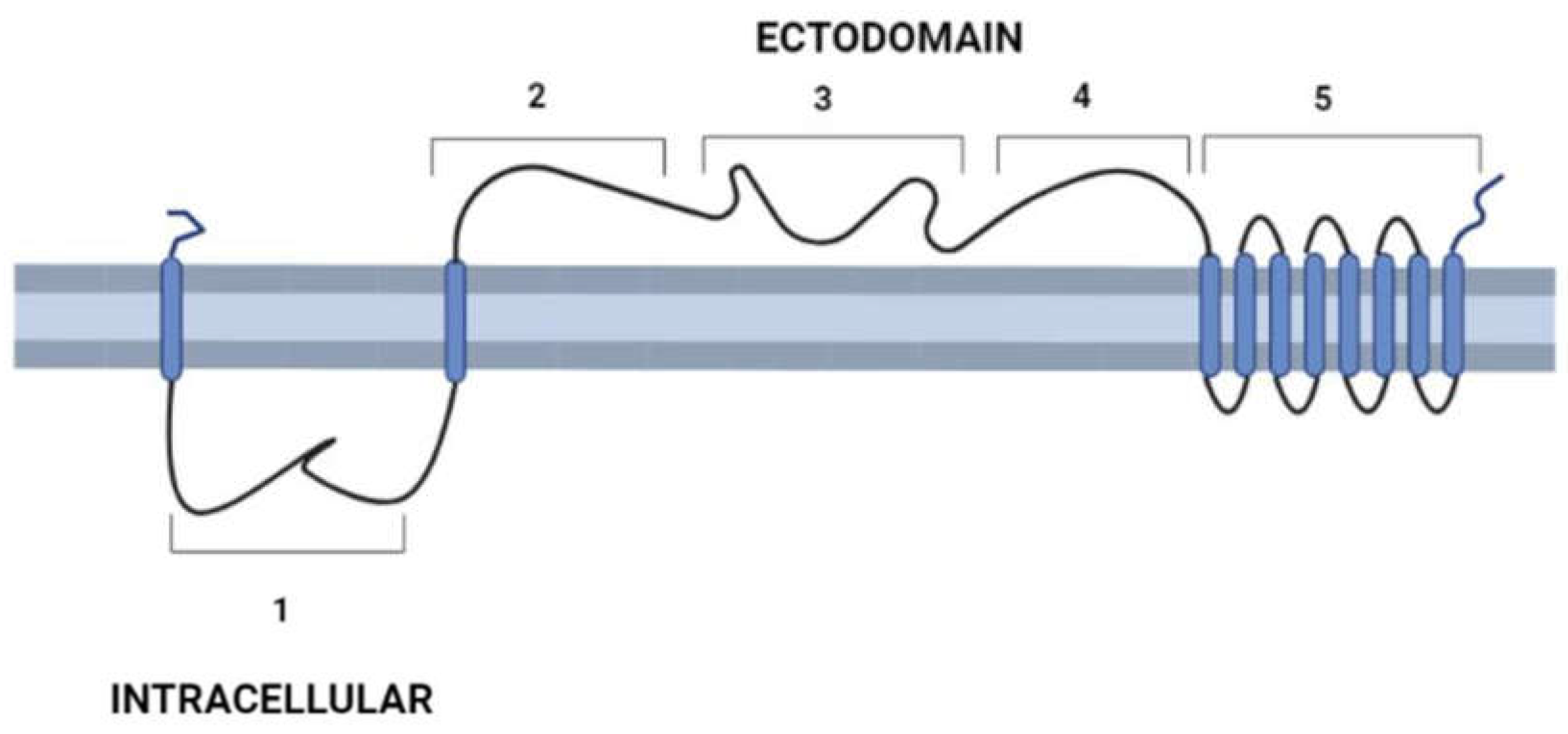
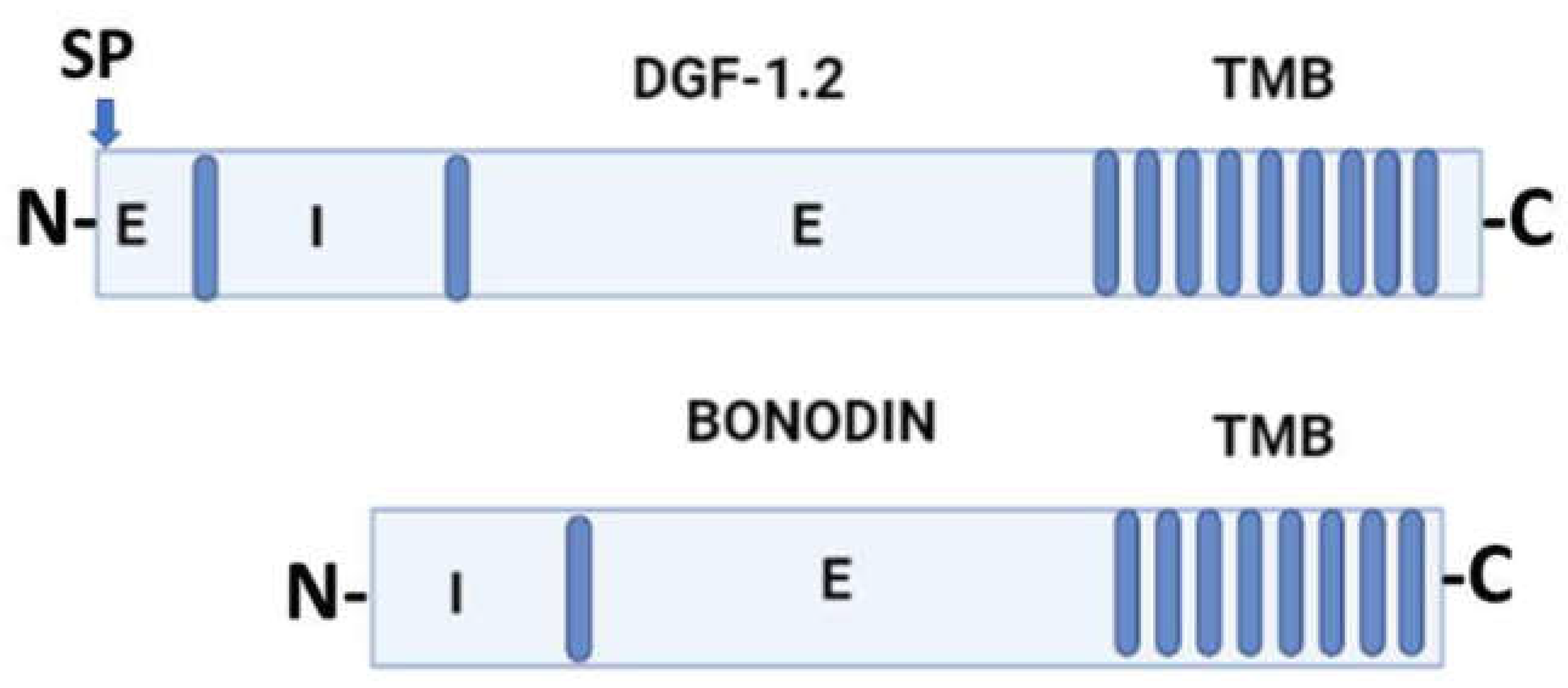
3. General molecular characteristics of DGF-1 proteins
4. DGF-1 genealogy
5. Chromosomal distribution and DGF-1 copy numbers
6. Possible roles for DGF-1 genes
In silico studies in DGF-1
7. Expression of DGF-1 genes
Translation
Transcription
8. Conclusions
9. Future Directions
Acknowledgments
References
- Tibayrenc, M., Ayala, F.J. (2022). Microevolution and subspecific taxonomy of Trypanosoma cruzi. Infect. Genet. Evol. 103:105344. [CrossRef]
- Https://www.who.int/news-room/fact-sheets/detail/chagas-disease-(american-trypanosomiasis).
- Gómez-Ochoa, S.A., Rojas, L.Z., Echeverría, L.E., Muka, T. and Franco, O.H., 2022. Global, Regional, and National Trends of Chagas Disease from 1990 to 2019: Comprehensive Analysis of the Global Burden of Disease Study. Global Heart, 17(1), p.59. [CrossRef]
- García-Huertas, P., Cardona-Castro, N. (2021) Advances in the treatment of Chagas disease: Promising new drugs, plants and targets. Biomedicine & Pharmacotherapy, Volume 142:112020. [CrossRef]
- El-Sayed, N.M., Myler, P.J., Bartholomeu, D.C. Nilsson, D., Aggarwal, G., Tran, Anh-Nhi, Ghedin, E, Worthey, E.A, Delcher, A.L, Blandin, G.et. al. (2005). The genome sequence of Trypanosoma cruzi, etiologic agent of Chagas disease. Science. 309:409-415. [CrossRef]
- Wincker, P., Roizes, G. Goldenberg, S. (1990). Characterization of a Trypanosoma cruzi specific nuclear repeated sequence, Mol. Biochem. Parasitol. 41:147-152. [CrossRef]
- Chávez, S., Urbaniak, M.D., Benz, C., Smircich, P., Garat, B., Sotelo-Silveira, J.R., Duhagon, M.A. (2021). Extensive Translational Regulation through the Proliferative Transition of Trypanosoma cruzi Revealed by Multi-Omics. mSphere. 6(5):e0036621. [CrossRef]
- Wincker, P., Cristina Murto-Dovales, A., Goldenberg, S. (1992). Nucleotide sequence of a representative member of a Trypanosoma cruzi dispersed gene family, Mol. Biochem. Parasitol. 55:217-220. [CrossRef]
- Stoco, P.H., Wagner, G., Talavera-Lopez, C., Gerber, A., Zaha, A., Thompson, C.E., Bartholomeu, D.C, Lückemeyer, D.D, Bahia, D, Loreto, E, et al., (2014). Genome of the avirulent human-infective trypanosome Trypanosoma rangeli. PLoS Negl Trop Dis. 18;8(9): e3176. [CrossRef]
- Bradwell, K.R., Koparde, V.N., Matveyev, A.V. Serraneo, M.G, Joao, J.M.P., Parikh, H., Huang, B., Lee, V., Espinosa-Alvarez O, et al. (2018). Genomic comparison of Trypanosoma conorhini and Trypanosoma rangeli to Trypanosoma cruzi strains of high and low virulence. BMC Genomics. 19(1):770. [CrossRef]
- Kelly, S., Ivens, A., Mott, G.A. O’Neill, M., Emms, D., Macleod, O., Voorheis, P., Tyler, K., Clark, M., Matthews J, et. al. (2017). An Alternative Strategy for Trypanosome Survival in the Mammalian Bloodstream Revealed through Genome and Transcriptome Analysis of the Ubiquitous Bovine Parasite Trypanosoma (Megatrypanum) theileri. Genome Biol Evol. 9:2093-2109. [CrossRef]
- Kelly, S., Ivens, A., Manna, P., Gibson, W. Field, M.C. (2014). A draft genome for the African crocodilian trypanosome Trypanosoma grayi. Sci Data 1, 140024. [CrossRef]
- Kawashita, S. Y., da Silva, C.V., Mortara, R.A., Burleigh, B., Briones, M.R.S. (2009). Homology, paralogy and function of DGF-1, a highly dispersed Trypanosoma cruzi specific gene family and its implications for information entropy of its encoded proteins, Mol. Biochem. Parasitol. 165:19-31. [CrossRef]
- Kim, D., Chiurillo, M.A., El-Sayed, N., Santos, M.R.M., Johns, K., Porcile, P.E., Andersson, B., Myler, P., da Silveira, J.F., Ramírez, J.L. (2005). Telomere and subtelomere of Trypanosoma cruzi chromosomes are enriched in (pseudo)genes of retrotransposon hot spot and trans-sialidase-like gene families: the origins of T. cruzi telomeres. Gene 346:153-61. [CrossRef]
- Lander, N., Bernal, C., Diez, N., Añez, N., Docampo, R., Ramirez, J.L. (2010). Localization and developmental regulation of a disperse gene family 1 protein in Trypanosoma cruzi. Infect. Immun 78:231-241.
- Ouaissi, M.A. (1988) Role of the RGD sequence in parasite adhesion to host cells. Parasitol. Today 4:169-173.
- Jumper, J., Evans, R., Pritzel, A., Green, T., Figurnov, M., Ronneberger, O.,Tunyasuvunakool, K., Bates, R., Žídek, A., Potapenko A, et. al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589. [CrossRef]
- Baek, M., DiMaio, F., Anishchenko, I., Bauparas, J., Ovchinnikov, S. Lee, G.R., Wang, J., Cong, Q., Kinch, L.N., Schaeffer, R.D. (2021). Accurate prediction of protein structures and interactions using a three-track neural network. Science 373:871-876. [CrossRef]
- Bhave, G., Nadin, B.M, Brasier, D.J., Glauner, K.S., Heinemann, G.F, Farzana, K. Karim, F., Gereau, R.W. (2003). Membrane Topology of a Metabotropic Glutamate Receptor J. Biol. Chem. 278: 30294–30301.
- . [CrossRef]
- Jackson, A.P., Otto, T.D., Aslett, M. Amstrong, S.D., Bringaud, F., Schlasch, A., Hartley, C., Sanders, M., Wastling, J.M., Dacks, J.B. et. al. (2016). Kinetoplastid Phylogenomics Reveals the Evolutionary Innovations Associated with the Origins of Parasitism Current Biology 26:161–172. [CrossRef]
- Olivares, M., Thomas, M.C., Lopez-Barajas, A., Requena, J.M., Garcias-Perez, J.L., Alonso, C, Lopez, M.C. (2000). Genome clustering of the Trypanosoma cruzi non-long terminal L1Tc retrotransposon with defined intersperse repeated DNA elements. Electrophoresis 21:2973-2982.
- Callejas-Hernández, F., Rastrojo, A., Poveda, C., Girones, N., Fresno, M. (2018). Genomic assemblies of newly sequenced Trypanosoma cruzi strains reveal new genomic expansion and greater complexity. Sci. Rep. 8, 14631. [CrossRef]
- Talavera-Lopez, C., Messenger, L.A., Lewis, M.D., Yeo, M., Reis-Cunha, J.L., Machado Matos, G., Bartholomeu, D.C., Calzada, J.E., Saldaña, A., Ramírez, J.D., Guhl, F, et. al. (2021). Repeat-driven Generation of antigenic diversity in a major human pathogen, Trypanosoma cruzi Front. Cell. Infect Microbiol. 11:614665. [CrossRef]
- Berná, L., Rodriguez, M., Chiribao, M.L., Parodi-Talice, A., Pita, S. Rijo, G., Alvarez-Valin, F., Robello, C. (2018). Expanding an expanded genome: long-read sequencing of Trypanosoma cruzi. Microb. Genomics. 4(5). [CrossRef]
- Ramirez, J.L. (2020) An Evolutionary View of Trypanosoma cruzi Telomeres. Front. Cell. Infect. Microbiol. 9:439. [CrossRef]
- Bezerra de Araujo, C., Pinheiro Chagas da Cunha, J., Inada, T., Damasceno, J., Lima, A.R.J., Iraiwa, P., Marques, C., Gonçalves, E., Nishiyama-Junior, M.Y., McCulloch, R., et. al. (2020) Replication origin location might contribute to genetic variability in Trypanosoma cruzi BMC Genomics 21:414. [CrossRef]
- Saha, A., Nanavaty, V.P., Li, B. (2019). Telomere and Subtelomere R-loops and Antigenic variation in Trypanosomes, J. Mol. Biol. [CrossRef]
- Sunil Laxman, S., Riechers, A., Sadilek, M. (2006). Hydrolysis products of cAMP analogs cause transformation of Trypanosoma brucei from slender to stumpy-like forms. PNAS 103: 19194–19199. [CrossRef]
- Bachmaier, S., Volpato Santos, Y., Kramer, S., Githure, G.B., Klockner, T, Pepperi, J., Baums, C., Schenk, R., Schwede, F., Genieser, H.G. et al. (2019). Nucleoside analogue activators of cyclic AMP-independent protein kinase A of Trypanosoma. Nat Commun 10, 1421. [CrossRef]
- Bao, Y., Weiss, L.M., Braunstein, V.L. (2008) The Role of Protein Kinase A in Trypanosoma cruzi. Infect Imm. 76:4757-4763 76. [CrossRef]
- Atwood, J.A. 3rd, Minning, T., Ludolf, F., Nuccio, A., Wheatherly, D.B., Alvarez-Manilla, G. Tarleton, R., Orlando, R. (2006) Glycoproteomics of Trypanosoma cruzi trypomastigotes using subcellular fractionation, lectin affinity, and stable isotope labeling. J. Proteome Res. 5:3376-84. [CrossRef]
- Azuaje, F.J., Ramirez, J.L., Da Silveira, J.F. (2007). In silico, biologically-inspired modelling of genomic variation generation in surface proteins of Trypanosoma cruzi. Kinetoplastid Biol Dis. 10; 6:6. [CrossRef]
- Azuaje, F.J., Ramirez, J.L., Da Silveira, J.F. (2007). An exploration of the genetic robustness landscape of surface protein families in the human protozoan parasite Trypanosoma cruzi. IEEE Trans Nanobioscience. 6:223-8. [CrossRef] [PubMed]
- Gonzalez, A. M., Azuaje, F. J., Ramirez, J.L., Dorronsoro, J.R. (2009). Machine Learning Techniques for the Automated Classification of Adhesin-Like Proteins in the Human Protozoan Parasite Trypanosoma cruzi, IEEE/ACM Transactions on Computational Biology and Bioinformatics 6;695-702. [CrossRef]
- Ramirez, M. I., de Cassia Ruiz, R., Araya, J.E, J.F.Da S ilveira, Yoshida, N. (1993). Involvement of the Stage-Specific 82-Kilodalton Adhesion Molecule of Trypanosoma cruzi Metacyclic Trypomastigotes in Host Cell Invasion. Infect. Imm. 61: 3636-3641.
- Wen, Y.Z., Zheng, L.L., Qu, L.H., Ayala, F.J., Lun, Z-R (2012). Pseudogenes are not pseudo any more. RNA Biol. 9:27-32.
- Harrison, P., Zheng, D., Zhang, Z. Carriero, N., Gerstein, M. (2005). Transcribed processed pseudogenes in the human genome: an intermediate form of expressed retrosequence lacking protein-coding ability. Nucleic Acids Res. 33: 2374-83. [CrossRef]
- Sasidharan, R., Gerstein, M. (2008). Protein fossils live on as RNA. Nature. 453:729-31. [CrossRef]
- Devor, E.J. (2006) Primate MicroRNAs miR-220 and miR-492 Lie within Processed Pseudogenes J. Hered. 97:186-190. [CrossRef]
- Garcia Silva, M.R., Tosar, J.P., Frugier, M., Pantano, S., Bonilla, B., Esteban, L., Serra, E., Rovira, C., Robello, C., Cayota, A. (2010) Cloning, characterization and subcellular localization of a Trypanosoma cruzi argonaute protein defining a new subfamily distinctive of trypanosomatids. Gene. 466:26-35. [CrossRef]
- Zheng, Y., Cai, X., and Bradley, J.E. (2013) microRNAs in parasites and parasite infection, RNA Biology, 10:371-379. [CrossRef]
- Chandaa, I., Panb, A., Kumar Saha, S. (2007). Comparative codon and amino acid composition analysis of Tritryps-conspicuous features of Leishmania major FEBS Letters 581 5751–5758.
- Ulrich, P.N., Jimenez, V., Park, M., Martins, V.P., Altwood II, J., Collins, D., Rohloff, P., Tarleton, R., Moreno, S.N., Orlando, R. et. al. (2011) Identification of Contractile Vacuole Proteins in Trypanosoma cruzi. PLoS ONE 6(3): e18013. [CrossRef]
- Brossas, J.Y., Gulin, J.E.N., Bisio, M.M.C., Chapelle, M., Marinach-Patrice, C., Bordessoules, M., Palazon Ruiz, G., Vion, J., Paris, L., Altcheh, J., et. al. (2017). Secretome analysis of Trypanosoma cruzi by proteomics studies. PLoS One.;12(10):e0185504. [CrossRef]
- Atwood, J.A. 3rd, Weatherly, D.B., Minning, T.A., Bundy, B., Cavola, C., Opperdoes, F.R. Orlando, R., Tarleton, R.L. (2005). The Trypanosoma cruzi proteome. Science 309:473-6. [CrossRef]
- Ghazalpour, A., Bennett, B., Petyuk, V.A., Orozco, L., Agopian, R., Mungrue, I.N., Farber, C.R., Sinsheimer, J., Kang, H.M., Furlotte, N. et. al. (2011) Comparative Analysis of Proteome and Transcriptome Variation in Mouse. PLoSGenet 7(6): e1001393. [CrossRef]
- Schwanhäusser, B., Busse, D., Li, N., Dittmar, G., Schuchhardt, J., Wolf, A, Chen, W., Selbach, M. (2011) Global quantification of mammalian gene expression control. Nature 473:337–342. [CrossRef]
- Cortazzo da Silva, L., Aoki, J.I. and Floeter-Winter, L.M. (2022). Finding Correlations Between mRNA and Protein Levels in Leishmania Development: Is There a Discrepancy? Front. Cell. Infect. Microbiol. 12:852902. [CrossRef]
- Callejas-Hernández, F., Gutierrez-Nogues, A., Rastrojo, A. (2019). Analysis of mRNA processing at whole transcriptome level, transcriptomic profile and genome sequence refinement of Trypanosoma cruzi. Sci. Reports 9:17376. [CrossRef]
- Ibarrola-Vannucci, A.K., De Pablos, L.M., Moreira, R.L., Cornet-Gomez, A., Cruz-Bustos, T., Schijman, A.G., Ramírez, J.L., Vílchez, S., Osuna, A. (2021). Characterization and functional analysis of the proteins Prohibitin 1 and 2 in Trypanosoma cruzi. PLoS Negl. Trop. Dis. 15(4): e0009322.
- Pastro, L., Smircich, P., Di Paolo, A., Becco, L., Duhagon, L.A, Sotello-Silveira, J, Garat, B. (2017) Nuclear Compartmentalization Contributes to Stage-Specific Gene Expression Control in Trypanosoma cruzi. Front. Cell Dev. Biol. 5:8. [CrossRef]
- Clayton, C. (2019). Regulation of gene expression in trypanosomatids: living with polycistronic transcription. Open Biol. 9:190072. [CrossRef]
- Kramer, S. and Carrington, M. (2011). Trans-acting proteins regulating mRNA maturation, stability and translation in trypanosomatids Trends Parasitol. 27:1471-1492.
- de Pablos, L.M., Ferreira, T.R., Dowle, A.A., Forrester, S., Parry, E., Newlin, K., Walrat, P.B. (2019). The mRNA-bound Proteome of Leishmania mexicana: Novel Genetic Insight into an Ancient Parasite. Mol. Cell. Proteomics 18:1271–1284.
- Smircich, P., Eastman, G., Bispo, S., Duhagon, M.A., Guerra-Slompo, E.P., Garat, B., Goldenberg, S., Munroe, D.J., Dallagiovanna, B., Holetz, F. et. al. (2015). Ribosome profiling reveals translation control as a key mechanism generating differential gene expression in Trypanosoma cruzi. BMC Genomics 16:443. [CrossRef]
- Berná, L., Chiribao, M. L., Greif, G. (2017). Transcriptomic analysis reveals metabolic switches and surface remodeling as key processes for stage transition in Trypanosoma cruzi. Peer J., 5, e3017. [CrossRef]
- Kahn, S., Van Voorhis, W.C. and Eisen, H. (1990) The major 85-kd surface antigen of the mammalian form of Trypanosoma cruzi is encoded by a large heterogeneous family of simultaneously expressed genes, J. Exp. Med. 172:589-597.
- Macleod, O.J.S., Cook, A.D., Webb, H., Crow, M., Burns, R., Redpath, M., Seisenberger, S., Trevor, C.E., Peacock, L., Schwede, A. et. al. (2022). Invariant surface glycoprotein 65 of Trypanosoma brucei is a complement C3 receptor. Nat. Commun.13:5085. [CrossRef]
- Iida, K., Whitlow, M. B., Nussenzweig, V. (1989). Amastigotes of Trypanosoma cruzi escape destruction by the terminal complement components. J. Exp. Med. 169:881-891. [CrossRef]
- Ramírez-Toloza, G., Ferreira, A. (2016). Trypanosoma cruzi evades the Complement System as an Efficient Strategy to Survive in the Mammalian Host: The Specific Roles of Host/Parasite Molecules and Trypanosoma cruzi Calreticulin. Frontiers in Microbiology, 8. [CrossRef]
- Rawal, K., Sinha, R., Abbasi, B.A., Chaudary, A., Nath, S.K., Kumari, P., Preeti, P., Saraf, D., Singh, S., Mishra, K., et. al. (2021). Identification of vaccine targets in pathogens and design of a vaccine using computational approaches. Sci Rep. 2021;11(1):17626. [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




