Submitted:
22 February 2023
Posted:
23 February 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Identification of CBL and CIPK family genes in A. littoralis
2.2. Phylogenetic analysis and classification of AlCBL and AlCIPK gene family
2.3. Motif analysis and gene structure of AlCBLs and AlCIPKs
2.4. Plant materials growth conditions and salt treatments
2.5. RNA extraction and cDNA synthesis
2.6. Real-time PCR
3. Results
3.1. Physicochemical properties of AlCBLs and AlCIPKs
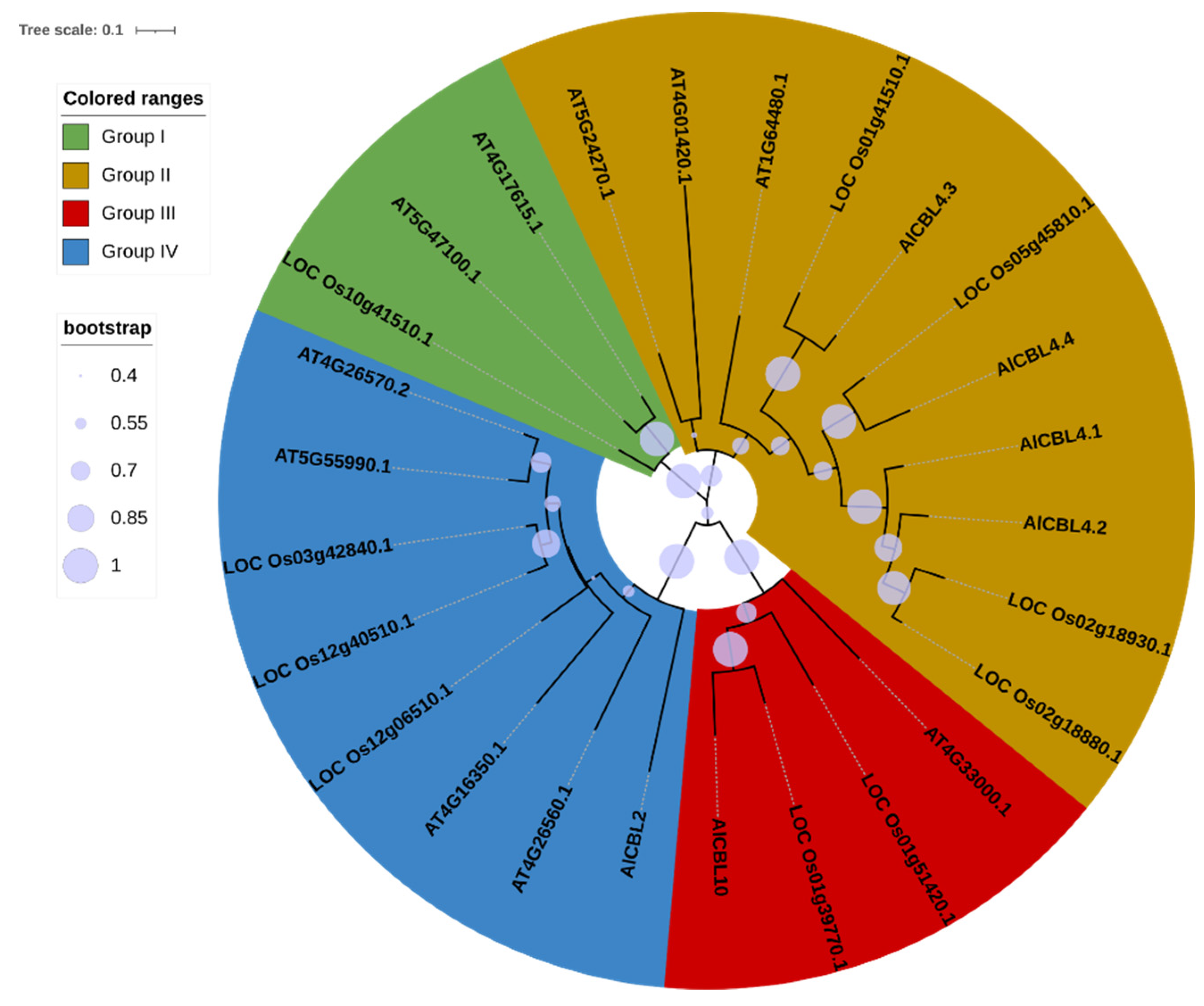
3.2. Phylogenetic analysis of AlCBLs
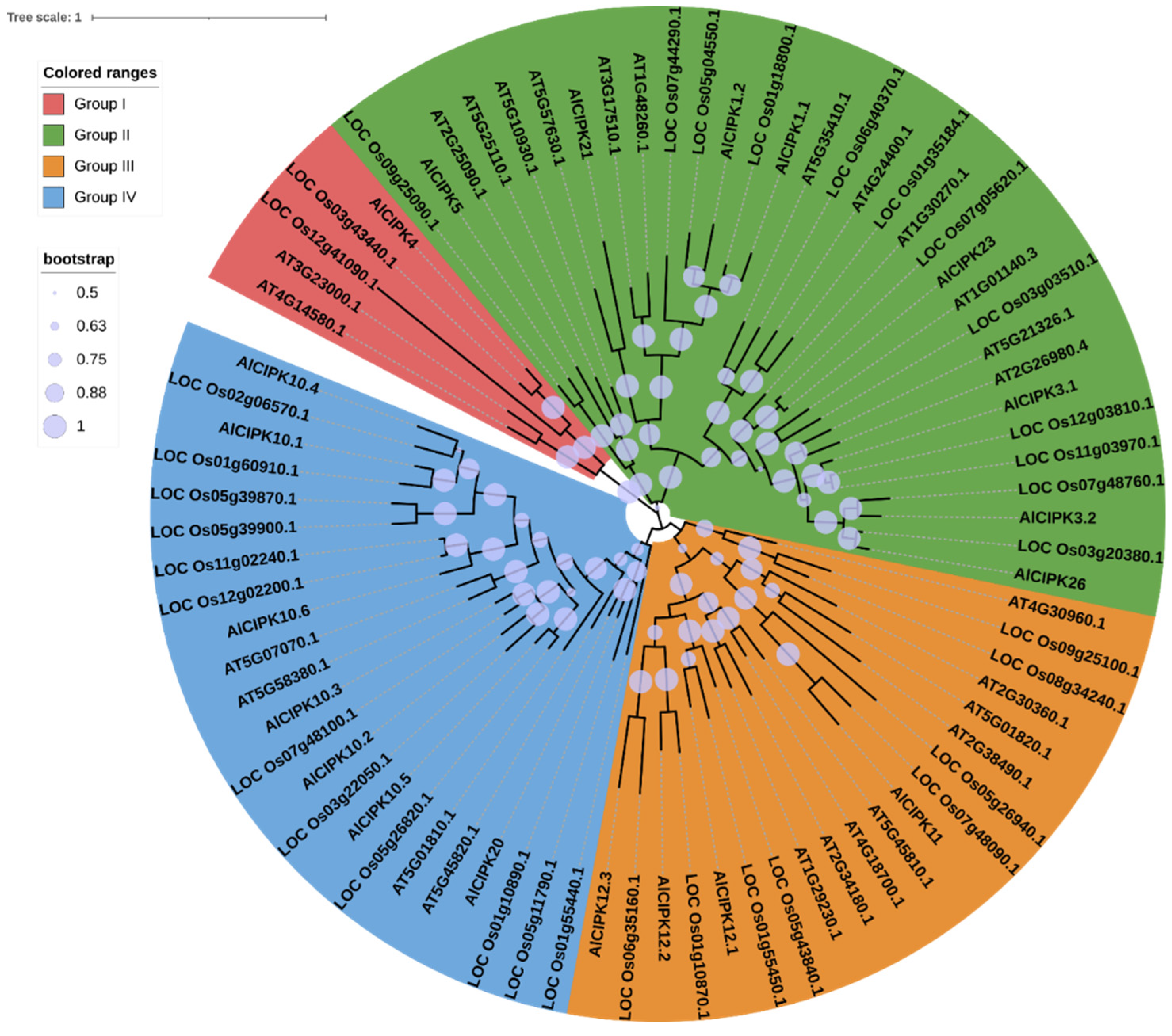
3.3. Phylogenetic analysis of AlCIPKs
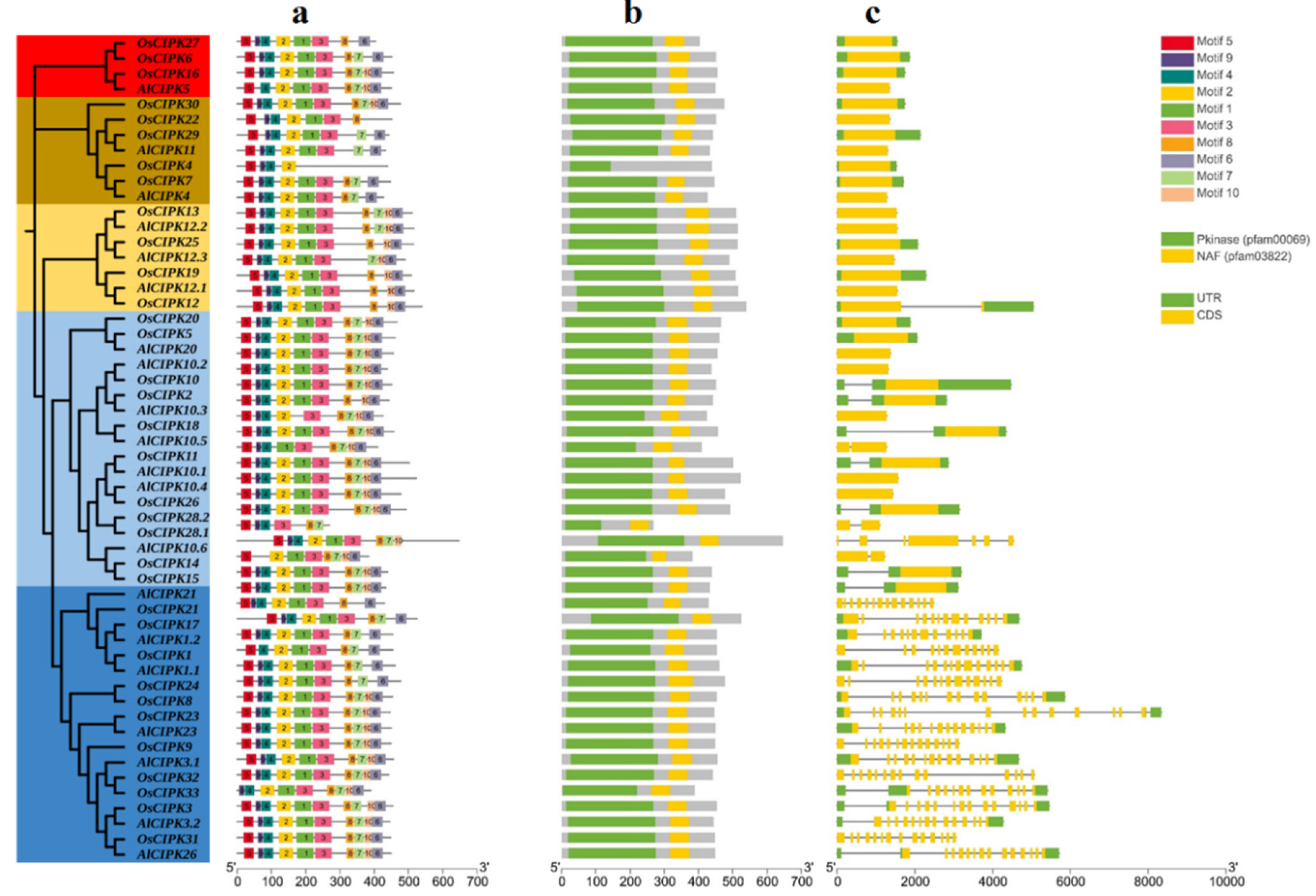
3.4. Gene structure and conserved motifs of AlCBLs
3.5. Gene structure and conserved motifs of AlCIPKs
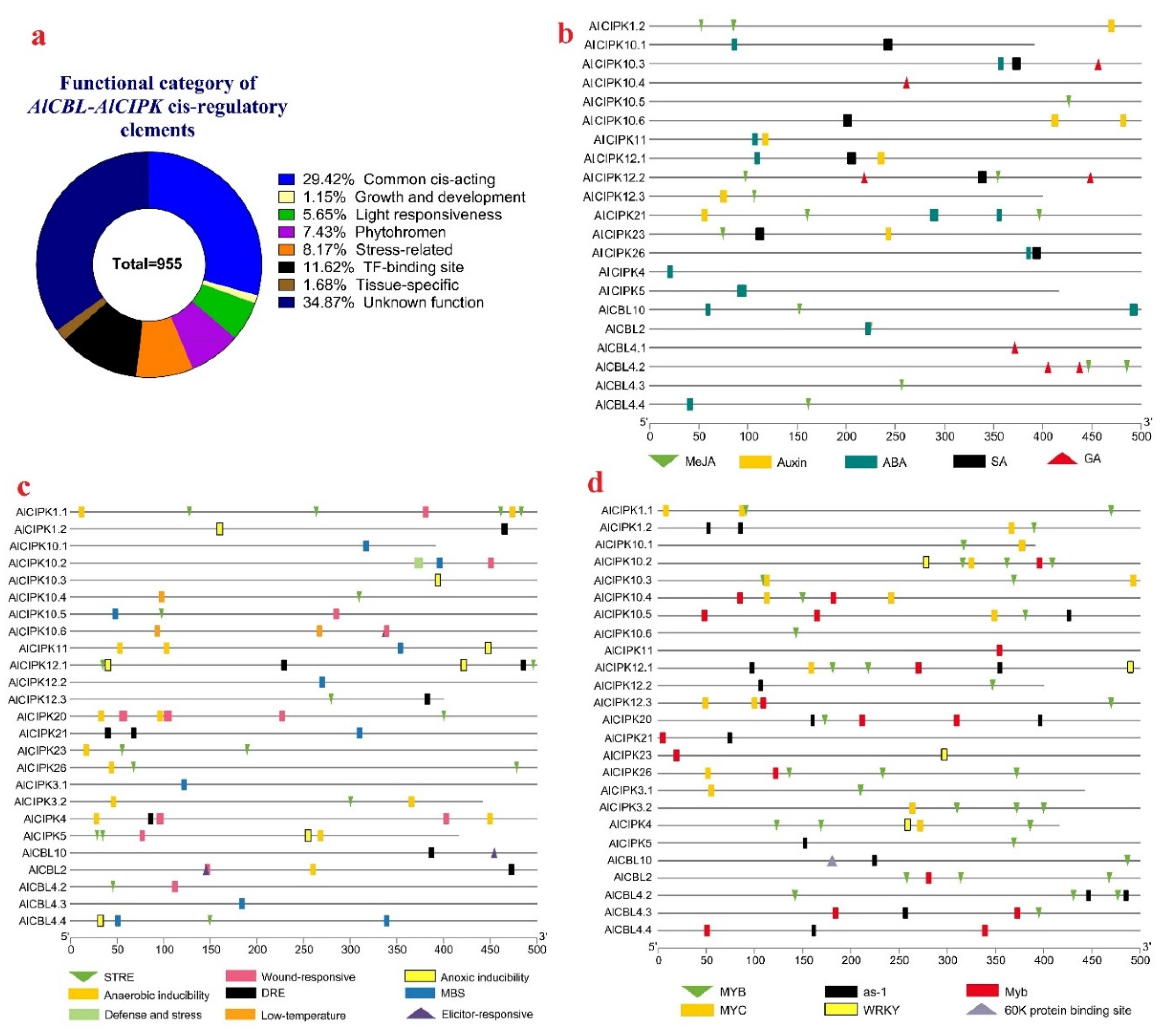
3.6. Promoter analysis
3.7. Expression profile of AlCBL genes in response to salinity
3.8. Expression profile of AlCIPK genes in response to salinity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Dodd, A.N.; Kudla, J.; Sanders, D. The language of calcium signaling. Annu. Rev. Plant Biol. 2010, 61, 593–620. [Google Scholar] [CrossRef] [PubMed]
- Tang, R.-J.; Luan, S. Regulation of calcium and magnesium homeostasis in plants: from transporters to signaling network. Curr. Opin. Plant Biol. 2017, 39, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Aslam, M.; Fakher, B.; Jakada, B.H.; Zhao, L.; Cao, S.; Cheng, Y.; Qin, Y. Genome-Wide Identification and Expression Profiling of CBL-CIPK Gene Family in Pineapple (Ananas comosus) and the Role of Ac CBL1 in Abiotic and Biotic Stress Response. Biomolecules 2019, 9, 293. [Google Scholar] [CrossRef] [PubMed]
- Costa, A.; Navazio, L.; Szabo, I. The contribution of organelles to plant intracellular calcium signalling. J. Exp. Bot. 2018, 69, 4175–4193. [Google Scholar] [CrossRef]
- Kudla, J.; Becker, D.; Grill, E.; Hedrich, R.; Hippler, M.; Kummer, U.; Parniske, M.; Romeis, T.; Schumacher, K. Advances and current challenges in calcium signaling. New Phytol. 2018, 218, 414–431. [Google Scholar] [CrossRef]
- Ding, Y.; Shi, Y.; Yang, S. Advances and challenges in uncovering cold tolerance regulatory mechanisms in plants. New Phytol. 2019, 222, 1690–1704. [Google Scholar] [CrossRef]
- Behera, S.; Xu, Z.; Luoni, L.; Bonza, M.C.; Doccula, F.G.; De Michelis, M.I.; Morris, R.J.; Schwarzländer, M.; Costa, A. Cellular Ca2+ signals generate defined pH signatures in plants. Plant Cell 2018, 30, 2704–2719. [Google Scholar] [CrossRef] [PubMed]
- Michard, E.; Simon, A.A.; Tavares, B.; Wudick, M.M.; Feijó, J.A. Signaling with ions: the keystone for apical cell growth and morphogenesis in pollen tubes. Plant Physiol. 2017, 173, 91–111. [Google Scholar] [CrossRef] [PubMed]
- De Vriese, K.; Himschoot, E.; Dünser, K.; Nguyen, L.; Drozdzecki, A.; Costa, A.; Nowack, M.K.; Kleine-Vehn, J.; Audenaert, D.; Beeckman, T. Identification of novel inhibitors of auxin-induced Ca2+ signaling via a plant-based chemical screen. Plant Physiol. 2019, 180, 480–496. [Google Scholar] [CrossRef] [PubMed]
- Meena, M.K.; Prajapati, R.; Krishna, D.; Divakaran, K.; Pandey, Y.; Reichelt, M.; Mathew, M.K.; Boland, W.; Mithöfer, A.; Vadassery, J. The Ca2+ channel CNGC19 regulates Arabidopsis defense against Spodoptera herbivory. Plant Cell 2019, 31, 1539–1562. [Google Scholar] [CrossRef] [PubMed]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef] [PubMed]
- Bender, K.W.; Zielinski, R.E.; Huber, S.C. Revisiting paradigms of Ca2+ signaling protein kinase regulation in plants. Biochem. J. 2018, 475, 207–223. [Google Scholar] [CrossRef] [PubMed]
- Ranty, B.; Aldon, D.; Cotelle, V.; Galaud, J.-P.; Thuleau, P.; Mazars, C. Calcium sensors as key hubs in plant responses to biotic and abiotic stresses. Front. Plant Sci. 2016, 7, 327. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Lv, Y.; Jahan, N.; Chen, G.; Ren, D.; Guo, L. Sensing of abiotic stress and ionic stress responses in plants. Int. J. Mol. Sci. 2018, 19, 3298. [Google Scholar] [CrossRef] [PubMed]
- Batistic, O.; Kudla, J. Integration and channeling of calcium signaling through the CBL calcium sensor/CIPK protein kinase network. Planta 2004, 219, 915–924. [Google Scholar] [CrossRef] [PubMed]
- Halfter, U.; Ishitani, M.; Zhu, J.-K. The Arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. Proc. Natl. Acad. Sci. 2000, 97, 3735–3740. [Google Scholar] [CrossRef] [PubMed]
- Kolukisaoglu, U.; Weinl, S.; Blazevic, D.; Batistic, O.; Kudla, J. Calcium sensors and their interacting protein kinases: genomics of the Arabidopsis and rice CBL-CIPK signaling networks. Plant Physiol. 2004, 134, 43–58. [Google Scholar] [CrossRef] [PubMed]
- Pandey, G.K.; Cheong, Y.H.; Kim, K.-N.; Grant, J.J.; Li, L.; Hung, W.; D’Angelo, C.; Weinl, S.; Kudla, J.; Luan, S. The calcium sensor calcineurin B-like 9 modulates abscisic acid sensitivity and biosynthesis in Arabidopsis. Plant Cell 2004, 16, 1912–1924. [Google Scholar] [CrossRef] [PubMed]
- Mähs, A.; Steinhorst, L.; Han, J.-P.; Shen, L.-K.; Wang, Y.; Kudla, J. The calcineurin B-like Ca2+ sensors CBL1 and CBL9 function in pollen germination and pollen tube growth in Arabidopsis. Mol. Plant 2013, 6, 1149–1162. [Google Scholar] [CrossRef] [PubMed]
- Ligaba-Osena, A.; Fei, Z.; Liu, J.; Xu, Y.; Shaff, J.; Lee, S.; Luan, S.; Kudla, J.; Kochian, L.; Piñeros, M. Loss-of-function mutation of the calcium sensor CBL 1 increases aluminum sensitivity in Arabidopsis. New Phytol. 2017, 214, 830–841. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.-L.; Ren, H.-M.; Chen, L.-Q.; Wang, Y.; Wu, W.-H. A protein kinase, calcineurin B-like protein-interacting protein Kinase9, interacts with calcium sensor calcineurin B-like Protein3 and regulates potassium homeostasis under low-potassium stress in Arabidopsis. Plant Physiol. 2013, 161, 266–277. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Ding, S.; Zhang, H.; Du, H.; An, L. CIPK7 is involved in cold response by interacting with CBL1 in Arabidopsis thaliana. Plant Sci. 2011, 181, 57–64. [Google Scholar] [CrossRef]
- Yang, Y.; Wu, Y.; Ma, L.; Yang, Z.; Dong, Q.; Li, Q.; Ni, X.; Kudla, J.; Song, C.P.; Guo, Y. The Ca2+ sensor SCaBP3/CBL7 fine tunes arabidopsis alkali tolerance and modulats plasma membrane H+-ATPase activity. Plant Cell 2019, 31, 1367–1384. [Google Scholar] [CrossRef] [PubMed]
- Held, K.; Pascaud, F.; Eckert, C.; Gajdanowicz, P.; Hashimoto, K.; Corratgé-Faillie, C.; Offenborn, J.N.; Lacombe, B.; Dreyer, I.; Thibaud, J.-B. Calcium-dependent modulation and plasma membrane targeting of the AKT2 potassium channel by the CBL4/CIPK6 calcium sensor/protein kinase complex. Cell Res. 2011, 21, 1116–1130. [Google Scholar] [CrossRef] [PubMed]
- Albrecht, V.; Weinl, S.; Blazevic, D.; D’Angelo, C.; Batistic, O.; Kolukisaoglu, Ü.; Bock, R.; Schulz, B.; Harter, K.; Kudla, J. The calcium sensor CBL1 integrates plant responses to abiotic stresses. Plant J. 2003, 36, 457–470. [Google Scholar] [CrossRef] [PubMed]
- Nozawa, A.; Koizumi, N.; Sano, H. An Arabidopsis SNF1-related protein kinase, AtSR1, interacts with a calcium-binding protein, AtCBL2, of which transcripts respond to light. Plant Cell Physiol. 2001, 42, 976–981. [Google Scholar] [CrossRef] [PubMed]
- Gong, D.; Guo, Y.; Jagendorf, A.T.; Zhu, J.-K. Biochemical characterization of the Arabidopsis protein kinase SOS2 that functions in salt tolerance. Plant Physiol. 2002, 130, 256–264. [Google Scholar] [CrossRef] [PubMed]
- Du, W.; Yang, J.; Ma, L.; Su, Q.; Pang, Y. Identification and characterization of abiotic stress responsive CBL-CIPK family genes in Medicago. Int. J. Mol. Sci. 2021, 22, 4634. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.; Liu, B.; Liu, H.; Sun, X.; Sun, X.; Wang, W.; Zheng, C. A new CIPK gene CmCIPK8 enhances salt tolerance in transgenic chrysanthemum. Sci. Hortic. (Amsterdam). 2023, 308, 111562. [Google Scholar] [CrossRef]
- Ma, R.; Liu, W.; Li, S.; Zhu, X.; Yang, J.; Zhang, N.; Si, H. Genome-Wide Identification, Characterization and Expression Analysis of the CIPK Gene Family in Potato (Solanum tuberosum L.) and the Role of StCIPK10 in Response to Drought and Osmotic Stress. Int. J. Mol. Sci. 2021, 22, 13535. [Google Scholar] [CrossRef]
- Wu, G.-Q.; Xie, L.-L.; Wang, J.-L.; Wang, B.-C.; Li, Z.-Q. Genome-Wide Identification of CIPK Genes in Sugar Beet (Beta vulgaris) and Their Expression Under NaCl Stress. J. Plant Growth Regul. 2022, 1–15. [Google Scholar] [CrossRef]
- Su, W.; Ren, Y.; Wang, D.; Huang, L.; Fu, X.; Ling, H.; Su, Y.; Huang, N.; Tang, H.; Xu, L. New insights into the evolution and functional divergence of the CIPK gene family in Saccharum. BMC Genomics 2020, 21, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Xiao, C.; Zhang, H.; Xie, F.; Pan, Z.-Y.; Qiu, W.-M.; Tong, Z.; Wang, Z.-Q.; He, X.-J.; Xu, Y.-H.; Sun, Z.-H. Evolution, gene expression, and protein‒protein interaction analyses identify candidate CBL-CIPK signalling networks implicated in stress responses to cold and bacterial infection in citrus. BMC Plant Biol. 2022, 22, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Mao, J.; Mo, Z.; Yuan, G.; Xiang, H.; Visser, R.G.F.; Bai, Y.; Liu, H.; Wang, Q.; van der Linden, C.G. The CBL-CIPK network is involved in the physiological crosstalk between plant growth and stress adaptation. Plant. Cell Environ. 2022. [Google Scholar] [CrossRef] [PubMed]
- Xiaolin, Z.; Baoqiang, W.; Xian, W.; Xiaohong, W. Identification of the CIPK-CBL family gene and functional characterization of CqCIPK14 gene under drought stress in quinoa. BMC Genomics 2022, 23, 1–18. [Google Scholar] [CrossRef]
- Saad, R. Ben; Romdhan, W. Ben; Zouari, N.; Azaza, J.; Mieulet, D.; Verdeil, J.-L.; Guiderdoni, E.; Hassairi, A. Promoter of the AlSAP gene from the halophyte grass Aeluropus littoralis directs developmental-regulated, stress-inducible, and organ-specific gene expression in transgenic tobacco. Transgenic Res. 2011, 20, 1003–1018. [Google Scholar] [CrossRef] [PubMed]
- Hashemi, S.H.; Nematzadeh, G.; Ahmadian, G.; Yamchi, A.; Kuhlmann, M. Identification and validation of Aeluropus littoralis reference genes for Quantitative Real-Time PCR Normalization. J. Biol. Res. 2016, 23, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Hashemi-Petroudi, S.H.; Arab, M.; Dolatabadi, B.; Kuo, Y.-T.; Baez, M.A.; Himmelbach, A.; Nematzadeh, G.; Maibody, S.A.M.M.; Schmutzer, T.; Mälzer, M. Initial Description of the Genome of Aeluropus littoralis, a Halophile Grass. Front. Plant Sci. 2022, 13. [Google Scholar] [CrossRef]
- Arend, D.; Lange, M.; Chen, J.; Colmsee, C.; Flemming, S.; Hecht, D.; Scholz, U. e! DAL-a framework to store, share and publish research data. BMC Bioinformatics 2014, 15, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Marchler-Bauer, A.; Derbyshire, M.K.; Gonzales, N.R.; Lu, S.; Chitsaz, F.; Geer, L.Y.; Geer, R.C.; He, J.; Gwadz, M.; Hurwitz, D.I. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2015, 43, D222–D226. [Google Scholar] [CrossRef] [PubMed]
- Schultz, J.; Copley, R.R.; Doerks, T.; Ponting, C.P.; Bork, P. SMART: a web-based tool for the study of genetically mobile domains. Nucleic Acids Res. 2000, 28, 231–234. [Google Scholar] [CrossRef] [PubMed]
- Quevillon, E.; Silventoinen, V.; Pillai, S.; Harte, N.; Mulder, N.; Apweiler, R.; Lopez, R. InterProScan: protein domains identifier. Nucleic Acids Res. 2005, 33, W116–W120. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating Maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Apte, A.; Singh, S. AlleleID. PCR Prim. Des. 2007, 329–345. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Zhu, K.; Fan, P.; Liu, H.; Tan, P.; Ma, W.; Mo, Z.; Zhao, J.; Chu, G.; Peng, F. Insight into the CBL and CIPK gene families in pecan (Carya illinoinensis): identification, evolution and expression patterns in drought response. BMC Plant Biol. 2022, 22, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Wang, Y.; Wang, M.; Li, T.; Zhou, Y.; Wang, X.; Wei, S.; He, G.; Yang, G. Identification and comprehensive analyses of the CBL and CIPK gene families in wheat (Triticum aestivum L.). BMC Plant Biol. 2015, 15, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Yang, B.; Liu, W.-Z.; Li, H.; Wang, L.; Wang, B.; Deng, M.; Liang, W.; Deyholos, M.K.; Jiang, Y.-Q. Identification and characterization of CBL and CIPK gene families in canola (Brassica napus L.). BMC Plant Biol. 2014, 14, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Ahmadizadeh, M.; Rezaee, S.; Heidari, P. Genome-wide characterization and expression analysis of fatty acid desaturase gene family in Camelina sativa. Gene Reports 2020, 21, 100894. [Google Scholar] [CrossRef]
- Faraji, S.; Filiz, E.; Kazemitabar, S.K.; Vannozzi, A.; Palumbo, F.; Barcaccia, G.; Heidari, P. The AP2/ERF Gene Family in Triticum durum: Genome-Wide Identification and Expression Analysis under Drought and Salinity Stresses. Genes (Basel). 2020, 11, 1464. [Google Scholar] [CrossRef] [PubMed]
- Puresmaeli, F.; Heidari, P.; Lawson, S. Insights into the Sulfate Transporter Gene Family and Its Expression Patterns in Durum Wheat Seedlings under Salinity. Genes (Basel). 2023, 14, 333. [Google Scholar] [CrossRef] [PubMed]
- Yaghobi, M.; Heidari, P. Genome-Wide Analysis of Aquaporin Gene Family in Triticum turgidum and Its Expression Profile in Response to Salt Stress. Genes (Basel). 2023, 14, 202. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Yu, A.; Du, Y.; Wang, G.; Li, Y.; Zhao, G.; Wang, X.; Zhang, W.; Cheng, K.; Liu, X. Foxtail millet (Setaria italica (L.) P. Beauv) CIPKs are responsive to ABA and abiotic stresses. PLoS One 2019, 14, e0225091. [Google Scholar] [CrossRef] [PubMed]
- Nuruzzaman, M.; Manimekalai, R.; Sharoni, A.M.; Satoh, K.; Kondoh, H.; Ooka, H.; Kikuchi, S. Genome-wide analysis of NAC transcription factor family in rice. Gene 2010, 465, 30–44. [Google Scholar] [CrossRef]
- Heidari, P.; Puresmaeli, F.; Mora-Poblete, F. Genome-Wide Identification and Molecular Evolution of the Magnesium Transporter (MGT) Gene Family in Citrullus lanatus and Cucumis sativus. Agronomy 2022, 12, 2253. [Google Scholar] [CrossRef]
- Heidari, P.; Abdullah; Faraji, S. ; Poczai, P. Magnesium transporter Gene Family: Genome-Wide Identification and Characterization in Theobroma cacao, Corchorus capsularis and Gossypium hirsutum of Family Malvaceae. Agronomy 2021, 11, 1651. [Google Scholar] [CrossRef]
- Faraji, S.; Ahmadizadeh, M.; Heidari, P. Genome-wide comparative analysis of Mg transporter gene family between Triticum turgidum and Camelina sativa. BioMetals 2021, 4. [Google Scholar] [CrossRef] [PubMed]
- Jaradat, A.A. Genetic resources of energy crops: biological systems to combat climate change. Aust. J. Crop Sci. 2010, 4, 309–323. [Google Scholar]
- Li, J.; Jiang, M.; Ren, L.; Liu, Y.; Chen, H. Identification and characterization of CBL and CIPK gene families in eggplant (Solanum melongena L.). Mol. Genet. Genomics 2016, 291, 1769–1781. [Google Scholar] [CrossRef] [PubMed]
- Shu, B.; Cai, D.; Zhang, F.; Zhang, D.J.; Liu, C.Y.; Wu, Q.S.; Luo, C. Identifying citrus CBL and CIPK gene families and their expressions in response to drought and arbuscular mycorrhizal fungi colonization. Biol. Plant. 2020, 64, 773–783. [Google Scholar] [CrossRef]
- Ma, X.; Li, Q.-H.; Yu, Y.-N.; Qiao, Y.-M.; Haq, S. ul; Gong, Z.-H. The CBL–CIPK pathway in plant response to stress signals. Int. J. Mol. Sci. 2020, 21, 5668. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Zhang, C.; Tang, R.-J.; Xu, H.-X.; Lan, W.-Z.; Zhao, F.; Luan, S. Calcineurin B-Like proteins CBL4 and CBL10 mediate two independent salt tolerance pathways in Arabidopsis. Int. J. Mol. Sci. 2019, 20, 2421. [Google Scholar] [CrossRef] [PubMed]

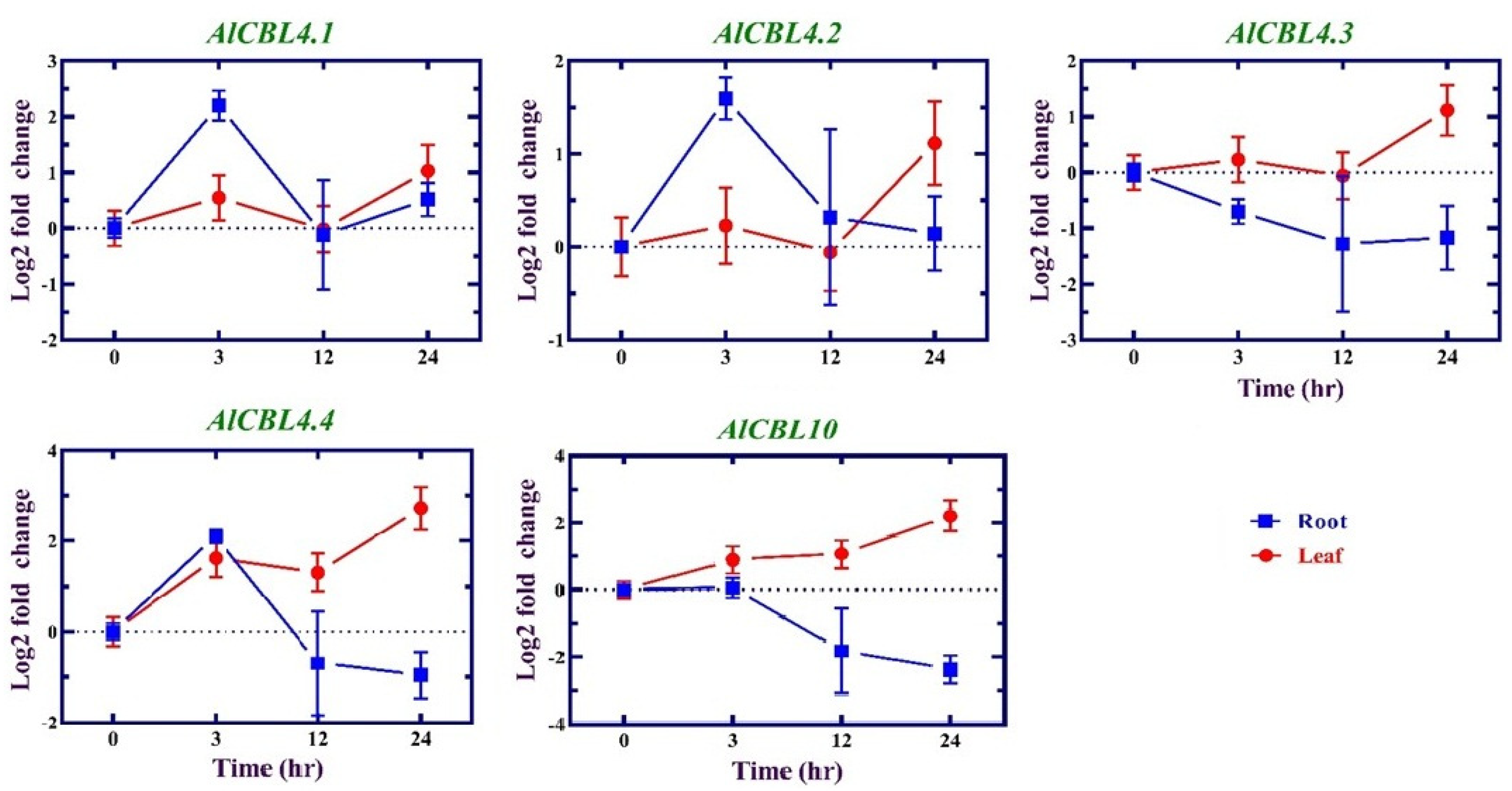
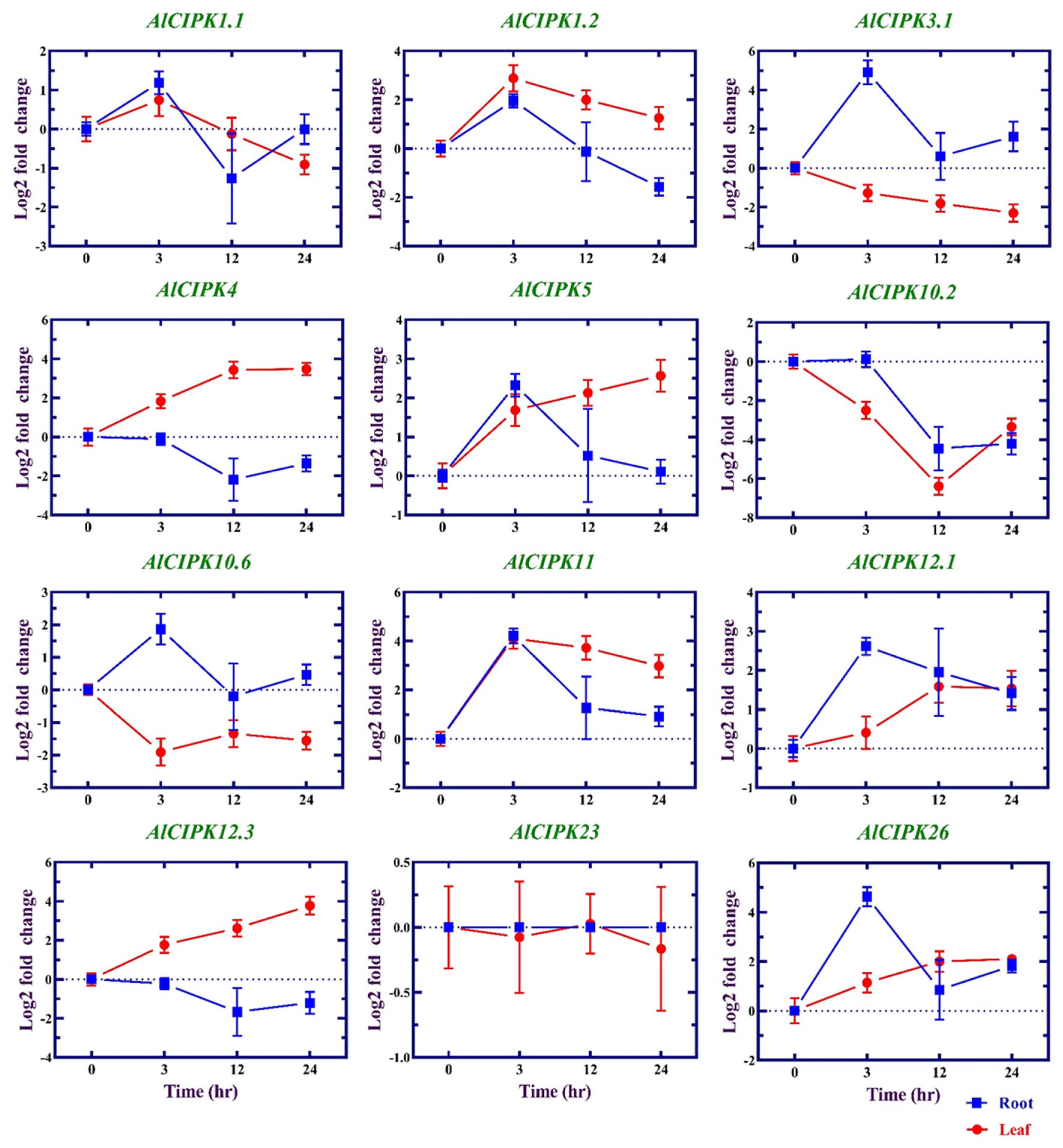
| Family | Gene ID | Gene Name | Length (aa) | Intron number | MW (kDa) | pI | GRAVY |
|---|---|---|---|---|---|---|---|
| CBL | Alg14121 | AlCBL2 | 226 | 7 | 25.87 | 4.98 | -0.219 |
| Alg15558 | AlCBL4.1 | 214 | 7 | 24.35 | 4.71 | -0.196 | |
| Alg11525 | AlCBL4.2 | 213 | 7 | 24.33 | 4.94 | -0.259 | |
| Alg8494 | AlCBL4.3 | 217 | 7 | 24.88 | 5.19 | -0.299 | |
| Alg13204 | AlCBL4. 4 | 166 | 5 | 18.70 | 4.78 | -0.341 | |
| Alg5886 | AlCBL10 | 303 | 8 | 34.67 | 5.28 | 0.133 | |
| CIPK | Alg4127 | AlCIPK1.1 | 473 | 12 | 53.48 | 6.52 | 0.372 |
| Alg7902 | AlCIPK1.2 | 454 | 11 | 50.69 | 6.62 | -0.320 | |
| Alg7566 | AlCIPK3.1 | 442 | 13 | 50.76 | 7.64 | -0.460 | |
| Alg12052 | ALCIPK3.2 | 448 | 13 | 50.63 | 8.23 | -0.407 | |
| Alg15044 | AlCIPK4 | 427 | 0 | 46.34 | 8.59 | -0.115 | |
| Alg5583 | AlCIPK5 | 450 | 0 | 48.19 | - | 0.054 | |
| Alg12300 | AlCIPK10.1 | 523 | 0 | 58.97 | 9.03 | 0.401 | |
| Alg9524 | ALCIPK10.2 | 438 | 0 | 49.72 | 9.28 | -0.260 | |
| Alg4701 | AlCIPK10.3 | 421 | 0 | 47.98 | 9.03 | 0.400 | |
| Alg3308 | AlCIPK10.4 | 478 | 0 | 54.66 | 9.13 | 0.514 | |
| Alg13906 | AlCIPK10.5 | 410 | 1 | 45.99 | 8.93 | -0.307 | |
| ALg9805 | AlCIPK10.6 | 383 | 1 | 42.04 | 8.99 | 0.480 | |
| Alg2698 | AlCIPK11 | 433 | 0 | 47.40 | 8.95 | -0.151 | |
| Alg8115 | AlCIPK12.1 | 516 | 0 | 57.36 | 8.64 | -0.341 | |
| Alg10559 | AlCIPK12.2 | 515 | 0 | 57.47 | 8.06 | -0.374 | |
| Alg11449 | AlCIPK12.3 | 490 | 0 | 54.06 | 8.84 | -0.254 | |
| Alg11347 | AlCIPK20 | 456 | 0 | 51.64 | 9.08 | -0.422 | |
| Alg8711 | AlCIPK21 | 430 | 13 | 48.54 | 6.21 | -0.303 | |
| Alg1003 | AlCIPK23 | 449 | 13 | 50.51 | 9.16 | -0.371 | |
| Alg7179 | AllCIPK26 | 448 | 13 | 50.44 | 8.41 | -0.395 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





