Introduction
The discovery and development of novel RNA therapy candidates has accelerated since the success of the COVID-19 mRNA vaccines 1. RNA therapeutics have been tapped to provide novel approaches to treat various dermatological conditions where conventional treatments have offered little success. For example, diabetic patients suffer from poor wound healing, which correlates with an increased chance of infections leading to amputation. In addition, metastatic melanoma is one of the deadliest forms of skin cancer with a 5-year survival rate of less than 5% 2. It is inherently resistant to radiotherapy and chemotherapy with median survival reduced to approximately 10 months 2. Novel therapies are vital to treat these patients, with several candidate RNA therapies showing promising results.
The rapid advancement of RNA-based therapies underlines their inherent advantages. For example, (1) they have a transient effect, (2) present no risk of insertional mutagenesis, (3) are easy to develop and manufacture, and (4) are cost-effective. These traits make them attractive options for development. Here we summarize the RNA-based therapies currently in clinical trials for skin conditions (
Table 1), described are the main categories of RNA therapeutics: RNAi (siRNA and miRNA), anti-sense oligonucleotides (ASO), messenger RNA (mRNA), and provide examples detailing their mechanisms of action (
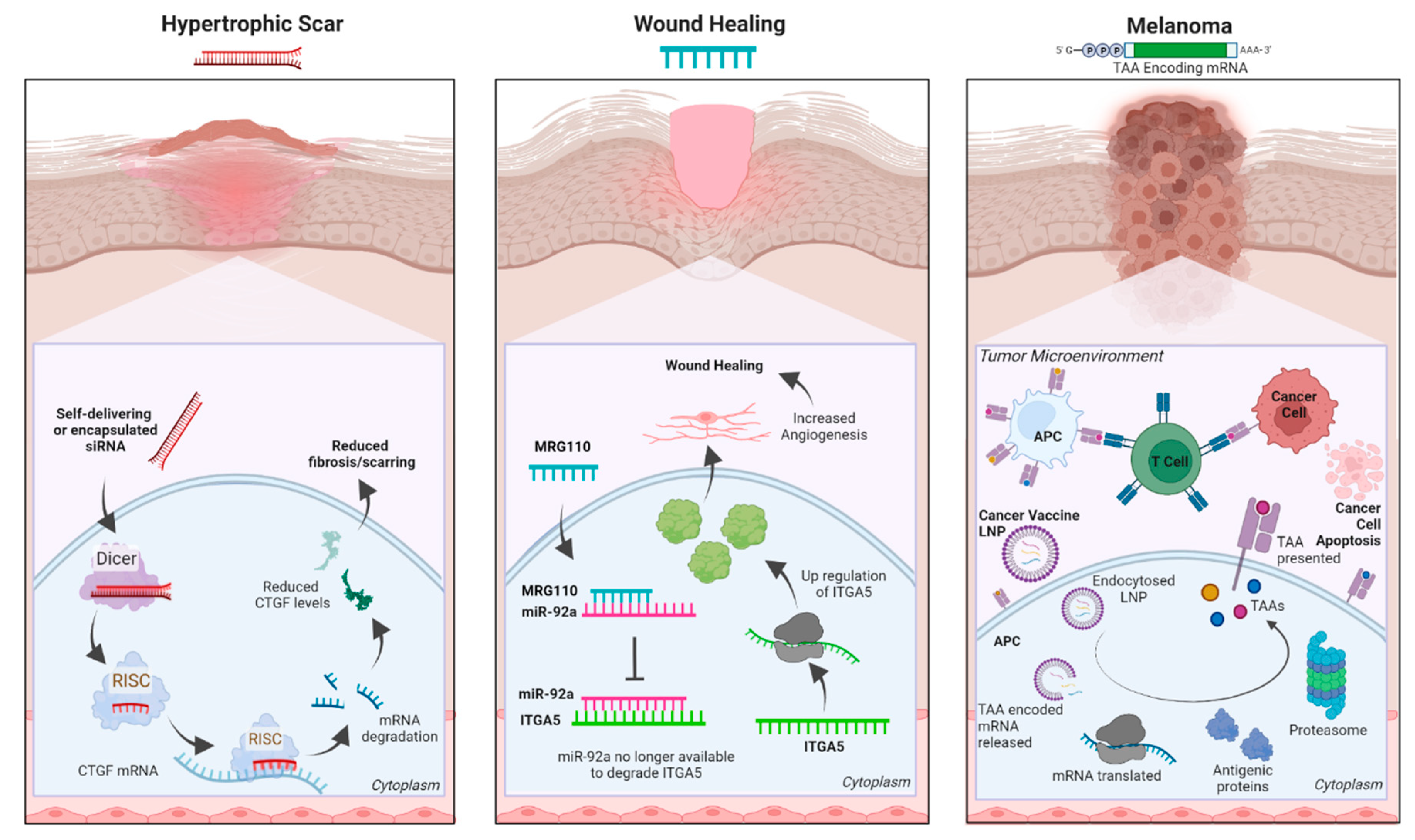
Figure 1).
RNA interference
The aberrant expression of a protein, a mutated protein, or a non-coding RNA 3; 4 can all cause skin disorders, and blocking such elements can be a beneficial therapeutic approach. RNA interference (RNAi) is one method to specifically target and lower the expression of pathological proteins. In RNAi, short double-stranded RNAs of exogenous or endogenous origins are processed by DICER and loaded into the RNA-induced silencing complex (RISC). Loaded RISC complexes recognize targeted mRNAs via perfect (siRNAs) or imperfect (miRNAs) complementarity and inhibit their translation and/or cause their degradation, thereby reducing the level of the encoded protein 5-7. siRNAs are designed to specifically target a single mRNA, whereas miRNAs (naturally occurring or synthetic) often have a broader array of targets 7.
The discovery of RNAi has greatly contributed to our understanding of the role(s) of countless proteins and their potential pathogenicity. More importantly to clinicians, RNAi provided a pathway to target disease-causing proteins that were otherwise impossible to treat. As with other RNA platforms, RNA stability, poor tissue penetration, and off-tissue targeting remain challenges; however, modifications to RNA bases and/or backbones plus the development of a wide variety of delivery systems enabled the FDA-approval of multiple siRNA therapeutics 8. Although several siRNA and miRNA mimics are currently advancing in clinical trials, no RNAi therapies have yet been approved for skin-related diseases.
Currently, at least three siRNAs (BMT101 (OLX101A), LEMS401, and RXI-109) targeting the connective tissue growth factor (CTGF) are in clinical trials (
Figure 1, left). High levels of CTGF have been linked to fibrotic disorders, and reducing CTGF levels is thought to be a viable approach for reducing cutaneous fibrosis
8-11. Further, CTGF is also an attractive target for treating keloids or hypertrophic scars, which occur in 40-70% of patients following surgery
12.
Antisense Oligonucleotides (ASOs)
Oligonucleotides are short chains of polymerized nucleotides, and antisense oligonucleotides (ASO) are short (generally 12-30 nucleotides) single-stranded synthetic nucleic acids whose reverse complement sequence allows for the targeting of specific RNA or DNA sequences 13-16. The use of oligonucleotides in clinical trials dates back to the late 1950s and early 1960s when methods to synthesize them were first established 17; 18. Although tested for decades, Fomivirsen, the first-in-class ASO, was only approved by the FDA to treat cytomegalovirus-induced retinitis in 1998 19.
ASOs can reduce or restore protein expression, inhibit 5' cap formation, or alter the splicing of targeted mRNAs. ASOs commonly enter the cell via endocytic pathways and recognize their targeted mRNAs through Watson-Crick base pairing 20; 21. ASOs predominantly function via two mechanisms. First, ASOs can employ an occupancy-mediated degradation mechanism by inducing the cell’s RNase H nuclease activity to degrade a targeted mRNA 22; 23. Second, ASOs can use an occupancy-only model and function via steric hindrance. This strategy can up or down-regulate target transcripts by altering their splicing patterns or by masking protein docking sites 24.
ASOs are usually trafficked to late endosomes and lysosomes which accounts for their slow release 25. Notably, the phosphodiester backbones of unmodified ASOs are prone to endonuclease degradation resulting in comparatively short half-lives; however, chemical modifications can overcome these shortcomings 26-29. ASOs are grouped into three generations based on their chemical modifications: (1) First-generation ASOs often replace a non-bridging oxygen atom in the phosphate group with either an amine (phosphoramidites), a methyl group (methyl phosphonates), or a sulfate group(phosphorothioates). When compared to phosphodiester oligonucleotides or unmodified ASOs, first-generation ASOs can resist nucleases and have longer half-lives in plasma 26. However, mRNA targeting affinity is slightly reduced due to the decreased melting temperature of these ASOs 15. (2) Second-generation ASOs usually have alkyl modifications at the 2' position of the ribose which improves binding affinity, tissue uptake, and nuclease resistance, while leading to both longer in vivo half-lives and lower toxicity 15; 30. (3) Third generation ASOs use chemical modifications to increase their stability, nuclease resistance, and hybridization affinity to the targeted RNA. The most commonly used third-generation ASOs usually incorporate peptide nucleic acids (PNA), locked nucleic acids (LNA), and morpholino phosphoramidite (MF) modifications 31. Notably, many third-generation ASOs function by causing steric hindrance of ribosomal machinery or altering the splicing of its targeted RNA 32-35.
MRG-110 (
Figure 1, center), a third generation LNA ASO developed by miRagen Therapeutics, Inc. (Boulder, CO), is currently in phase 1 clinical trial for wound healing. It blocks miR-92a, which then de-represses the integrin alpha 5 (ITGA5) gene
36. Higher levels of ITGA5 protein promotes angiogenesis which facilitates wound healing
37. Studies have shown significant results for MRG-110 reporting none or very low systemic toxicity and drug accumulation in distal tissues.
Messenger RNA (mRNA)
mRNAs are transient RNAs that encode a protein. Exogenous mRNAs were first used to elicit specific protein expression in vivo over three decades ago 38. Despite that initial success, nearly two decades passed until data were reported for the first clinical trial employing mRNA as a therapeutic 39. The development of mRNA-based therapeutics has seen a renaissance as the COVID-19 global pandemic demonstrated their versatility and power. The mRNA-based therapies currently undergoing clinical trials for dermatologically related diseases are listed in Table 1. Three main mRNA treatment modalities have emerged. First, cancer vaccines use mRNAs encoding tumor-specific antigens to stimulate a protective immune response 1; 40. Second, replacement therapies use mRNAs to produce therapeutic proteins or to counteract the phenotypes of a defective gene/protein. Third, cell-based therapies use mRNA transfected into cells ex vivo, with these cells being re-introduced into the patient to modify a specific diseased phenotype/function 1; 41. Despite the broad applicability of mRNAs, the constraints of this mini review restrict our discussion to mRNA cancer vaccines.
Therapeutic cancer vaccines generally encode tumor-associated antigens (TAAs), or unique markers expressed in cancerous, but not normal, cells. Targeting several TAAs in a single vaccine reduces the risk of tumor antigen escape as it triggers a broad immune response and it aids in the detection of poorly expressed antigens, thereby increasing the robustness of the vaccine and its antitumor response. BioNTech initially pursued a cancer vaccine targeting four melanoma-associated antigens in their phase 1 Lipo-MERIT monotherapy trial
42; 43. While this clinical trial is still ongoing, BioNTech has used the Lipo-MERIT data to steer the development of BioNTech’s FixVac (BNT111) therapy for melanoma, which shows a promising safety profile and anti-tumor immune response
44. BioNTech’s FixVac mRNA therapeutic platform is a fixed set of mRNA-encoded TAAs known to be expressed in particular cancer types (e.g., melanoma), therefore, prompting a strong and precise immune response against the particular cancer (
Figure 1, right). BNT111 was developed to treat patients with anti-PD-1-refractory/relapsed unresectable stage III or IV melanoma and is one of the most promising cancer immunotherapies in development
42; 43; 45; 46.
Cancerous cells are characterized by their rapid proliferation and expansion which often generates somatic mutations, each of which becomes a potentially targetable neoantigen. A patient’s specific tumor neoantigen profile (mutanome) could be analyzed and incorporated into personalized neoantigen-encoding mRNA vaccines. As neoantigen expression is restricted to tumor cells, this feature could easily be exploited to treat melanomas as they have a high mutation burden 47. Recently, Moderna Inc. and Merck & Co. released promising new data regarding their mRNA-based personalized (neoantigen) cancer vaccine, mRNA-4157/V940, for the treatment of melanoma. When the vaccine is administered in concert with Keytruda, a cancer immunotherapy, reports show a 44% reduction in the risk of recurrence or patient death when compared to Keytruda alone over a period of 1 year 42; 43; 48; 49. These data are a major breakthrough in the field of RNA therapeutics, demonstrating the power of mRNA to “train” a patient’s immune system to recognize and attack their specific tumor mutanome, hopefully, translating into durable remission.
mRNA cancer vaccines have many advantages. First, mRNA-based modalities elicit a potent yet reliable immune response to selected antigens in vivo and are customizable to a patient’s particular neoantigen profile. Second, mRNA therapies are safe compared to viral vector vaccines as they do not incorporate into the host genome, thus avoiding the risk of insertional mutagenesis. Lastly, mRNA vaccines are relatively inexpensive and rapid to synthesize 40. Altogether, mRNA therapeutics hold great promise for the treatment of various dermatological diseases.
Summary
As mentioned above, RNA therapeutics are a rapidly evolving class of therapies that have the potential to change the face of personalized medicine and revolutionize healthcare 1. A number of RNA therapeutics have been approved by FDA and many more are in clinical trials for a broad array of indications. In this review, we focused exclusively on RNA therapeutics in clinical trials for dermatological disorders, such as those used to treat melanoma, psoriasis, hypertrophic scars, wound healing, alopecia, epidermolysis bullosa, keloids, etc. Our research uncovered 35 different RNA therapeutics currently undergoing clinical trials and dozens more are in the discovery or preclinical stages of development for skin conditions as well. In closing, many RNA therapies are rapidly progressing through clinical trials and this evolving and growing class of drug candidates offers much promise to improve existing treatment regimens or to become standalone treatments themselves.
Acknowledgments and Funding
REK, SK, and NB contributed equally to this work and all authors acknowledge that all three authors can list themselves first on career materials such as curriculum vitae. REK, SK, NB, and DLK all performed the literature searches and wrote and revised sections of the manuscript. REK, SK, and NB constructed and edited the table and figure. DLK reviewed and edited the entire manuscript incorporating revisions from all authors. All authors approved the final manuscript. All authors apologize for primary works that were not cited due to citation limits for minireviews. This work was supported by a Houston Methodist Research Institution (HMRI) Career Cornerstone Award, an award from the Houston Methodist Foundation (both to DLK) and Cancer Prevention and Research Institute of Texas (RP150611 and RP200619) grants to the RNA Core at the HMRI. DLK is supported in part by an R35 grant from the NIH (R35GM137819). REK is supported by an NIH postdoctoral supplement (R35GM137819-03S3). The content presented here is solely the responsibility of the authors and does not represent the official views of the HMRI, CPRIT, or the NIH.
Conflicts of Interest
Dr. Kiss runs an externally funded laboratory (American Heart Association [20CDA35310329], and NIH [R35GM137819-03, -03S1, -03S2, -03S3], plus two subcontracts to NIH Contract #: 75N93019C00045) that is actively designing and testing different candidate RNA therapeutics. All authors anticipate seeking appropriate intellectual property protection for promising candidates that emerge from the lab's work. Dr. Kiss and Dr. Bejar have recently filed a patent application concerning a new RNA therapy technology. Dr. Kiss also has patents planned for several RNA therapeutics and an inducible stable HUVEC cell line. Dr. Bejar is named as a co-inventor on a patent filing for a candidate RNA therapeutic (no.17969496). Further, Dr. Kiss serves as an ad hoc consultant for different consulting companies and to the RNA Core at the Houston Methodist Research Institute. Dr. Kiss also reports private stock for BioLife Solutions Inc.
References
- Damase, T.R., Sukhovershin, R., Boada, C., Taraballi, F., Pettigrew, R.I., and Cooke, J.P. (2021). The Limitless Future of RNA Therapeutics. Front Bioeng Biotechnol 9, 628137. [CrossRef]
- Kalal, B.S., Upadhya, D., and Pai, V.R. (2017). Chemotherapy Resistance Mechanisms in Advanced Skin Cancer. Oncol Rev 11, 326. [CrossRef]
- Li, X., Ponandai-Srinivasan, S., Nandakumar, K.S., Fabre, S., Xu Landen, N., Mavon, A., and Khmaladze, I. (2021). Targeting microRNA for improved skin health. Health Sci Rep 4, e374. [CrossRef]
- Banerjee, J., Chan, Y.C., and Sen, C.K. (2011). MicroRNAs in skin and wound healing. Physiol Genomics 43, 543-556. [CrossRef]
- Bartel, D.P. (2018). Metazoan MicroRNAs. Cell 173, 20-51. [CrossRef]
- Hu, B., Zhong, L., Weng, Y., Peng, L., Huang, Y., Zhao, Y., and Liang, X.-J. (2020). Therapeutic siRNA: state of the art. Signal Transduction and Targeted Therapy 5, 101. [CrossRef]
- Lam, J.K., Chow, M.Y., Zhang, Y., and Leung, S.W. (2015). siRNA Versus miRNA as Therapeutics for Gene Silencing. Mol Ther Nucleic Acids 4, e252. [CrossRef]
- Zhu, Y., Zhu, L., Wang, X., and Jin, H. (2022). RNA-based therapeutics: an overview and prospectus. Cell Death Dis 13, 644. [CrossRef]
- Bejar, N., Tat, T.T., and Kiss, D.L. (2022). RNA Therapeutics: the Next Generation of Drugs for Cardiovascular Diseases. Curr Atheroscler Rep 24, 307-321. [CrossRef]
- Makino, K., Makino, T., Stawski, L., Lipson, K.E., Leask, A., and Trojanowska, M. (2017). Anti-connective tissue growth factor (CTGF/CCN2) monoclonal antibody attenuates skin fibrosis in mice models of systemic sclerosis. Arthritis Res Ther 19, 134. [CrossRef]
- Kang, S., Kim, J., Ahn, M., Kim, J., Heo, M.G., Min, D.H., and Won, C. (2020). RNAi nanotherapy for fibrosis: highly durable knockdown of CTGF/CCN-2 using siRNA-DegradaBALL (LEM-S401) to treat skin fibrotic diseases. Nanoscale 12, 6385-6393.
- Carswell, L., and Borger, J. (2022). Hypertrophic Scarring Keloids. In StatPearls. (Treasure Island (FL).
- Roberts, T.C., Langer, R., and Wood, M.J.A. (2020). Advances in oligonucleotide drug delivery. Nat Rev Drug Discov 19, 673-694. [CrossRef]
- Crooke, S.T., Baker, B.F., Crooke, R.M., and Liang, X.H. (2021). Antisense technology: an overview and prospectus. Nat Rev Drug Discov 20, 427-453. [CrossRef]
- Quemener, A.M., Bachelot, L., Forestier, A., Donnou-Fournet, E., Gilot, D., and Galibert, M.D. (2020). The powerful world of antisense oligonucleotides: From bench to bedside. Wiley Interdiscip Rev RNA 11, e1594. [CrossRef]
- Di Fusco, D., Dinallo, V., Marafini, I., Figliuzzi, M.M., Romano, B., and Monteleone, G. (2019). Antisense Oligonucleotide: Basic Concepts and Therapeutic Application in Inflammatory Bowel Disease. Front Pharmacol 10, 305. [CrossRef]
- Reese, C.B. (1978). The chemical synthesis of oligo-and poly-nucleotides by the phosphotriester approach. Tetrahedron 34, 3143-3179. [CrossRef]
- Schaller, H., Weimann, G., Lerch, B., and Khorana, H.G. (1963). Studies on Polynucleotides. XXIV.1 The Stepwise Synthesis of Specific Deoxyribopolynucleotides (4).2 Protected Derivatives of Deoxyribonucleosides and New Syntheses of Deoxyribonucleoside-3″ Phosphates3. Journal of the American Chemical Society 85, 3821-3827. [CrossRef]
- Orr, R.M. (2001). Technology evaluation: fomivirsen, Isis Pharmaceuticals Inc/CIBA vision. Curr Opin Mol Ther 3, 288-294.
- Chan, J.H., Lim, S., and Wong, W.S. (2006). Antisense oligonucleotides: from design to therapeutic application. Clin Exp Pharmacol Physiol 33, 533-540. [CrossRef]
- Juliano, R.L., and Carver, K. (2015). Cellular uptake and intracellular trafficking of oligonucleotides. Adv Drug Deliv Rev 87, 35-45. [CrossRef]
- Liang, X.H., Sun, H., Nichols, J.G., and Crooke, S.T. (2017). RNase H1-Dependent Antisense Oligonucleotides Are Robustly Active in Directing RNA Cleavage in Both the Cytoplasm and the Nucleus. Mol Ther 25, 2075-2092. [CrossRef]
- Mulhbacher, J., St-Pierre, P., and Lafontaine, D.A. (2010). Therapeutic applications of ribozymes and riboswitches. Curr Opin Pharmacol 10, 551-556. [CrossRef]
- Desterro, J., Bak-Gordon, P., and Carmo-Fonseca, M. (2020). Targeting mRNA processing as an anticancer strategy. Nat Rev Drug Discov 19, 112-129. [CrossRef]
- Juliano, R.L. (2016). The delivery of therapeutic oligonucleotides. Nucleic Acids Res 44, 6518-6548. [CrossRef]
- Younis, H.S., Templin, M.V., Whiteley, L.O., Kornbrust, D.J., Kim, T.-W., and Henry, S.P. (2013). Overview of the Nonclinical Development Strategies and Class-Effects of Oligonucleotide-Based Therapeutics. In. (. [CrossRef]
- Wan, J., Bauman, J.A., Graziewicz, M.A., Sazani, P., and Kole, R. (2013). Oligonucleotide therapeutics in cancer. Cancer Treat Res 158, 213-233. [CrossRef]
- Crooke, S.T., Vickers, T.A., and Liang, X.-h. (2020). Phosphorothioate modified oligonucleotide–protein interactions. Nucleic Acids Research 48, 5235-5253. [CrossRef]
- Sikes, R.A. (2007). Chemistry and pharmacology of anticancer drugs. British Journal of Cancer 97, 1713-1713. [CrossRef]
- Agrawal, S., Jiang, Z., Zhao, Q., Shaw, D., Cai, Q., Roskey, A., Channavajjala, L., Saxinger, C., and Zhang, R. (1997). Mixed-backbone oligonucleotides as second generation antisense oligonucleotides: in vitro and in vivo studies. Proc Natl Acad Sci U S A 94, 2620-2625. [CrossRef]
- Sardone, V., Zhou, H., Muntoni, F., Ferlini, A., and Falzarano, M.S. (2017). Antisense Oligonucleotide-Based Therapy for Neuromuscular Disease. Molecules 22. [CrossRef]
- Wahlestedt, C., Salmi, P., Good, L., Kela, J., Johnsson, T., Hokfelt, T., Broberger, C., Porreca, F., Lai, J., Ren, K., et al. (2000). Potent and nontoxic antisense oligonucleotides containing locked nucleic acids. Proc Natl Acad Sci U S A 97, 5633-5638. [CrossRef]
- Havens, M.A., and Hastings, M.L. (2016). Splice-switching antisense oligonucleotides as therapeutic drugs. Nucleic Acids Res 44, 6549-6563. [CrossRef]
- Prakash, T.P. (2011). An overview of sugar-modified oligonucleotides for antisense therapeutics. Chem Biodivers 8, 1616-1641. [CrossRef]
- Hagedorn, P.H., Persson, R., Funder, E.D., Albæk, N., Diemer, S.L., Hansen, D.J., Møller, M.R., Papargyri, N., Christiansen, H., Hansen, B.R., et al. (2018). Locked nucleic acid: modality, diversity, and drug discovery. Drug Discov Today 23, 101-114. [CrossRef]
- Gallant-Behm, C.L., Piper, J., Dickinson, B.A., Dalby, C.M., Pestano, L.A., and Jackson, A.L. (2018). A synthetic microRNA-92a inhibitor (MRG-110) accelerates angiogenesis and wound healing in diabetic and nondiabetic wounds. Wound Repair Regen 26, 311-323. [CrossRef]
- Huang, C.K., Kafert-Kasting, S., and Thum, T. (2020). Preclinical and Clinical Development of Noncoding RNA Therapeutics for Cardiovascular Disease. Circ Res 126, 663-678. [CrossRef]
- Wolff, J.A., Malone, R.W., Williams, P., Chong, W., Acsadi, G., Jani, A., and Felgner, P.L. (1990). Direct gene transfer into mouse muscle in vivo. Science 247, 1465-1468. [CrossRef]
- Weide, B., Carralot, J.P., Reese, A., Scheel, B., Eigentler, T.K., Hoerr, I., Rammensee, H.G., Garbe, C., and Pascolo, S. (2008). Results of the first phase I/II clinical vaccination trial with direct injection of mRNA. J Immunother 31, 180-188. [CrossRef]
- Gu, Y., Duan, J., Yang, N., Yang, Y., and Zhao, X. (2022). mRNA vaccines in the prevention and treatment of diseases. MedComm (2020) 3, e167. [CrossRef]
- van Dulmen, M., and Rentmeister, A. (2020). mRNA Therapies: New Hope in the Fight against Melanoma. Biochemistry 59, 1650-1655. [CrossRef]
- National Library of Medicine. In. (National Library of Medicine.
- (2023). AdisInsight. In. (Springer.
- Sahin, U., Oehm, P., Derhovanessian, E., Jabulowsky, R.A., Vormehr, M., Gold, M., Maurus, D., Schwarck-Kokarakis, D., Kuhn, A.N., Omokoko, T., et al. (2020). An RNA vaccine drives immunity in checkpoint-inhibitor-treated melanoma. Nature 585, 107-112. [CrossRef]
- BioNTech. (2023). In.
- BioNTech. (2021). In. (BioNTech.
- Kang, K., Xie, F., Mao, J., Bai, Y., and Wang, X. (2020). Significance of Tumor Mutation Burden in Immune Infiltration and Prognosis in Cutaneous Melanoma. Front Oncol 10, 573141. [CrossRef]
- Vitale, G. (2023). Chemical & Engineering News. In. (Chemical & Engineering News, American Chemical Society.
- Moderna. (2022). In. (Moderna Inc.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).