Submitted:
18 February 2023
Posted:
03 March 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
“Such a dream of transforming an agro-based economy into an information society must either be a flight of fancy or thinking hardly informed by the industrial economic background of developed economies that are in transition to informational economies. For an economy with about half of its adult population engaged in the food production sector, and about 70% of its development budget sourced from donor support, any talk of transition into an information society sounds like a far-fetched dream [8]”
- What are the recent precision agriculture research developments particularly on the disease/pest/weed detection systems?
- What are the found limitations and gaps in the literature review?
- Lastly, what are the arising opportunities for further research?
- What topological amendments can be made to the traditional precision agricultural systems to make them more economical to employ in rural farms and make them more accessible?
2. Literature Review: Precision Agriculture Research Developments
2.1. Plant Disease Detection System Basic Principles
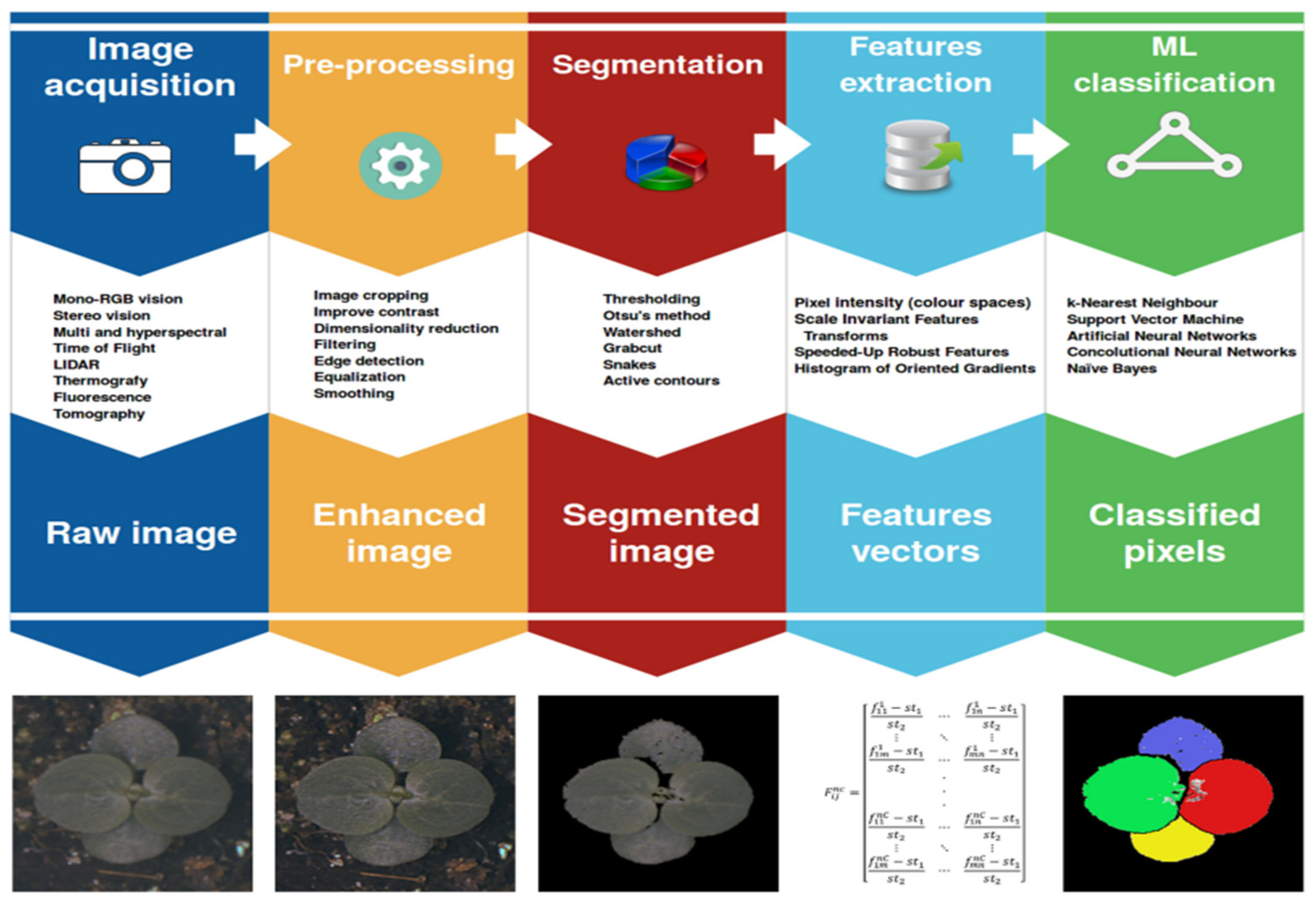
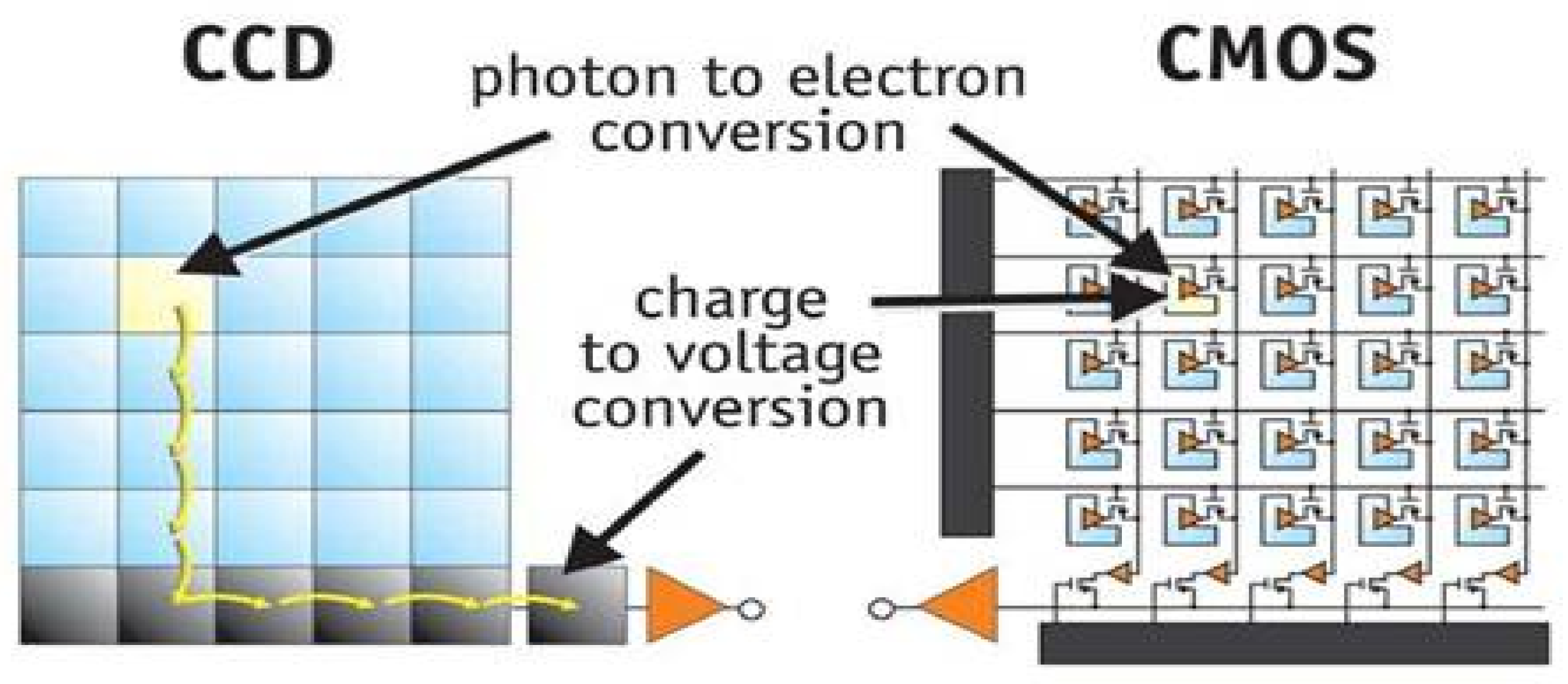
2.1.1. Image Acquisition.

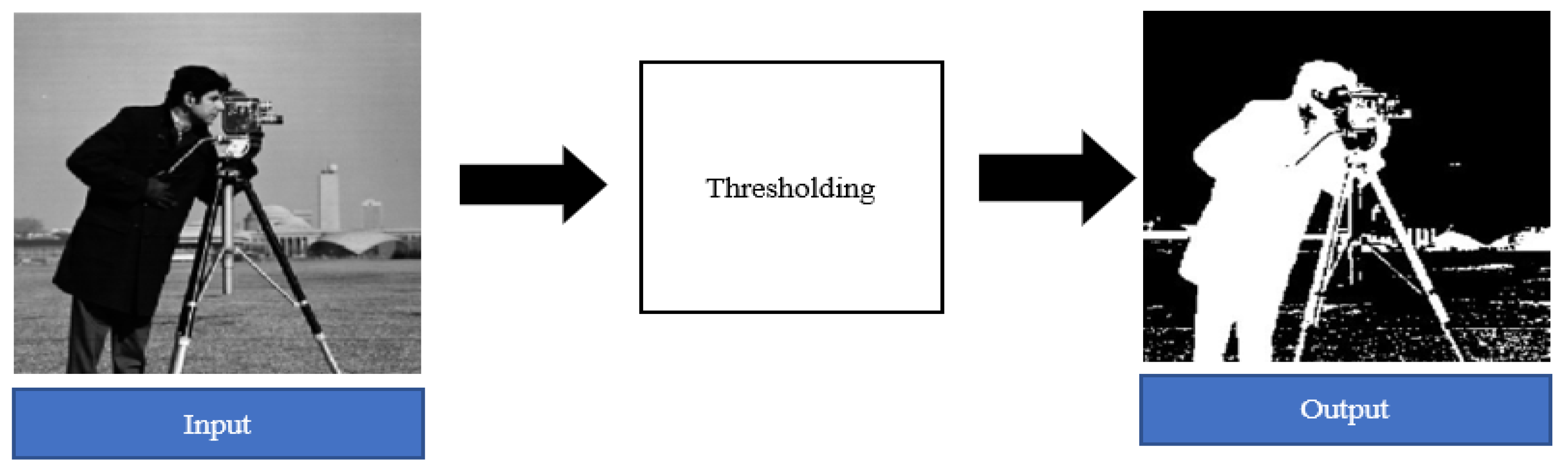
2.1.2. Image Pre-processing.
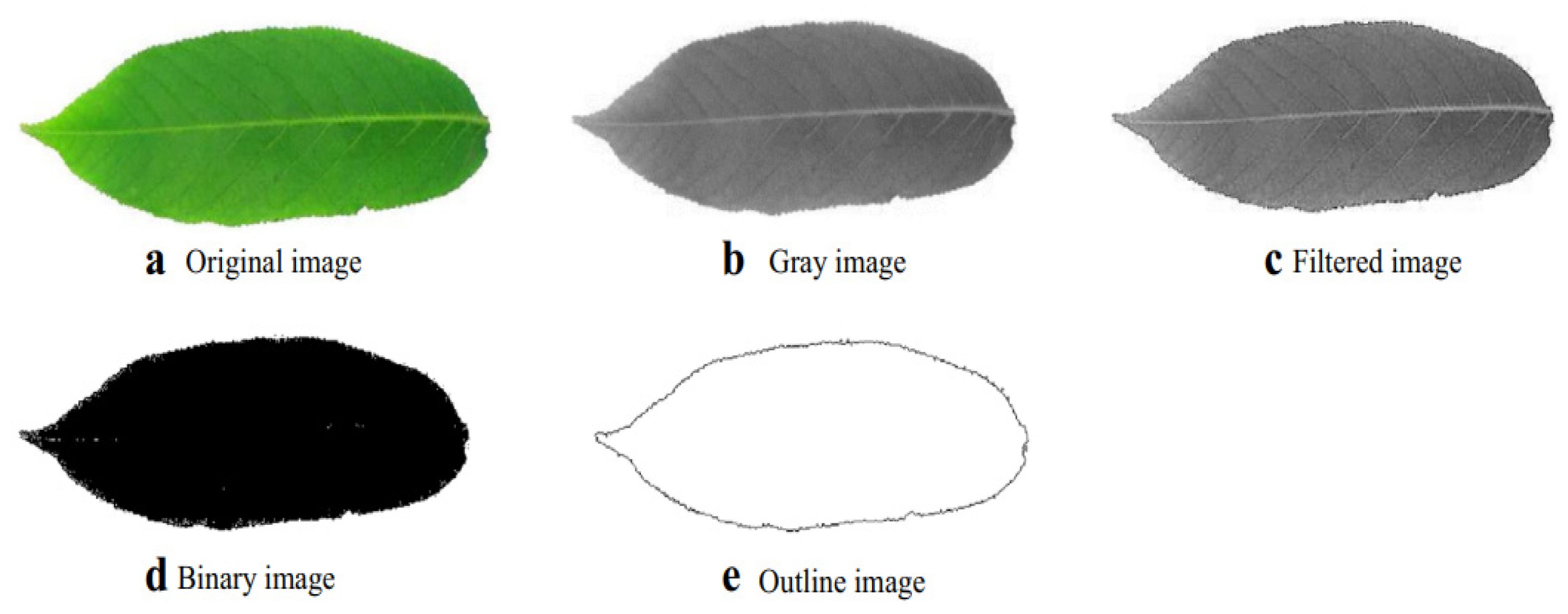
2.1.3. Image Segmentation
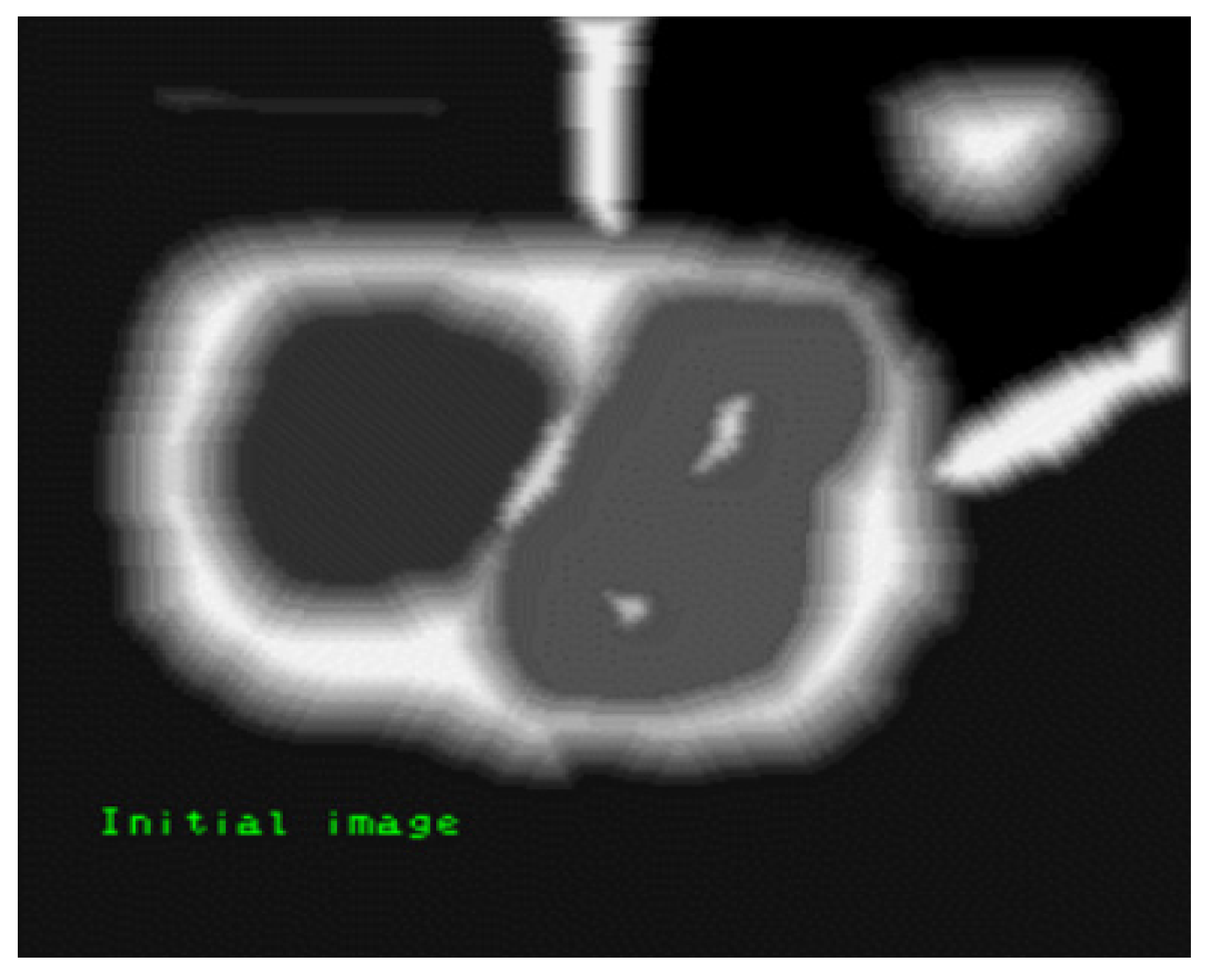
2.1.4. Feature Extraction
2.1.4.1. Shape Features
- Circularity (C) – A feature defining the degree to which a leaf conforms to a perfect circle. It is defined as [60]:
- Rectangularity (R) – A feature defining the degree to which a leaf conforms to a rectangle. It is defined as [55]:
- Aspect Ratio (AS) – Ratio of width to length of a leaf. It is defined as [55]:
- Smooth factor (SF) – Ratio of leaf picture area when a 5x5 and 2x2 regular smoothing filters have been used [58].
- Perimeter to diameter ratio (PDr) – Ratio of the perimeter to the diameter of a leaf. It is defined as [64]:
- Perimeter to length plus width ratio (PLWr) – Ratio of the perimeter to length plus width of a leaf. It is defined as [64]:
- Narrow factor (NFr) – Ratio of diameter to length of a leaf [60]:
- Area convexity (ACr) – Area ratio between the area of a leaf and the area of its convex hull [59].
- Perimeter convexity (ACr) – The ratio between the perimeter of a leaf to that of its convex hull [60].
- Eccentricity (Ar) – The degree to which a leaf shape is a centroid [64].
- Irregularity (Ir) – Ratio of the diameters of an inscribed to the circumscribed circles on the image of a leaf [59].
2.1.4.2. Colour Features
- Colour standard deviation (σ) – A major of how much the different colours found in an image match one another or are rather different from one another [60]. Say an image is differentiated into an array of its basic building blocks, the pixels, then i is a pointer moving across the rows of pixels in an array from the origin to the very last row M while j is a pointer moving across the columns of pixels in an array from the origin to the very last column N. At any point, a pixel colour intensity is defined by p(i, j) where i and j denote the coordinate position of a pixel in an image array. Therefore, the colour standard deviation is mathematically defined as follows:
- Colour mean (μ) – A major to identify a dominant colour in a leaf image. This feature is normally used to identify the leaf type [63]. It is mathematically defined as follows:
- Colour skewness (φ) – A major to identify a colour symmetry in a leaf image [21, 46]:
- Colour Kurtosis (φ) – A major to identify a colour shape dispersion in a leaf image [65]:
2.1.4.3. Texture Features
- Entropy (Entr) – This is a measure of how complex and uniforms a texture of a leaf image [68]:
- Contrast (Con) – This is a measure of how clear the features are in a leaf image, it is also referred to as the moment of inertia[69, 70]:
- Energy (En) – This is a major of the degree of how the uniform is a grey image. It is also called the second moment [69]:
- Correlation (Cor) – This is a major of whether there is a similar element in a sample picture, which corresponds to the re-occurrence of a similar matrix within a large array of pixels [68].Where:
- Difference moment inverse (DMI) – This is a major of the degree of how homogenous an image [69]:
2.1.5. Feature Classification Through Machine Learning Algorithms
2.1.5.1. SVM Classifier
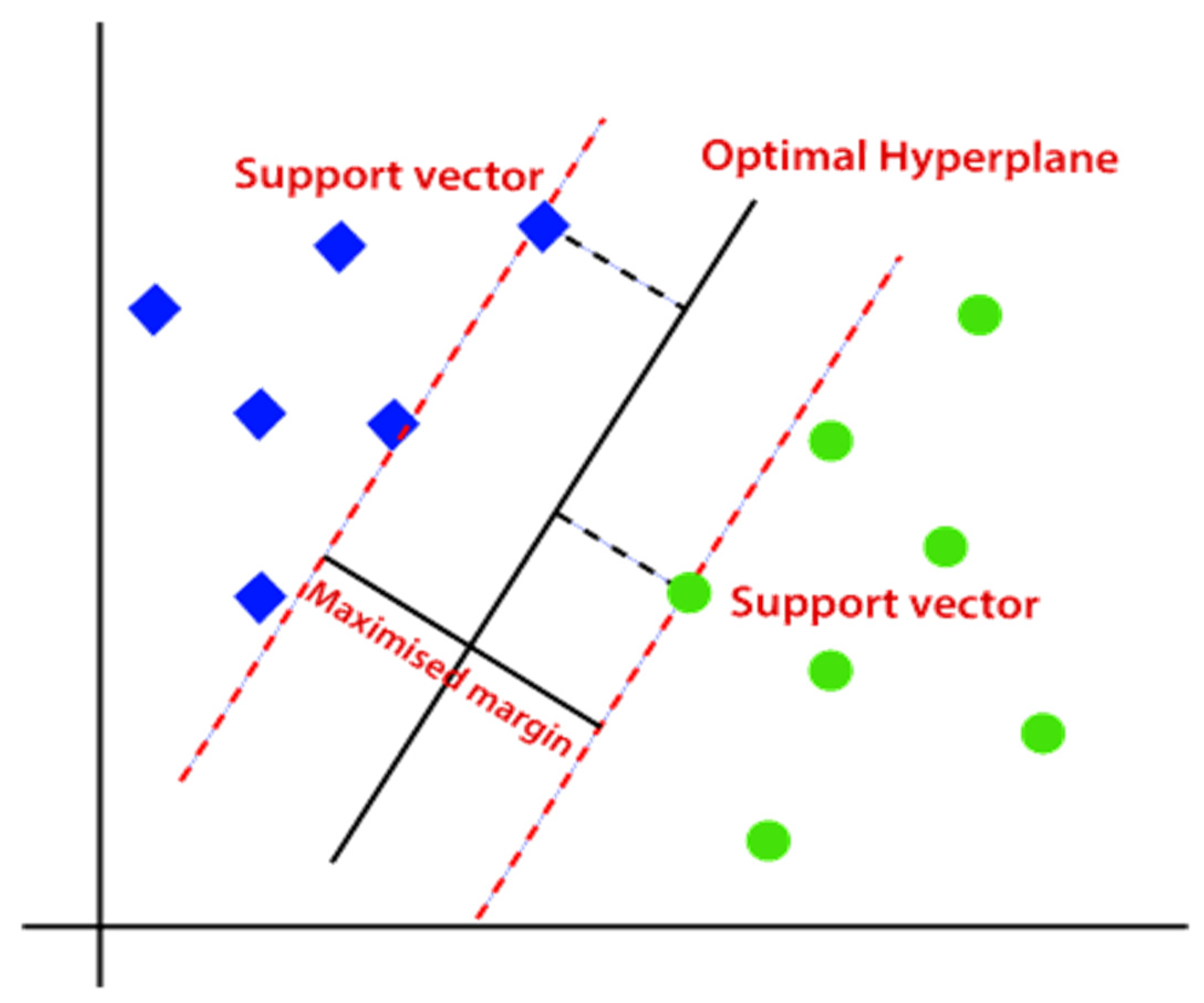
2.1.5.2. ANN Classifier
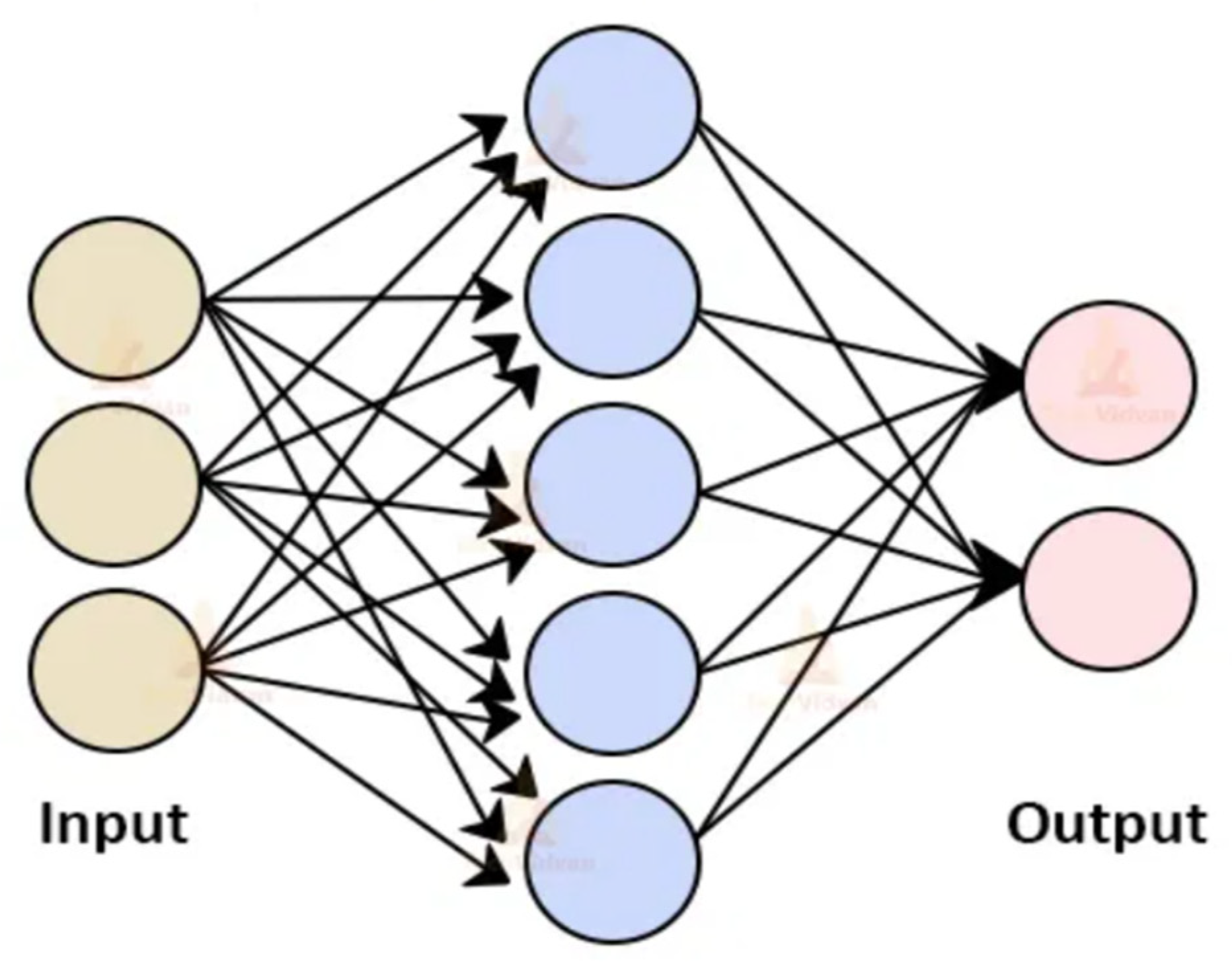
2.1.5.3. kNN Classifier
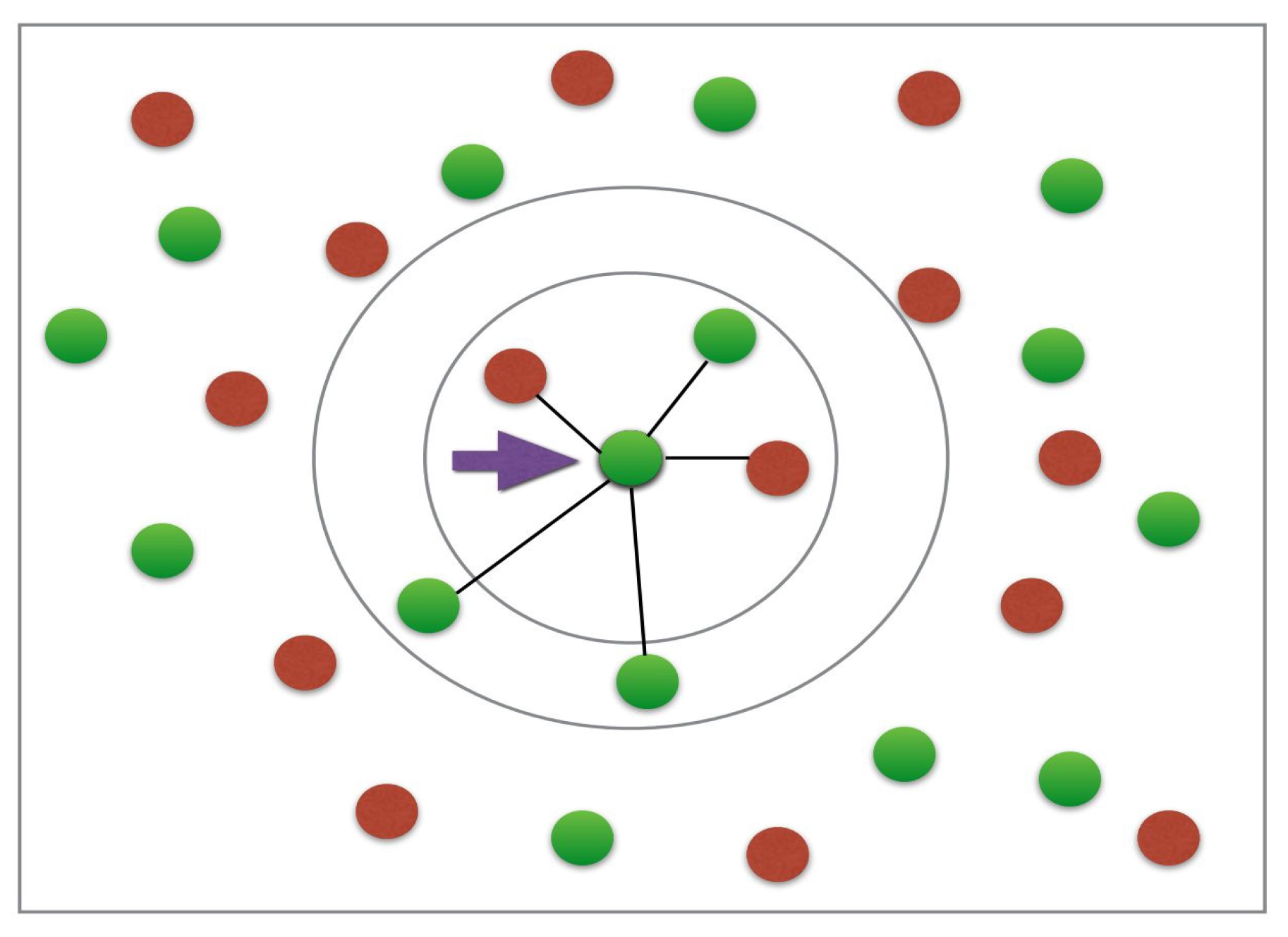
- Take the uncategorized data point as input to a model
- Measure the spatial distance between this unclassified point to all the other already classified points. The distance can be computed via Euclidean, Minkowski or Manhattan formulae [79].
- Check the points with the shortest displacement from the unknown data point to be classified for a certain K value (K is defined by the supervisor of the algorithm) and separate these points by class of belonging [79].
- Select the correct class of membership as the one with the most frequent vectors as the neighbours of the unknown data point [79].
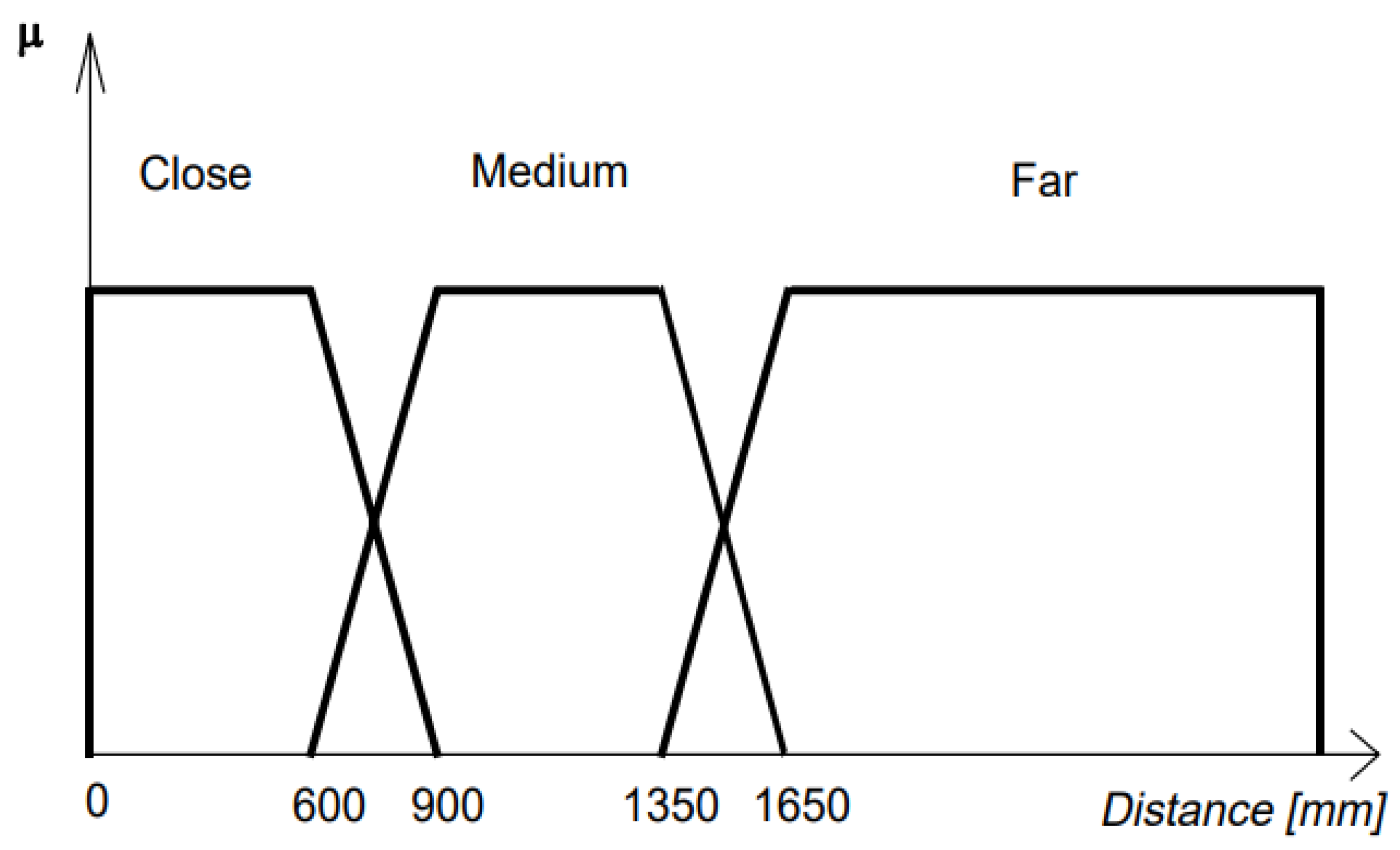
2.1.5.4. FUZZY Classifier
2.2. Literature Survey: Plant Disease/Nutrient Deficiency Monitoring Systems
2.2.1. Tabulated summary of Plant Disease/Nutrient Deficiency Monitoring Systems publications
| Classification Method | Plant/Crop | Reference | Number of diseases | Disease | Results |
| SVM Classification | Maize | [83] | 1 | Not Specified | 79% accuracy |
| Grapefruit, Lemon, lime | [84] | 2 | Canker And Anthracnose Diseases | 95% accuracy for both | |
| Grape | [85] | 2 | Downy Mildew And Powdery Mildew | 88.89% accuracy for both | |
| Oil palm | [3] | 2 | Chimaera And Anthracnose | 97% and 95% accuracy respectively | |
| Potato | [86] | 4 | Late Blight And Early Blight | 95% for both | |
| Grape | [10] | 3 | Black Rot, Esca And Leaf Blight | Not specified | |
| Tea | [87] | 3 | Not Specified | 90% accuracy | |
| Soybean | [84] | 3 | Downy Mildew, Frog Eye, And Septories Leaf | 90% accuracy average | |
| Tomato | [88] | 6 | Not Specified | 96% accuracy | |
| Rice | [89] | Not specified | Pests Diseases | 92% accuracy | |
| Soybean | [90] | 1 | Charcoal Rot | 90% accuracy | |
| Cucumber | [91] | 1 | Downy Mildew | Not specified | |
| Rice | [92] | 1 | Rice Blast | 93% accuracy | |
| Rice | [93] | 1 | Rice Blight | 80% accuracy | |
| Tea | [94] | 1 | Not Specified | 90% accuracy | |
| ANN Classification | zucchini | [95] | 1 | Soft-Rot | Not specified |
| Not specified | [96] | 4 | Alternaria Alternata, Anthracnose, Bacterial Blight, Cercospora Leaf Spot | 96% accuracy average | |
| Grapefruit | [97] | 3 | Grape-Black Rot, Powdery Mildew, Downy Mildew | 94% accuracy average | |
| Apple | [98] | 3 | Apple Scab, Apple Rot, Apple Blotch | 81% accuracy average | |
| Pomegranate | [99] | 3 | Bacterial Blight, Aspergillus Fruit Rot, Gray Mold | 99% accuracy average | |
| Not specified | [100] | 4 | Early Scorch, Cottony Mould, Late Scorch, Tiny Whiteness | 93% accuracy average | |
| Cucumber | [101] | 2 | Downy Mildew, Powdery Mildew | 99% accuracy average | |
| Pomegranate | [102] | 4 | Leaf Spot, Bacterial Blight, Fruit Spot, Fruit Rot | 90% accuracy average | |
| Groundnut | [103] | 1 | Cercospora | 97% accuracy | |
| Pomegranate | [104] | 1 | Not Specified | 90% accuracy | |
| Cucumber | [105] | 1 | Downy Mildew | 80% accuracy | |
| Rice | [106] | 3 | Bacterial Leaf Blight, Brown Spot, Leaf Smut | 96% accuracy average | |
| Citrus | [107] | 5 | Anthracnose, Black Spot, Canker, Citrus Scab, Melanose | 90% accuracy average | |
| Wheat | [108] | 4 | Powdery Mildew, Rust Puccinia Triticina, Leaf Blight, Puccinia Striifomus | Not specified | |
| k-NN Classification | Not specified | [109] | 5 | (YS) the yellow spotted, (WS) white spotted, (RS) red spotted, (N) Normal and (D) discoloured spotted | 86% Accuracy |
| Groundnut | [77] | 5 | Early leaf spot, Late leaf spot, Rust, early and late spot Bud Necrosis | 96% Accuracy | |
| Tomato, Corn, Potato | [110] | Not Specified | No disease: Leaf Recognition | 94% Accuracy (Corn) 86% Accuracy (Potato) 80% Accuracy |
|
| Tomato | [111] | 3 | Rust, early and late spot Bud Necrosis | 95% Accuracy | |
| Banana | [112] | 2 | bunchy top, sigatoka | 99% Accuracy | |
| Tomato | [113] | 3 | Rust, early and late spot Bud Necrosis | 97% Accuracy | |
| Rice | 4 | Bacterial Blight of rice, Rice Blast disease, Rice Tungro, False smut | 88% Accuracy Average | ||
| Fuzzy Classification | Mango | [82] | 3 | Powdery Mildew, Phoma blight, Bacterial canker | 90% Accuracy Average |
| strawberry | [114] | 1 | Iron deficiency | 97% Accuracy | |
| Cotton, Wheat | [115] | 18 | Bacterial blight, Leaf Curl, Root Rot, Verticillium wilt, Anthracnose, Seed rot, Tobacco streak virus, Tropical rust, Fusarium wilt, Black stem rust, leaf rust, stripe rust, Loose smut, Flag smut, complete bunt, partial bunt, Ear_cockle, Tundo |
99% Accuracy Average | |
| Soybean | [19] | 1 | Foliar | 96% Accuracy | |
| Cotton | 3 | Bacteria blight, Foliar, Alternaria | 95% Accuracy Average |
2.2.2. Research Opportunities Identified
- Little or no literature discussed the real-time monitoring of the onset signs of diseases before they spread throughout the whole plant part.
- Few papers discussed real-time monitoring and real-time mitigation measures such as actuation operations, spraying pesticides, and spraying fertilizers, to name a few examples.
- Very little research discusses the combination of these monitoring and phenotyping tasks into 1 system to reduce costs and improve technology availability to farmers and add convenience.
3. Conclusion
References
- A. Badage, “Crop disease detection using machine learning: Indian agriculture,” Int. Res. J. Eng. Technol, vol. 5, no. 9, pp. 866-869, 2018.
- U. Ukaegbu, L. Tartibu, T. Laseinde, M. Okwu, and I. Olayode, “A deep learning algorithm for detection of potassium deficiency in a red grapevine and spraying actuation using a raspberry pi3.” pp. 1-6.
- U. Shruthi, V. Nagaveni, and B. Raghavendra, “A review on machine learning classification techniques for plant disease detection.” pp. 281-284.
- S. Adekar, and A. Raul, “Detection of Plant Leaf Diseases using Machine Learning,” 2019 International Research Journal of Engineering and Technology (IRJET), 2019.
- R. S. Pachade, “A REVIEW ON COMPARATIVE STUDY OF MODERN TECHNIQUES TO ENHANCE INDIAN FARMING USING AI AND MACHINE LEARNING.”.
- M.-L. du Preez, “4IR and Water Smart Agriculture in Southern Africa: A Watch List of Key Technological Advances,” 2020.
- M. S. Hoosain, B. S. Paul, and S. Ramakrishna, “The impact of 4IR digital technologies and circular thinking on the United Nations sustainable development goals,” Sustainability, vol. 12, no. 23, pp. 10143, 2020. [CrossRef]
- B. Swaminathan, “Identification of Plant Disease and Wetness/Dryness Detection.”.
- M. Islam, K. A. Wahid, A. V. Dinh, and P. Bhowmik, “Model of dehydration and assessment of moisture content on onion using EIS,” Journal of food science and technology, vol. 56, no. 6, pp. 2814-2824, 2019. [CrossRef] [PubMed]
- S. Anju, B. Chaitra, C. Roopashree, K. Lathashree, and S. Gowtham, “Various Approaches for Plant Disease Detection,” 2021.
- S. Swain, S. K. Nayak, and S. S. Barik, “A review on plant leaf diseases detection and classification based on machine learning models,” Mukt shabd, vol. 9, no. 6, pp. 5195-5205, 2020.
- N. Prashar, “A Review on Plant Disease Detection Techniques.” pp. 501-506.
- N. Agrawal, J. Singhai, and D. K. Agarwal, “Grape leaf disease detection and classification using multi-class support vector machine.” pp. 238-244.
- Z. A. Dar, S. A. Dar, J. A. Khan, A. A. Lone, S. Langyan, B. Lone, R. Kanth, A. Iqbal, J. Rane, and S. H. Wani, “Identification for surrogate drought tolerance in maize inbred lines utilizing high-throughput phenomics approach,” Plos one, vol. 16, no. 7, pp. e0254318, 2021.
- F. Perez-Sanz, P. J. Navarro, and M. Egea-Cortines, “Plant phenomics: An overview of image acquisition technologies and image data analysis algorithms,” GigaScience, vol. 6, no. 11, pp. gix092, 2017.
- K. Padmavathi, and K. Thangadurai, “Implementation of RGB and grayscale images in plant leaves disease detection–comparative study,” Indian Journal of Science and Technology, vol. 9, no. 6, pp. 1-6, 2016.
- A. Kern, “CCD and CMOS detectors,” 2019.
- S. S. Magazov, “Image recovery on defective pixels of a CMOS and CCD arrays,” Informatsionnye Tekhnologii i Vychslitel'nye Sistemy, no. 3, pp. 25-40, 2019.
- D. Defrianto, M. Shiddiq, U. Malik, V. Asyana, and Y. Soerbakti, “Fluorescence spectrum analysis on leaf and fruit using the ImageJ software application,” Science, Technology & Communication Journal, vol. 3, no. 1, pp. 1-6, 2022.
- A. F. A. Netto, R. N. Martins, G. S. A. De Souza, F. F. L. Dos Santos, and J. T. F. Rosas, “Evaluation of a low-cost camera for agricultural applications,” J. Exp. Agric. Int, vol. 32, pp. 1-9, 2019.
- P. K. R. Maddikunta, S. Hakak, M. Alazab, S. Bhattacharya, T. R. Gadekallu, W. Z. Khan, and Q.-V. Pham, “Unmanned aerial vehicles in smart agriculture: Applications, requirements, and challenges,” IEEE Sensors Journal, vol. 21, no. 16, pp. 17608-17619, 2021. [CrossRef]
- J. Trivedi, Y. Shamnani, and R. Gajjar, “Plant leaf disease detection using machine learning.” pp. 267-276.
- H. Wang, S. Shang, D. Wang, X. He, K. Feng, and H. Zhu, “Plant disease detection and classification method based on the optimized lightweight YOLOv5 model,” Agriculture, vol. 12, no. 7, pp. 931, 2022. [CrossRef]
- P. B. Wakhare, S. Neduncheliyan, and K. R. Thakur, "Study of Disease Identification in Pomegranate Using Leaf Detection Technique." pp. 1-6.
- B. K. Ekka, and B. S. Behera, “Disease Detection in Plant Leaf Using Image Processing Technique,” International Journal of Progressive Research in Science and Engineering, vol. 1, no. 4, pp. 151-155, 2020.
- N. R. Kolhalkar, and V. Krishnan, “Mechatronics system for diagnosis and treatment of major diseases in grape vineyards based on image processing,” Materials Today: Proceedings, vol. 23, pp. 549-556, 2020.
- K. Takada, “A Study on Learning Algorithms of Value and Policy Functions in Hex,” 北海道大学, 2019.
- X. Contreras, N. Amberg, A. Davaatseren, A. H. Hansen, J. Sonntag, L. Andersen, T. Bernthaler, C. Streicher, A. Heger, and R. L. Johnson, “A genome-wide library of MADM mice for single-cell genetic mosaic analysis,” Cell reports, vol. 35, no. 12, pp. 109274, 2021. [CrossRef]
- C. Mazur, J. Ayers, J. Humphrey, G. Hains, and Y. Khmelevsky, "Machine Learning Prediction of Gamer’s Private Networks (GPN® S)." pp. 107-123.
- V. Vijayalakshmi, and K. Venkatachalapathy, “Comparison of predicting student’s performance using machine learning algorithms,” International Journal of Intelligent Systems and Applications, vol. 11, no. 12, pp. 34, 2019. [CrossRef]
- K. S. Adewole, A. G. Akintola, S. A. Salihu, N. Faruk, and R. G. Jimoh, “Hybrid rule-based model for phishing URLs detection.” pp. 119-135.
- D. Krivoguz, “Validation of landslide susceptibility map using ROCR package in R.” pp. 188-192.
- M. Sieber, S. Klar, M. F. Vassiliou, and I. Anastasopoulos, “Robustness of simplified analysis methods for rocking structures on compliant soil,” Earthquake Engineering & Structural Dynamics, vol. 49, no. 14, pp. 1388-1405, 2020.
- C. Aybar, Q. Wu, L. Bautista, R. Yali, and A. Barja, “rgee: An R package for interacting with Google Earth Engine,” Journal of Open Source Software, vol. 5, no. 51, pp. 2272, 2020. [CrossRef]
- M. Schweinberger, "Tree-based models in R," The University of Queensland. Retrieved from https://slcla dal. github. io …, 2021.
- S. Pölsterl, “scikit-survival: A Library for Time-to-Event Analysis Built on Top of scikit-learn,” J. Mach. Learn. Res., vol. 21, no. 212, pp. 1-6, 2020.
- N. Melnykova, R. Kulievych, Y. Vycluk, K. Melnykova, and V. Melnykov, “Anomalies Detecting in Medical Metrics Using Machine Learning Tools,” Procedia Computer Science, vol. 198, pp. 718-723, 2022. [CrossRef]
- E. J. Gómez-Hernández, P. A. Martínez, B. Peccerillo, S. Bartolini, J. M. García, and G. Bernabé, “Using PHAST to port Caffe library: First experiences and lessons learned,” arXiv preprint arXiv:2005.13076, 2020. arXiv:2005.13076.
- M. N. Gevorkyan, A. V. Demidova, T. S. Demidova, and A. A. Sobolev, “Review and comparative analysis of machine learning libraries for machine learning,” Discrete and Continuous Models and Applied Computational Science, vol. 27, no. 4, pp. 305-315, 2019.
- M. Weber, H. Wang, S. Qiao, J. Xie, M. D. Collins, Y. Zhu, L. Yuan, D. Kim, Q. Yu, and D. Cremers, “Deeplab2: A tensorflow library for deep labeling,” arXiv preprint arXiv:2106.09748, 2021. arXiv:2106.09748.
- G. DESHPANDE, “Deceptive security using Python.”.
- V. Patel, “Open Source Frameworks for Deep Learning and Machine Learning.”.
- A. Pocock, “Tribuo: Machine Learning with Provenance in Java,” arXiv preprint arXiv:2110.03022, 2021. arXiv:2110.03022.
- E. Schubert, and A. Zimek, “ELKI: A large open-source library for data analysis-ELKI Release 0.7. 5" Heidelberg",” arXiv preprint arXiv:1902.03616, 2019. arXiv:1902.03616.
- Y. Liu, X. S. Shan, W. Kan, B. Kong, H. Li, C. Wang, T. Yang, C. Li, Y. Tan, and H. Qu, “JSAT.”.
- A. Bhatia, and B. Kaluza, Machine Learning in Java: Helpful techniques to design, build, and deploy powerful machine learning applications in Java: Packt Publishing Ltd, 2018.
- H. Luu, Beginning Apache Spark 2: with resilient distributed datasets, Spark SQL, structured streaming and Spark machine learning library: Apress, 2018.
- M. K. Vanam, B. A. Jiwani, A. Swathi, and V. Madhavi, “High performance machine learning and data science based implementation using Weka,” Materials Today: Proceedings, 2021.
- T. Saha, N. Aaraj, N. Ajjarapu, and N. K. Jha, “SHARKS: Smart Hacking Approaches for RisK Scanning in Internet-of-Things and cyber-physical systems based on machine learning,” IEEE Transactions on Emerging Topics in Computing, 2021.
- R. R. Curtin, M. Edel, M. Lozhnikov, Y. Mentekidis, S. Ghaisas, and S. Zhang, “mlpack 3: a fast, flexible machine learning library,” Journal of Open Source Software, vol. 3, no. 26, pp. 726, 2018. [CrossRef]
- Z. Wen, J. Shi, Q. Li, B. He, and J. Chen, “ThunderSVM: A fast SVM library on GPUs and CPUs,” The Journal of Machine Learning Research, vol. 19, no. 1, pp. 797-801, 2018.
- K. Kolodiazhnyi, Hands-On Machine Learning with C++: Build, train, and deploy end-to-end machine learning and deep learning pipelines: Packt Publishing Ltd, 2020.
- A. Mohan, A. K. Singh, B. Kumar, and R. Dwivedi, “Review on remote sensing methods for landslide detection using machine and deep learning,” Transactions on Emerging Telecommunications Technologies, vol. 32, no. 7, pp. e3998, 2021. [CrossRef]
- R. Prasad, and V. Rohokale, “Artificial intelligence and machine learning in cyber security,” Cyber Security: The Lifeline of Information and Communication Technology, pp. 231-247: Springer, 2020.
- F. Garcia-Lamont, J. Cervantes, A. López, and L. Rodriguez, “Segmentation of images by color features: A survey,” Neurocomputing, vol. 292, pp. 1-27, 2018. [CrossRef]
- A. Wang, W. Zhang, and X. Wei, “A review on weed detection using ground-based machine vision and image processing techniques,” Computers and electronics in agriculture, vol. 158, pp. 226-240, 2019. [CrossRef]
- J. Ker, S. P. Singh, Y. Bai, J. Rao, T. Lim, and L. Wang, “Image thresholding improves 3-dimensional convolutional neural network diagnosis of different acute brain hemorrhages on computed tomography scans,” Sensors, vol. 19, no. 9, pp. 2167, 2019. [CrossRef]
- A. Kumar, and A. Tiwari, “A comparative study of otsu thresholding and k-means algorithm of image segmentation,” Int. J. Eng. Technol. Res, vol. 9, pp. 2454-4698, 2019.
- L. Zhang, L. Zou, C. Wu, J. Jia, and J. Chen, “Method of famous tea sprout identification and segmentation based on improved watershed algorithm,” Computers and Electronics in Agriculture, vol. 184, pp. 106108, 2021. [CrossRef]
- L. Xie, J. Qi, L. Pan, and S. Wali, “Integrating deep convolutional neural networks with marker-controlled watershed for overlapping nuclei segmentation in histopathology images,” Neurocomputing, vol. 376, pp. 166-179, 2020. [CrossRef]
- P. M. Anger, L. Prechtl, M. Elsner, R. Niessner, and N. P. Ivleva, “Implementation of an open source algorithm for particle recognition and morphological characterisation for microplastic analysis by means of Raman microspectroscopy,” Analytical Methods, vol. 11, no. 27, pp. 3483-3489, 2019. [CrossRef]
- S. Jadhav, and B. Garg, “Comparative Analysis of Image Segmentation Techniques for Real Field Crop Images.” pp. 1-17.
- C. Li, X. Zhao, and H. Ru, “GrabCut Algorithm Fusion of Extreme Point Features.” pp. 33-38.
- J. F. Randrianasoa, C. Kurtz, E. Desjardin, and N. Passat, “AGAT: Building and evaluating binary partition trees for image segmentation,” SoftwareX, vol. 16, pp. 100855, 2021. [CrossRef]
- N. Zhu, X. Liu, Z. Liu, K. Hu, Y. Wang, J. Tan, M. Huang, Q. Zhu, X. Ji, and Y. Jiang, “Deep learning for smart agriculture: Concepts, tools, applications, and opportunities,” International Journal of Agricultural and Biological Engineering, vol. 11, no. 4, pp. 32-44, 2018.
- Q. Zhang, Y. Liu, C. Gong, Y. Chen, and H. Yu, “Applications of deep learning for dense scenes analysis in agriculture: A review,” Sensors, vol. 20, no. 5, pp. 1520, 2020. [CrossRef]
- D. Ireri, E. Belal, C. Okinda, N. Makange, and C. Ji, “A computer vision system for defect discrimination and grading in tomatoes using machine learning and image processing,” Artificial Intelligence in Agriculture, vol. 2, pp. 28-37, 2019. [CrossRef]
- S. Singh¹, and P. P. Kaur, “A study of geometric features extraction from plant leafs,” 2019.
- M. Martsepp, T. Laas, K. Laas, J. Priimets, S. Tõkke, and V. Mikli, “Dependence of multifractal analysis parameters on the darkness of a processed image,” Chaos, Solitons & Fractals, vol. 156, pp. 111811, 2022.
- J. M. Ponce, A. Aquino, and J. M. Andújar, “Olive-fruit variety classification by means of image processing and convolutional neural networks,” IEEE Access, vol. 7, pp. 147629-147641, 2019. [CrossRef]
- N. R. Bhimte, and V. Thool, “Diseases detection of cotton leaf spot using image processing and SVM classifier.” pp. 340-344.
- G. K. Sandhu, and R. Kaur, “Plant disease detection techniques: a review.” pp. 34-38.
- P. Alagumariappan, N. J. Dewan, G. N. Muthukrishnan, B. K. B. Raju, R. A. A. Bilal, and V. Sankaran, “Intelligent plant disease identification system using Machine Learning,” Engineering Proceedings, vol. 2, no. 1, pp. 49, 2020.
- A. A. Bharate, and M. Shirdhonkar, “Classification of grape leaves using KNN and SVM classifiers.” pp. 745-749.
- S. Sivasakthi, and M. MCA, “Plant leaf disease identification using image processing and svm, ann classifier methods.” pp. 30-31.
- C. U. Kumari, S. J. Prasad, and G. Mounika, “Leaf disease detection: feature extraction with K-means clustering and classification with ANN.” pp. 1095-1098.
- M. Vaishnnave, K. S. Devi, P. Srinivasan, and G. A. P. Jothi, “Detection and classification of groundnut leaf diseases using KNN classifier.” pp. 1-5.
- E. Hossain, M. F. Hossain, and M. A. Rahaman, “A color and texture based approach for the detection and classification of plant leaf disease using KNN classifier.” pp. 1-6.
- J. Singh, and H. Kaur, “Plant disease detection based on region-based segmentation and KNN classifier.” pp. 1667-1675.
- A. Bakhshipour, and H. Zareiforoush, “Development of a fuzzy model for differentiating peanut plant from broadleaf weeds using image features,” Plant methods, vol. 16, no. 1, pp. 1-16, 2020.
- H. Sabrol, and S. Kumar, “Plant leaf disease detection using adaptive neuro-fuzzy classification.” pp. 434-443.
- P. Sutha, A. Nandhu Kishore, V. Jayanthi, A. Periyanan, and P. Vahima, “Plant Disease Detection Using Fuzzy Classification,” Annals of the Romanian Society for Cell Biology, pp. 9430-9441, 2021.
- K. P. Panigrahi, H. Das, A. K. Sahoo, and S. C. Moharana, “Maize leaf disease detection and classification using machine learning algorithms,” Progress in Computing, Analytics and Networking, pp. 659-669: Springer, 2020.
- Y. Kashyap, and S. Shrivastava, “ROLE OF VARIOUS SEGMENTATION AND CLASSIFICATION ALGORITHMS IN PLANT DISEASE DETECTION.”.
- A. Dwari, A. Tarasia, A. Jena, S. Sarkar, S. K. Jena, and S. Sahoo, “Smart Solution for Leaf Disease and Crop Health Detection,” Advances in Intelligent Computing and Communication, pp. 231-241: Springer, 2021.
- N. K. KUMAR, and A. PRATHYUSHA, “SOLANUM TUBEROSUM LEAF DISEASES DETECTION USING DEEP LEARNING.”.
- S. Hossain, R. M. Mou, M. M. Hasan, S. Chakraborty, and M. A. Razzak, “Recognition and detection of tea leaf's diseases using support vector machine.” pp. 150-154.
- S. Coulibaly, B. Kamsu-Foguem, D. Kamissoko, and D. Traore, “Deep neural networks with transfer learning in millet crop images,” Computers in Industry, vol. 108, pp. 115-120, 2019. [CrossRef]
- N. Cherukuri, G. R. Kumar, O. Gandhi, V. S. K. Thotakura, D. NagaMani, and C. Z. Basha, “Automated Classification of rice leaf disease using Deep Learning Approach.” pp. 1206-1210.
- E. Khalili, S. Kouchaki, S. Ramazi, and F. Ghanati, “Machine learning techniques for soybean charcoal rot disease prediction,” Frontiers in plant science, vol. 11, pp. 590529, 2020. [CrossRef]
- M. P. Patil, “Nearest neighborhood approach for identification of cucumber mosaic virus infection and yellow sigatoka disease on banana plants,” 2022.
- H. Orchi, M. Sadik, and M. Khaldoun, “On using artificial intelligence and the internet of things for crop disease detection: A contemporary survey,” Agriculture, vol. 12, no. 1, pp. 9, 2021. [CrossRef]
- D. Zhang, X. Zhou, J. Zhang, Y. Lan, C. Xu, and D. Liang, “Detection of rice sheath blight using an unmanned aerial system with high-resolution color and multispectral imaging,” PloS one, vol. 13, no. 5, pp. e0187470, 2018.
- G. Yashodha, and D. Shalini, “An integrated approach for predicting and broadcasting tea leaf disease at early stage using IoT with machine learning–a review,” Materials Today: Proceedings, vol. 37, pp. 484-488, 2021.
- A. V. Zubler, and J.-Y. Yoon, “Proximal methods for plant stress detection using optical sensors and machine learning,” Biosensors, vol. 10, no. 12, pp. 193, 2020.
- B. L. Thushara, and T. M. Rasool, “Analysis of plant diseases using expectation maximization detection with BP-ANN classification,” XIII (VIII), 2020.
- P. S. Thakur, P. Khanna, T. Sheorey, and A. Ojha, “Trends in vision-based machine learning techniques for plant disease identification: A systematic review,” Expert Systems with Applications, pp. 118117, 2022.
- A. I. Khan, S. Quadri, and S. Banday, “Deep learning for apple diseases: classification and identification,” International Journal of Computational Intelligence Studies, vol. 10, no. 1, pp. 1-12, 2021. [CrossRef]
- A. J. Das, R. Ravinath, T. Usha, B. S. Rohith, H. Ekambaram, M. K. Prasannakumar, N. Ramesh, and S. K. Middha, “Microbiome Analysis of the Rhizosphere from Wilt Infected Pomegranate Reveals Complex Adaptations in Fusarium—A Preliminary Study,” Agriculture, vol. 11, no. 9, pp. 831, 2021. [CrossRef]
- S. S. Gaikwad, “Fungi classification using convolution neural network,” Turkish Journal of Computer and Mathematics Education (TURCOMAT), vol. 12, no. 10, pp. 4563-4569, 2021.
- D. Priya, “Cotton leaf disease detection using Faster R-CNN with Region Proposal Network,” International Journal of Biology and Biomedicine, vol. 6, 2021.
- B. M. Joshi, and H. Bhavsar, “Plant leaf disease detection and control: A survey,” Journal of Information and Optimization Sciences, vol. 41, no. 2, pp. 475-487, 2020. [CrossRef]
- G. Gangadevi, and C. Jayakumar, “Review of Classifiers Used for Identification and Classification of Plant Leaf Diseases.” pp. 445-459.
- V. Vučić, M. Grabež, A. Trchounian, and A. Arsić, “Composition and potential health benefits of pomegranate: a review,” Current pharmaceutical design, vol. 25, no. 16, pp. 1817-1827, 2019. [CrossRef]
- V. Sahni, S. Srivastava, and R. Khan, “Modelling techniques to improve the quality of food using artificial intelligence,” Journal of Food Quality, vol. 2021, 2021.
- S. Patidar, A. Pandey, B. A. Shirish, and A. Sriram, “Rice plant disease detection and classification using deep residual learning.” pp. 278-293.
- M. Sharif, M. A. Khan, Z. Iqbal, M. F. Azam, M. I. U. Lali, and M. Y. Javed, “Detection and classification of citrus diseases in agriculture based on optimized weighted segmentation and feature selection,” Computers and electronics in agriculture, vol. 150, pp. 220-234, 2018. [CrossRef]
- T. Hayit, H. Erbay, F. Varçın, F. Hayit, and N. Akci, “Determination of the severity level of yellow rust disease in wheat by using convolutional neural networks,” Journal of Plant Pathology, vol. 103, no. 3, pp. 923-934, 2021. [CrossRef]
- S. S. Jasim, and A. A. M. Al-Taei, “A Comparison Between SVM and K-NN for classification of Plant Diseases,” Diyala Journal for Pure Science, vol. 14, no. 2, pp. 94-105, 2018. [CrossRef]
- P. DAYANG, and A. S. K. MELI, “AS: Evaluation of image segmentation algorithms for plant disease detection,” Int. J. Image Graph. Signal Process.(IJIGSP), vol. 13, no. 5, pp. 14-26, 2021. [CrossRef]
- M. Agarwal, S. K. Gupta, and K. Biswas, “Development of Efficient CNN model for Tomato crop disease identification,” Sustainable Computing: Informatics and Systems, vol. 28, pp. 100407, 2020.
- R. D. Devi, S. A. Nandhini, R. Hemalatha, and S. Radha, “IoT enabled efficient detection and classification of plant diseases for agricultural applications.” pp. 447-451.
- S. S. Harakannanavar, J. M. Rudagi, V. I. Puranikmath, A. Siddiqua, and R. Pramodhini, “Plant Leaf Disease Detection using Computer Vision and Machine Learning Algorithms,” Global Transitions Proceedings, 2022.
- H. Altıparmak, M. Al Shahadat, E. Kiani, and K. Dimililer, “Fuzzy classification for strawberry diseases-infection using machine vision and soft-computing techniques.” pp. 429-436.
- M. Toseef, and M. J. Khan, “An intelligent mobile application for diagnosis of crop diseases in Pakistan using fuzzy inference system,” Computers and Electronics in Agriculture, vol. 153, pp. 1-11, 2018. [CrossRef]
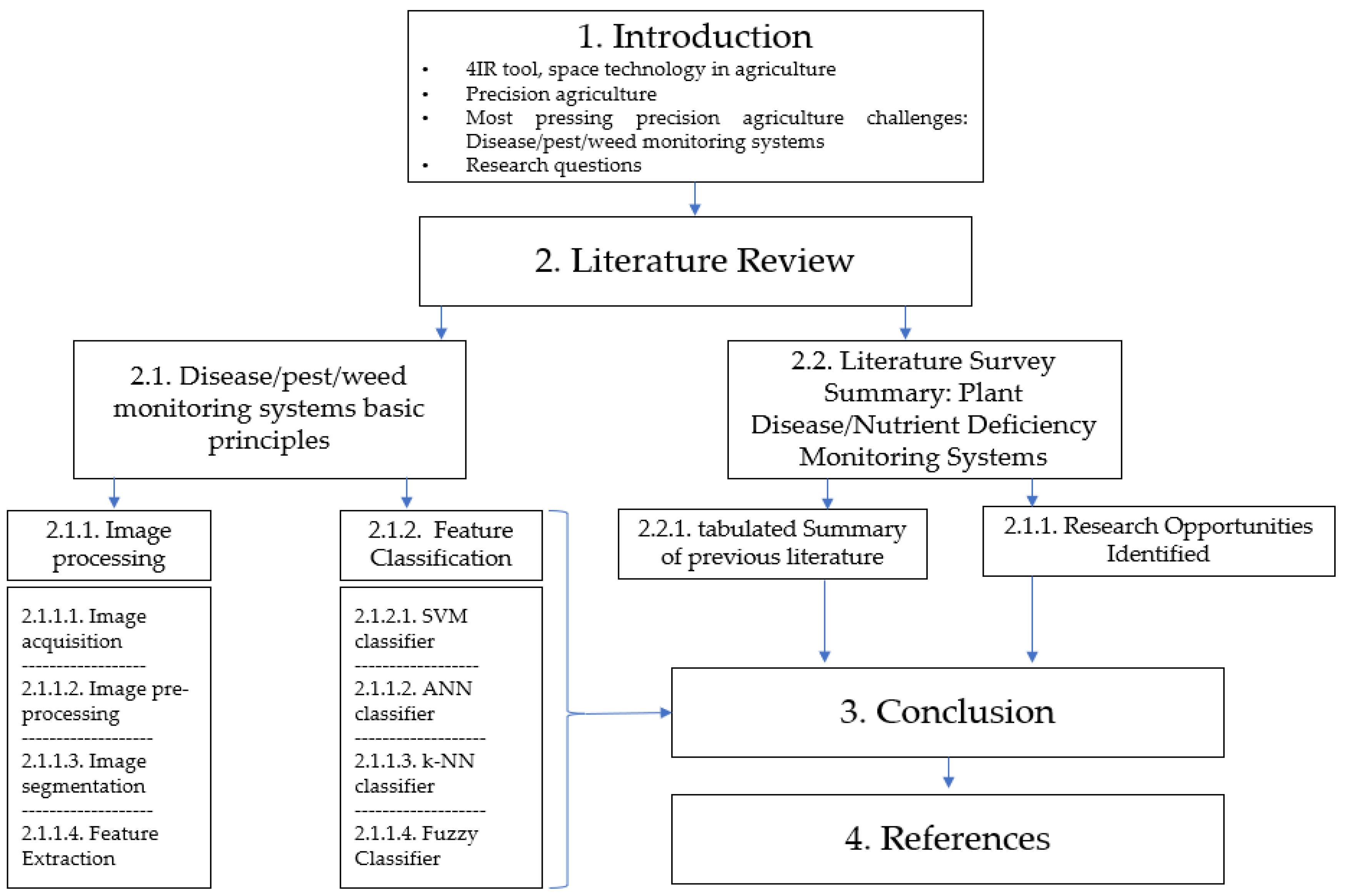
| A typical general plant disease detection system | |
| Summary of image processing steps | Different classification techniques |
| Image processing: Image acquisition Image pre-processing Image segmentation Feature extraction Machine learning classification |
SVM Classifier |
| ANN Classifier | |
| KNN Classifier | |
| FUZZY Classifier | |
| Software language of implementation | Library | Description | Open source |
| R | Kern-Lab | Mechanisms for segmentation, modelling, grouping, uniqueness identification, and feature matching using kernel-based deep learning [27]. | https://cran.r-project.org/ |
| MICE | This method can deal with data sets with missing data by computing estimates and filling in the missing data values [28]. | ||
| e1071 | Programming package containing functions for types of statistical methods, i.e., probability and statistics [29]. | ||
| CA-RET | Offers a wide range of tools for creating forecasting analytics utilizing R's extensive model library. it contains techniques for the pre-processing learning algorithm, determining the relevance of parameters, and presenting networks [30]. | ||
| Rweka | Data pre-processing, categorization, analysis, grouping, clustering algorithms, and image processing methods for all Java-based machine learning methods [31]. | ||
| ROCR | A tool for assessing and displaying the accuracy of rating classifiers [32]. | ||
| KlaR | various categorization and display functions [33]. | ||
| Earth | Utilize the methods from Friedman's publications "Multivariate Adaptive Regression Splines" and "Fast MARS" to create a prediction model [34]. | ||
| TREE | A library containing functions designated to work with trees [35]. | ||
| R, C | Igraph | Contains functions manipulating large graphs, and displaying [34]. | |
| Python, R | Scikit-learn | Offers a standardized interface for putting the machine into learning algorithms practice. It comprises various auxiliary tasks like data preprocessing operations, information resampling methods, assessment criteria, and search portals for adjusting and performance optimization of methods [36]. | |
| Python | NuPIC | Software for artificial intelligence that supports Hypertext Markup Language (HTML) learning models purely based on the neocortex's neurobiology [37]. | http://numenta.org/ |
| Caffe | Deep learning framework that prioritizes modularity, performance, and expression [38] | http://caffe.berkeleyvision.org/ | |
| Theano | A toolkit and processor that is optimized for working with and assessing equations, particularly those using array value [39]. | http://deeplearning.net/software/theano | |
| Tensorflow | Toolkit for quick computation of numbers in artificial intelligence and machine learning [40]. | https://www.tensorflow.org/ | |
| PyBrain | A versatile, powerful, and user-friendly machine learning library which offers algorithms that may be used for a range of machine learning tasks [41]. | http://pybrain.org/ | |
| Pylearn2 | A specially created library for machine learning to make learning much easier for developers. It is quick and gives a researcher a lot of versatility [42]. | http://deeplearning.net/software/pylearn2 | |
| Java | Java-ML | A collection of machine learning and data mining techniques that aim to offer a simple-to-use and extendable API. Algorithms rigorously adhere to their respective interfaces, which are maintained basic for each type of algorithm's interface [43]. | http://java-ml.sourceforge.net/ |
| ELKI | A data mining software that intends to make it possible to create and evaluate sophisticated data mining algorithms and study how they interact with database search architecture [44]. | http://elki.dbs.ifi.lmu.de/ | |
| JSAT | A library designed to fill the need for a general-purpose, reasonably high-efficiency, and versatile library in the Java ecosystem that is not sufficiently satisfied by Weka and Java-ML [45]. | https://github.com/EdwardRaff/JSAT | |
| Mallet | Toolkit for information extraction, text categorization, grouping, quantitative natural language processing, as well as other deep learning uses to text [46]. | http://mallet.cs.umass.edu/ | |
| Spark | Offers a variety of machine learning techniques such as grouping, categorization, extrapolation, and data aggregation, along with auxiliary features like simulation assessment and data acquisition [47]. | http://spark.apache.org/ | |
| Weka | Provides instruments for categorizing, forecasting, clustering, classification techniques, and visualization of information [48]. | http://www.cs.waikato.ac.nz/ml/weka/ | |
| C#, C++, C | Shark | Includes approach for neural networks, both linear and nonlinear programming, kernel-based learning algorithms, and other methods for machine learning [49]. | http://image.diku.dk/shark/ |
| mlpack | Gives the data processing techniques as simplified control scripts, Python bindings, and C++ objects that can be used in more extensive machine learning solutions [50]. | http://mlpack.org/ | |
| LibSVM | A Support Vector Machines (SVM) library [51]. | http://www.csie.ntu.edu.tcjlin/libsvm/ | |
| Shogun | Provides a wide range of data types and techniques for deep learning issues. It utilizes SWIG to provide interfaces for Octave, Python, R, Java, Lua, Ruby, and C# [52]. | http://shogun-toolbox.org/ | |
| Multiboot | offers a quick C++ solution for enhancing methods for many classes, labels, and tasks[53]. | http://www.multiboost.org/ | |
| MLC++ | Supervised machine learning methods and functions in a C++ ecosystem [52]. | http://www.sgi.com/tech/mlc/source.html | |
| Accord | Fully C#-written machine learning platform with audio and picture analysis libraries [54]. | http://accord-framework.net/ |
| PROS & CONS OF DIFFERENT CLASSIFICATION METHODS MOST USED IN PLANT PHENOMICS AND DISEASE MONITORING | |
| 1. Support Vector Machine (SVM) | |
| Advantages | Disadvantages |
| Works very accurately when there is a clear formation of a hyperplane [74]. | Accuracy difficulties with a large amount of training data [71]. |
| Works more accurately on high-dimension spaces like 3-D and 4-D [51]. | Susceptibility to noise and overlapping data classes [75]. |
| Saves memory space [71]. | The number of characteristics for a single data set must not exceed the number of data points in the training set [74]. |
| 2. Artificial Neural Network (ANN) | |
| Capable of multitasking [76]. | Complex programming algorithms [75]. |
| The machine is learning continuously, and the accuracy is improving iterable [50]. | Accuracy is data dependent; more training data translate to more accurate classification and the opposite is true [75]. |
| Have many applications (e.g., mining, agriculture, medicine, and engineering) [59]. | Hardware reliance (cost, complexity and maintenance) [33]. |
| 3. k-Nearest Neighbor (kNN) | |
| No initial training period [74]. | Accuracy difficulties with a large amount of training data [78]. |
| Simple to add new data to the model to extend its scope [79]. | Not suitable for high-dimensional space [79]. |
| Relatively easy to implement with only the two parameters to work out, the k value and the geometric distance between the points [77]. | Susceptibility to noise and outliers [74]. |
| 4. Fuzzy classifier | |
| Unclear, distorted, degraded or vague input data is accommodated by the model [80]. | Depending on people’s experience and expertise [81]. |
| More flexibility and ease to change the rules [82]. | Require excessive supervision in a form of testing and validation [81]. |
| Robust in applications with no exact input format [81]. | The is no universal approach to implementing fuzzy classification models which adds to their inaccuracy [82]. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





