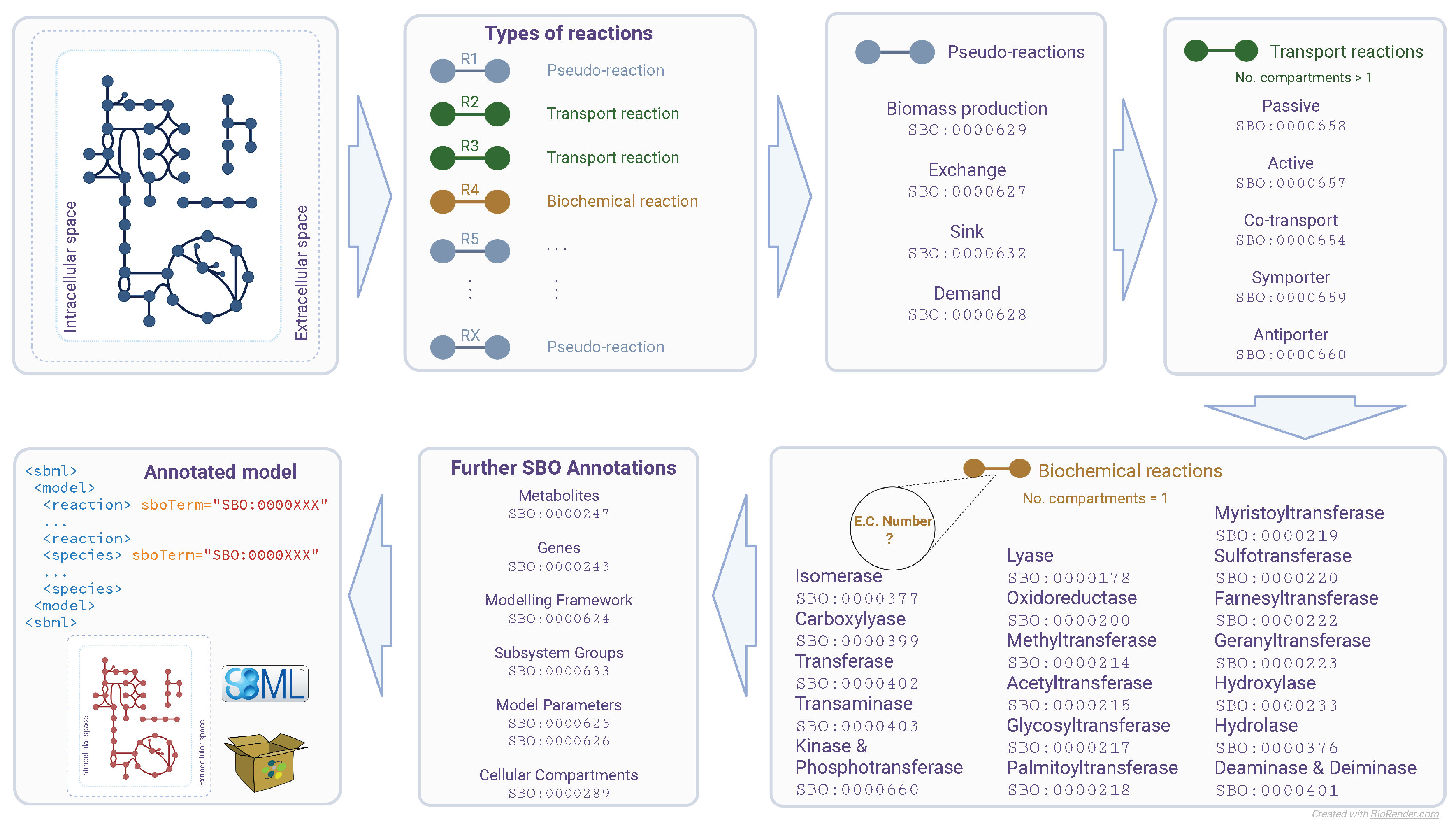
The SBOannotator workflow comprises six main steps (
Figure 1). At first, all reactions found within the model are labeled as either (a)
transporters that move molecules across different compartments, (b)
simple biochemical reactions that only take place in the cytosol, or (c)
pseudo-reactions that import or export metabolites and serve modeling purposes. Then, the pseudo-reactions are detected and analyzed. Pseudo-reactions in systems biology modeling do not correspond to any actual physical process and should not be confused with the pseudo-first-order reactions from the field of chemical kinetics. They are subdivided into demand, exchange, and sink reactions. The biomass objective function also belongs to this class. Exchange (
SBO:0000627) and sink (
SBO:0000632) reactions are reversible reactions that add or remove metabolites, with the latter one to be specific for intracellular compounds.
SBOannotator processes further by examining the transport reactions and assigning appropriate SBO terms. The classification mechanism in this step is comparably advanced since several types of transporters exist. The decision relies on the main characteristics of the different classes, such as the presence of one (passive transport) or more reaction participants, and the consumption of adenosine triphosphate (ATP) or phosphoenolpyruvate (PEP) (active transport). If reversible reactions are labeled as active transporters, a warning is printed to the user indicating that these reactions are thermodynamically infeasible. Subsequently, the total number of cellular compartments is derived to enable the distinction between symporters/antiporters and co-transporters. Reactions with metabolites from more than two compartments are characterized as co-transporters (SBO:0000654), while the rest is divided and either labeled as symporter (SBO:0000659, reactants, and products are from the same compartment) or antiporter (SBO:0000660, reactants, and products are from different compartments).
The remaining biochemical reactions are processed in the next step to enable more detailed labeling. For this purpose, the SBOannotator employs an Structured Query Language (SQL) database that contains mappings between Enzyme Commission (EC) numbers and the respective SBO terms. As the model’s size increases, using an already-defined database accelerates the computational time needed for their annotation. To create this database, we browsed all children nodes of the
biological reaction node in the SBO’s directed acyclic graph. Our mappings could be divided into three main categories: (a) one-to-one mapping; one SBO term represents EC numbers from a single sub-subclass (e.g., transamination), (b) one-to-few mapping; one SBO term maps only a subset of EC numbers belonging in a single sub-subclass (e.g., myristoylation), and (c) and one-to-many mapping; one SBO term covers a large subset of EC numbers within one sub-subclass (e.g., acetylation). The
supplementary Table S1 lists all mappings in detail. The SBOannotator assigns the general SBO term of a biochemical reaction (
SBO:0000176) if the reaction has multiple EC numbers assigned and all are from different classes resulting in an unambiguous description. Otherwise, the SBO terms are accredited based on the respective enzyme’s main class (e.g., oxidoreductases;
SBO:0000200, transferases;
SBO:0000402, hydrolases;
SBO:0000376). It is important to note that a proper term that describes the ligases (EC class 6) is currently missing from the SBO graph. This would be necessary to describe, for instance, reactions involving the formation of deoxyribonucleic acid (DNA), ribonucleic acid (RNA), and protein fragments. We included an appropriate SBO term for ligases that describes the general modification of covalent bonds (
SBO:0000182). The metabolic reactions that do not fall in any of the already mentioned cases and do not have any EC number assigned are given the general SBO term of a biochemical reaction (
SBO:0000176). The SBOannotator is designed to handle models with or without EC numbers assigned. However, they should either be annotated with Biochemical, Genetical, and Genomical (BiGG) [
7] identifiers or include any intonations. If the input model provides no EC numbers, an integrated Application Programming transfer Interface (API) call requests the necessary information from the BiGG database and adds all missing annotations into the model. Depending on the model’s size, this step may increase the computational time. Hence, we recommend the prior use of an annotation tool, such as ModelPolisher [
8]. We have tested the performance of SBOannotator in assigning descriptive and more precise terms to biochemical reactions using 108 metabolic models from the BiGG database. All downloaded models contained only five types of SBO annotation representing only top-level terms. Nevertheless, all biochemical reactions had a single generic term without specifying the exact type of reaction. However, our tool annotated the models with 31 different terms considering the underlying enzymatic properties (see
Tables S2 and S3). The biochemical reactions made up the largest reaction group before and after the SBOannotator. However, their coverage was reduced from
to
, meaning a large percentage of the initial reactions got a more specific term (see
Figures S1 and S2). The second most common term in the downloaded models described translocations. Our annotated models contain more specific terms based on the respective transport mechanisms. Across all models, decarbonylations (
SBO:0000400) occurred most rarely and only ten times.
Finally, SBOannotator assigns SBO terms to the remaining model entities. These include metabolites, genes, cellular compartments, and defined parameters. If subsystem groups are declared, the SBOannotator allocates the term subsystem (SBO:0000633), while the respective modeling framework is also assigned an appropriate term. The final annotated SBML model is stored in the current working directory with the name tag _SBOannotated in an Extensible Markup Language (XML) format.