3.1. Half-Space Proximal Network Model
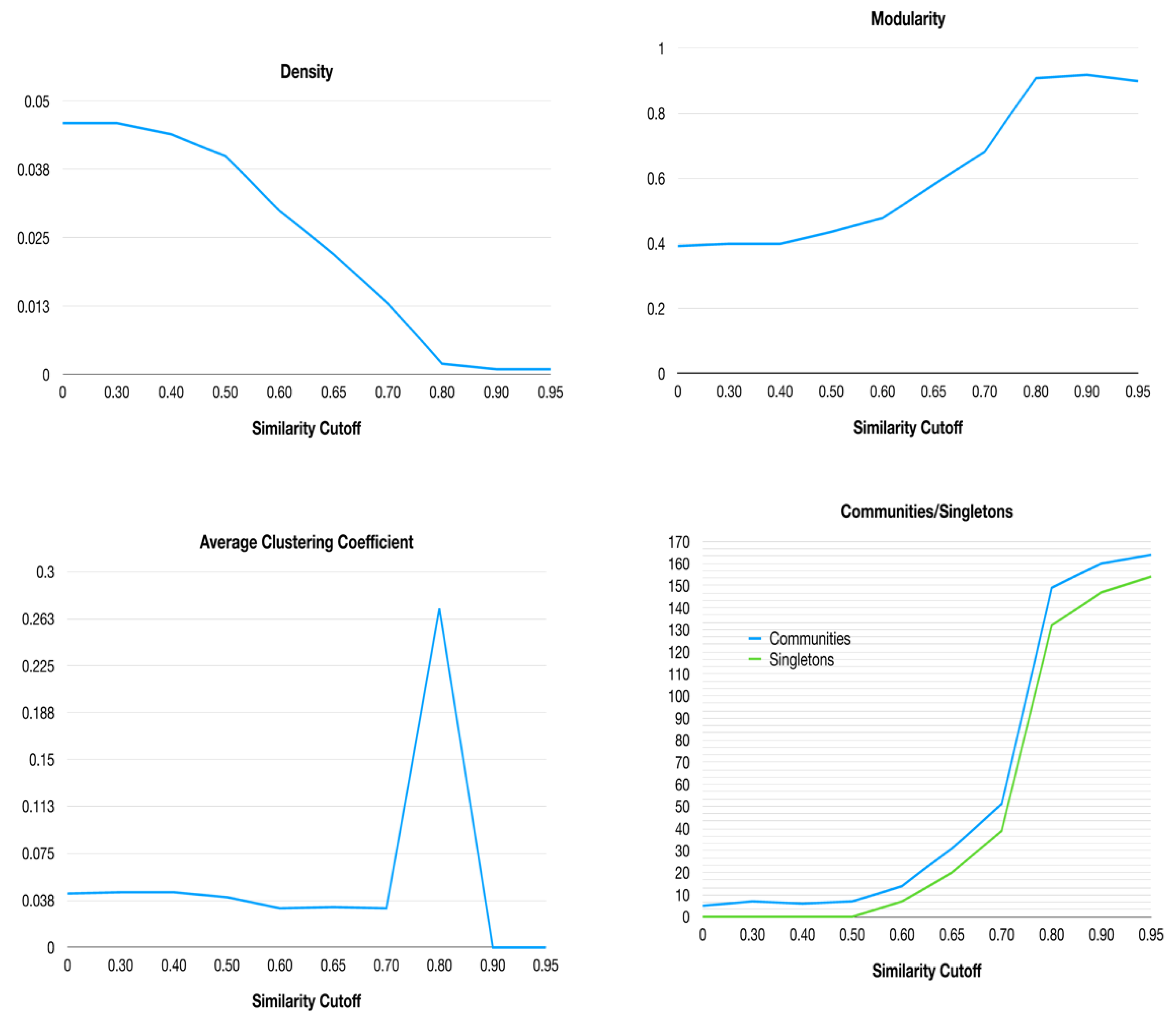
HSPNs of ABFPs were built with the Euclidean distance metric but changing similarity threshold from 0.0 to 0.95. As long the cutoff increases, the network density decreases due to the loss of edges satisfying the similarity cutoff. Therefore, several nodes get disconnected from the giant component of the network and appear as isolated communities or singletons representing atypical sequences. The original HSPNs (with no similarity cutoff) have the particularity that all nodes are fully connected like in the giant components of Half-Space Graphs, however at applying increased similarity cutoffs, increasingly sparser graphs with not connected nodes (singletons) are displayed.
Then, some network parameters were retrieved at different similarity cutoffs in order to determine the optimal one for determining the most informative network topology. In relevance order, network parameters such as density, modularity, average clustering coefficient and number of communities were analysed (Figure 1). The network density is the actual number of edges over the maximum number of possible edges in a network. If the density is too high, the understanding of network topological features gets complicated and by contrary it gets lose useful information; therefore, a compromise between both extremes is need. Generally, network density around 0.1 is acceptable, however as HSPNs show much lower density values (< 0.01) because their node connections are conditioned by a predefined proximal/adjacent space. At increasing the similarity cutoff, networks density tends to drop since only edges weighted with high similarity are retained.
The modularity of the networks was also analysed at each similarity cutoff. This is a network parameter that compares the density within a community with the expected one for the same group of nodes on a random network. We calculated modularity and the number of communities using the modularity optimization clustering algorithm (based on the Louvain method [
37]). Unlike the network density, both the modularity and the number of communities/singletons get significantly increased, especially at applying similarity cutoffs from 0.5 (
Figure 1).
On the other hand, the average clustering coefficient (ACC) is a global measure of nodes neighbourhood connectivity and can be also used for evaluating network topology changes against similarity thresholds. Although at similarity cutoff of 0.80 all network parameters displayed a dramatical change (
Figure 1), the optimal value was selected by jointly analysing all HSPN parameters (
File 1SM - Supplementary Materials). The point where the network density dramatically drops but at the same time the modularity up while taking care the trade-off between the number of communities/singletons generated is a good starting criterium for the selection. In this sense, the cutoff value should be between 0.6 – 0.7 where the density and modularity have an inverse behaviour while the ACC did not suffer any dramatical change and the number of communities and singletons are reasonable in order to display informative network topology (
Figure 1). Thus, the optimal cutoff of 0.65 was selected by analysing all HSPN parameters displayed in
Figure 1 and File 1SM.
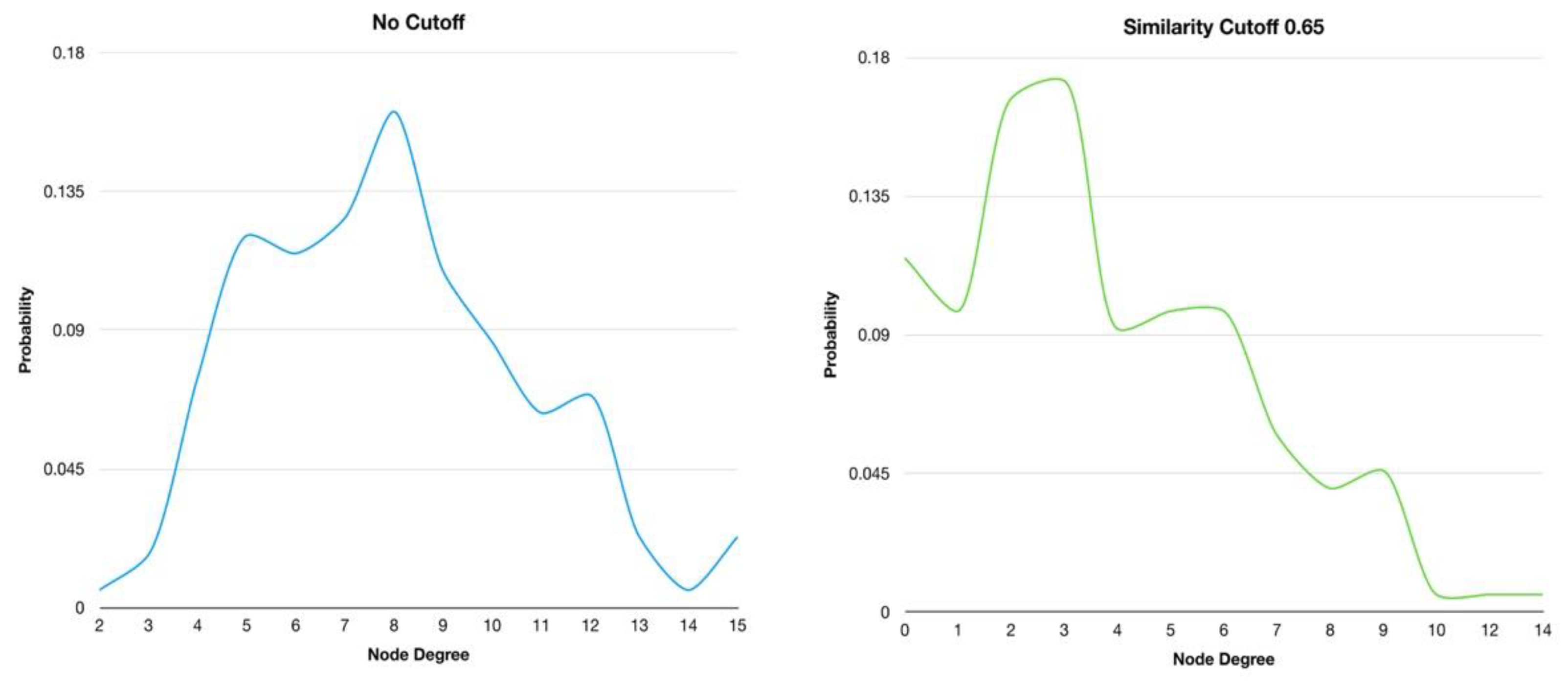
In addition, the degree distributions of the HSPN with no similarity cutoff and at 0.65 were plotted to explore the behaviour of these networks as generic models [
45]. The node degree distribution of HSPN with not cutoff shows a bell-shaped distribution (
Figure 2, File 1SM), revealing the random behaviour of the HSPNs, similarly to the random models. However, when the optimal similarity cutoff is applied the classical normal distribution get loses and several small bell-shape patterns appears along different node degree ranges. Therefore, the HSPN model has not an evident random behaviour which indicates that could be used as a topological network model (
Figure 2).
3.2. Network Visual Mining
3.2.1. Visual Mining of HSPNs, the Most Central and Atypical ABFPs
In addition to the numerical characterization of HSPNs by the calculation of network parameters, their visualization also provides new and simple insights to unravel the complex relationships of the objects they represent. In our case, they were used to represent and analyse the chemical space of 174 non-redundant ABFPs by applying AF similarity networks with the application of the half-proximal space graphs [
31,
33,
34]. Network visualization can mirror several network parameters like density, communities, nodes size can be ranked according to different centrality measures e.g. node degree, harmonic, hub-bridge, betweenness, etc. Thus, the most important or central peptides can be highlighted as well as the edges weighed with high or low similarities. This work is aimed to exploit at the top the visual representation of complex networks representing ABFPs for analysing their structural space and their associated metadata that are relevant for the discovery and design of antibiofilm agents [
28].
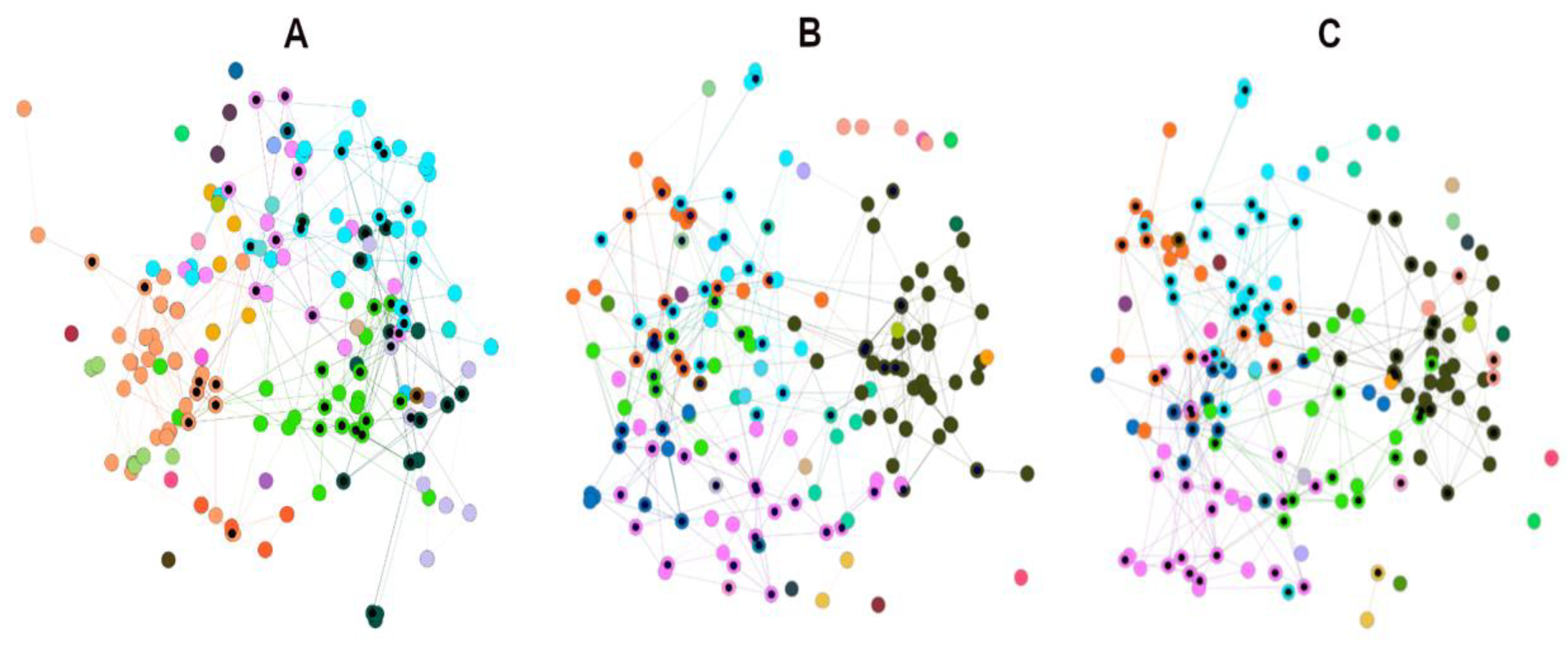
Both the original HSPN (no cutoff) and the HSPN model (cutoff at 0.65) representing the structural space occupied by the 174 ABFPs are depicted in Figure 3, and the File 2SM complement the network projection by numerical characterization. Networks communities are highlighted with different colours and nodes importance was represented by the node degree centrality. The original HSPN shows 5 communities clearly identified by different colour while in the HSPN model bearing 30 communities (20 singletons included), it is more difficult their delineation by colours (see details in File 2SM). However, since the HSPN model is a low-density network with a smaller number of edges (325) than the original HSPN (689) allows a better depiction of the node’s relationships (File 1SM).
Figure 3.
A – HSPNs visualization by using the Fruchterman-Reingold layout without similarity cutoff and B – with a similarity cutoff of 0.65. Peptides communities are represented by different colours while the nodes size was scaled according to node degree. .
Figure 3.
A – HSPNs visualization by using the Fruchterman-Reingold layout without similarity cutoff and B – with a similarity cutoff of 0.65. Peptides communities are represented by different colours while the nodes size was scaled according to node degree. .
The most relevant nodes according to the node degree were clearly identified in the HSPN with no cutoff (e.g. starPep_07526, starPep_02281, starPep_00048, starPep_03668, starPep_08958) whereas at applying the similarity cutoff a significant fraction of graph edges gets lost and the nodes degree decreases. That’s why only two of the most connected nodes were highlighted; the starPep_00048 and starPep_03668. As these two peptides also brought up in the original HSPN in similar locations and clusters, it would be figured it out that changes in the HSPN topology at applying similarity cutoffs do not alter the most popular peptides in both networks. In order to address this question, the top 10 most relevant peptides according to each four centrality measures (node degree, harmonic, betweenness and hub-bridge) were extracted from the HSPNs with and without similarity cutoff (File 2SM). Then, the intersection of the resulting four peptide sets was analysed for each HSPN. Table 1 displays the common peptides identified from the 10-top ranked by four, three and two centrality measures from HSPNs with and without similarity cutoff. The last four rows at each HSPN represent singular peptides identified for each of the four centralities. The cluster/community containing the 10-top ranked ABFPs is also displayed.
Table 1 shows a different composition of common and singular peptides from the 10-top ranked ABFPs by centrality measures at HSPNs with and without similarity cutoff. This observation supports that changes in the HSPN topology by removing edges (similarity relations) not only produce sparser networks with an increased number of clusters, but also lead to a variation of peptides centrality measures and therefore on its topological distribution. However, a small set made up of probably the most relevant peptides were identified for four and three centrality measures in both HSPNs. That are the cases of starPep_00048, starPep_03668, starPep_10922 and starPep_00000. These peptides seem to be very important within the ABF chemical space. The starPep_00048 is a human defensin derivative (HNP1) of 30 amino acid length with several reported interesting bioactivities (antiviral, anti-Gram+/ –, antifungal, anticancer, enzymatic inhibitor) besides its antibiofilm action which probably is exerted by its ability to disrupt membranes and interfere biological process involved in interspecies interaction [
46,
47]. The starPep_03668 and starPep_10922 are synthetic constructions of 35 and 12 aa length, respectively. The starPep_03668 was designed as pathogen-selective peptide, based on the fusion of a species-specific targeting peptide domain with a wide-spectrum antimicrobial peptide domain. Thus, it showed activity against several communities of
Streptococcus species [
48]. By contrast, starPep_10922 was designed as a D-enantiomeric peptide aimed to resist proteases degradation and also to prevent the accumulation of (p)ppGpp, which is a key messenger for biofilm formation. StarPep_10922 was able to prevent biofilm formation from
P. aeruginosa as well as to disperse and eradicate the bacteria in the resulting mature biofilm [
49]. So far, these three central peptides have not been reported as toxic for mammalian cells being lead peptides for developing ABFP drugs. The starPep_00000, a 26 aa length ABFP that was derived from Melittin (bee venom) show up as a promising candidate since several pharmacological activities has been assigned to it besides the antibiofilm one, and has also been extensively evaluated against many targets. However, starPep_00000 has also shown haemolytic activity and toxicity against to eukaryotic cells which may straightforward limit its therapeutic potential, unless its toxicity would be relieved by optimization procedures [
50].
The ABFP space is not only represented by the central peptides, there also exist disconnected peptides to the giant component of the network that bear low values of node degree. These peptides are categorized as atypical because they share very low sequence similarities with the central or popular ones, that prevent the estimation of their properties. However; they are remote members of the ABFP family and they also account for the antibiofilm chemical space. In this sense the HSPN with similarity cutoff is very useful to uncover atypical peptides. The cutoff at 0.65 of AF similarity was the optimal to retain a reasonable trade-off between the number of communities and singletons; where the simplest community is considered when 2 nodes (peptides) are connected, and singletons are those that are not connected with any other in the network. Atypical peptides represent singular structures that may represent privilege scaffolds for designing antibiofilm agents.
Since the HSPN with no cutoff is fully connected, no isolated communities and singletons can be identified.
Table 2 displays the atypical peptides from the HSPN at similarity cutoff of 0.65, the 20 singletons where all centrality measures reached 0 value and two isolated communities made up of 2 peptides interconnected with node degree 1.
The atypical peptides could have been similarly analysed than the central peptides, but as all singletons displayed 0 values for all centrality measures; then the analysis was carried out by exploring the metadata associated to each peptide by using the StarPep toolbox. Atypical peptides with no reported toxicity to mammalian cells are marked with an asterisk while those that also have a diversity of desired functions contributing to the antibiotic/antibiofilm activity were highlighted in bold. That’s the case of starPep_04044 which is a synthetic peptide of 13 aa length representing a singular structure that can coat successfully titanium surfaces and also can target Gram+ and Gram- bacterial strains in both their planktonic and biofilm forms allowing its utilization for preventing infection-related implant failures in dentistry and orthopaedics. It has been effective on
Pseudomonas aeruginosa,
Streptococcus gordonii, Porphyromonas gingivalis,
Staphylococcus aureus and
Escherichia coli [
51,
52].
3.2.2. Metadata Analysis by Visual Mining
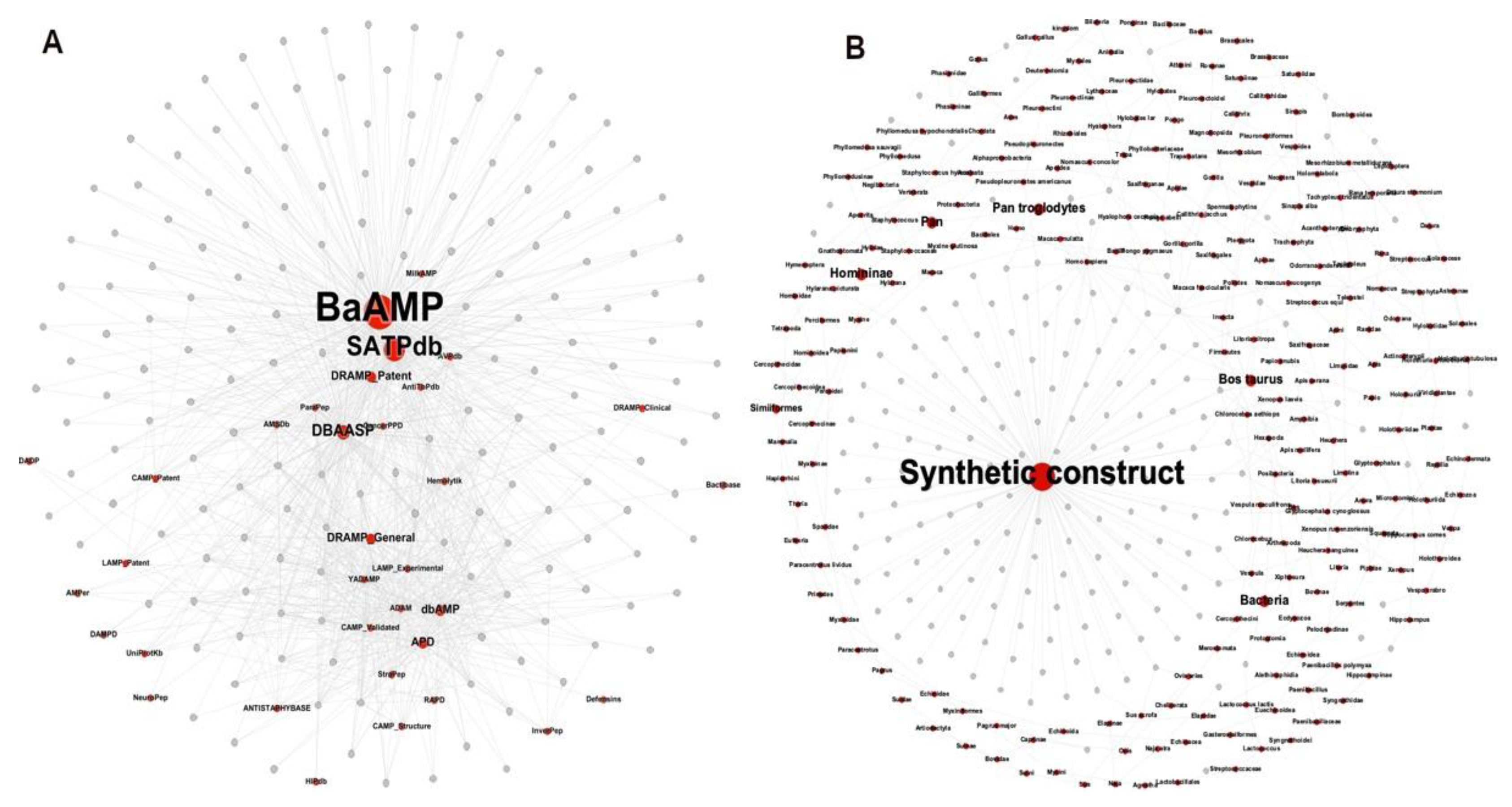
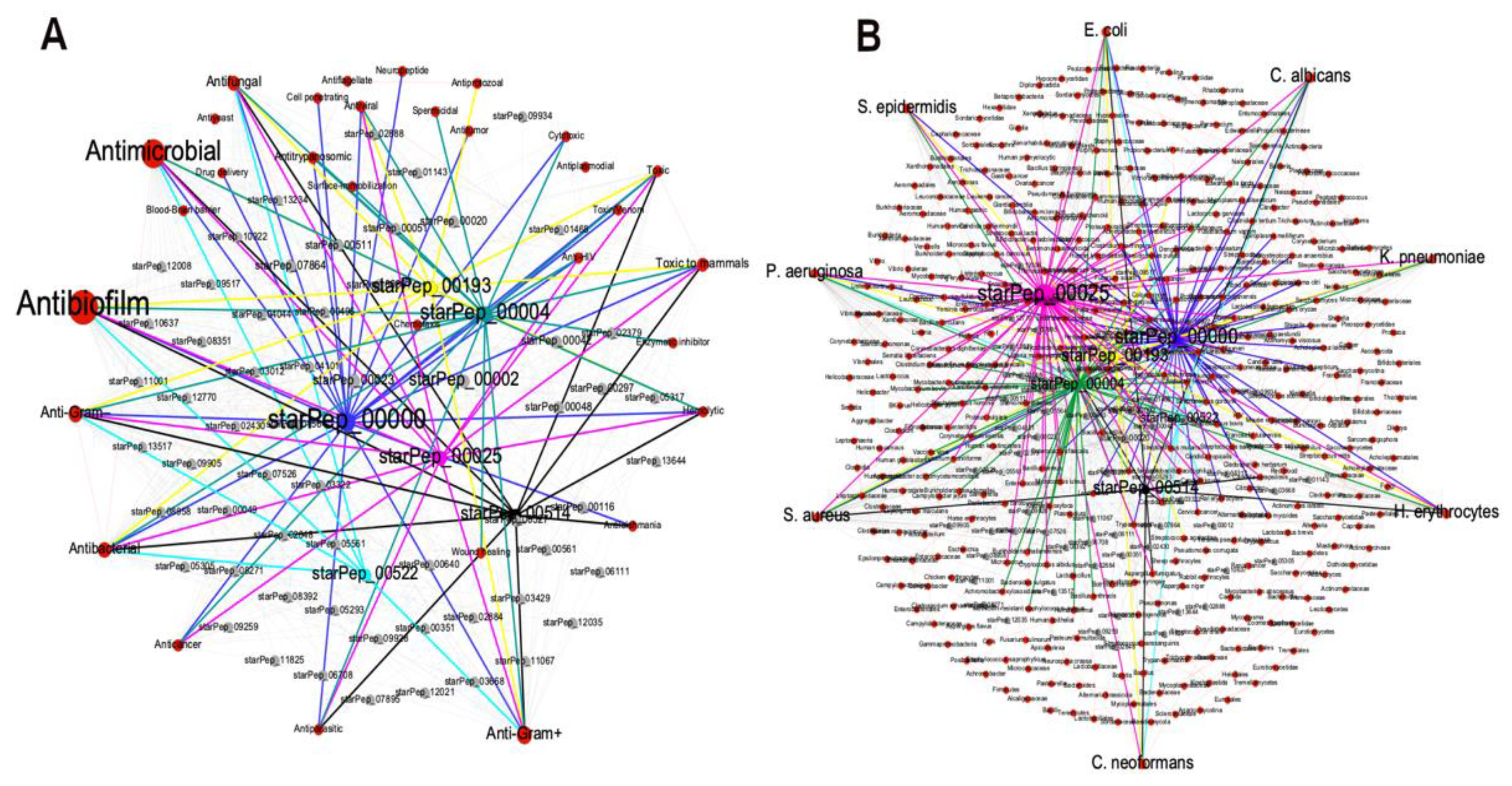
The METNs corresponding to the 174 ABFPs were constructed considering their source database and origin. Similar to the previous visualization, nodes were displayed by colors and size. Red nodes represent source databases and origin, respectively while the ABFPs were in grey (Figure 4A and 4B). Nodes size were scaled according to their betweenness centrality values, which is based on the shortest path between two nodes. Thus, this type of centrality is the number of shortest paths that pass through a target node, particularly on red nodes representing “database” and “origin”.
As shown in
Figure 4A, the 174 ABFPs registered in StarPepDB were mainly collected from BaAMP [
24], the Structurally Annotated Therapeutic database (SATPdb) [
53] and Da-tabase of Antimicrobial Activity and Structure of Peptides (DBAASP) [
54] which are represented by larger nodes with the highest connection with the ABFPs. While BaAMP and DBAASP entries were carefully collected from the literature, the SATPdb was built from 22 peptide databases that included BaAMP [
24], and others similarly dedicated to specific activities such as AVPdb (antiviral) [
55], ParaPep (anti parasitic) [
56], Hemolytik (hemolytic) [
57], CancerPPD (anticancer) [
58], etc; and also from generic AMP databases like DAMPD [
59], APD [
22], CAMP [
60], LAMP [
61], DRAMP [
62], etc. Such databases that integrated SATPdb were represented as smaller red nodes sharing less edges with the ABFPs.
Most of the ABFPs come from synthetic constructs, represented by the largest node or hub in the network (
Figure 4B). However, other natural sources have also provided ABFP scaffolds for further modification/optimization. Among the most contributing taxonomic groups are the Bacteria, Homininae, Similiformes, Pan and Bos (Bos Taurus). Therefore, this information can guide the discovery and design of antibiofilm agents from peptides. Particularly inform on what are the most relevant ABFPs databases and the main sources/origin where to find promising ABFP scaffolds.
3.3. Representing the ABFPs with a reduced subset
3.3.1. The selection of the best representative subset
The original space of 174 ABFPs, illustrated by the HSPN model at the optimal cutoff of 0.65 (Figure 3B), can be further simplified by applying the scaffold extraction algorithm implemented in the StarPep toolbox. This algorithm allows the topological simplification of the network by removing nodes with equal or similar values of centrality measures but retaining those that still share a local similarity below certain cutoff. As the reduction of ABFPs intends to keep the HSPN topology and properties, it is applied to all type of nodes, from the most central to the atypical ones (singletons). The reduction was performed by ranking the harmonic and hub-bridge centrality values of the nodes and by applying similarity cutoffs from relaxed criteria (retain all peptides sharing < 0.9) to more restrictive similarities (< 0.45 of sequence identity). This step produces several subsets for each centrality metrics (Table 3, File 3SM).
As mentioned before, the best subset representing the original space draw by the HSPN model should be composed by the minimum number of ABPFs with a coverage of the original space < 50%. However, the coverage is not the unique criterium to select a representative subset of the original space, its distribution on the bi-dimensional (2D) space occupied by the HSPN model should also be considered. An effective subset should have a topological representativeness over all the network, representing connected, isolated communities and even singletons. In this sense, a subset extracted under the criterium of only one centrality measure might not fulfil the expected 2D coverage. Thus, we decide to fuse the information of the HC and HB centralities since they rank the network nodes according to their position using different definitions. Thus, the union of the subsets 6, 7 and 8 which are highlighted in bold in Table 3 was evaluated. Subsets 6, 7 and 8 display a coverage <50% with a low of number of ABFPs, but promising for their union (HC ∪ HB) at the same cutoff value. It is noteworthy to say the union between two subsets include the common peptides (intersection) and the singular peptides from each subset. The union of subsets 6, 7 and 8 resulted in fasta files (File 3SM) containing 85, 66 and 52 ABFPs representing 49%, 38% and 30% of the HSPN model, respectively. The File 3SM also contain the 221 ABFPs registered in StarPepDB and the 174 used to generate the HSPNs.
Finally, the subsets 6, 7 and 8 from each centrality metric as well as their resulting fusion (HC ∪ HB) were overlapped on the HSPN model to evaluate their 2D coverage/spatial distribution (
Figure 1SM). In the three subsets, the union of the centrality measures (HC ∪ HB) at the evaluated similarity cutoffs displayed a better 2D spatial distribution on the HSPN model. Particularly, the HC ∪ HB from subset 7 showed the best trade-off considering the lowest number of peptides with the best 2D coverage (
Figure 1SM).
Figure 5 summarizes the overlapping of the HC ∪ HB from subset 6, 7 and 8 with the HSPN model. The overlapped subsets are represented by small black nodes over the coloured nodes that represents the communities in the HSPN model.
3.3.2. Visualizing/Analysing the best representative subset with HSPNs
The main goals of extracting a reduced subset representing the complex networks is to allow retrieving useful information from the visual inspection the networks. With reduced number of nodes and edges complex networks turn more legible for the human eyes and the representativeness of the subset result also useful for multi-reference similarity searches against unlabelled peptides. The
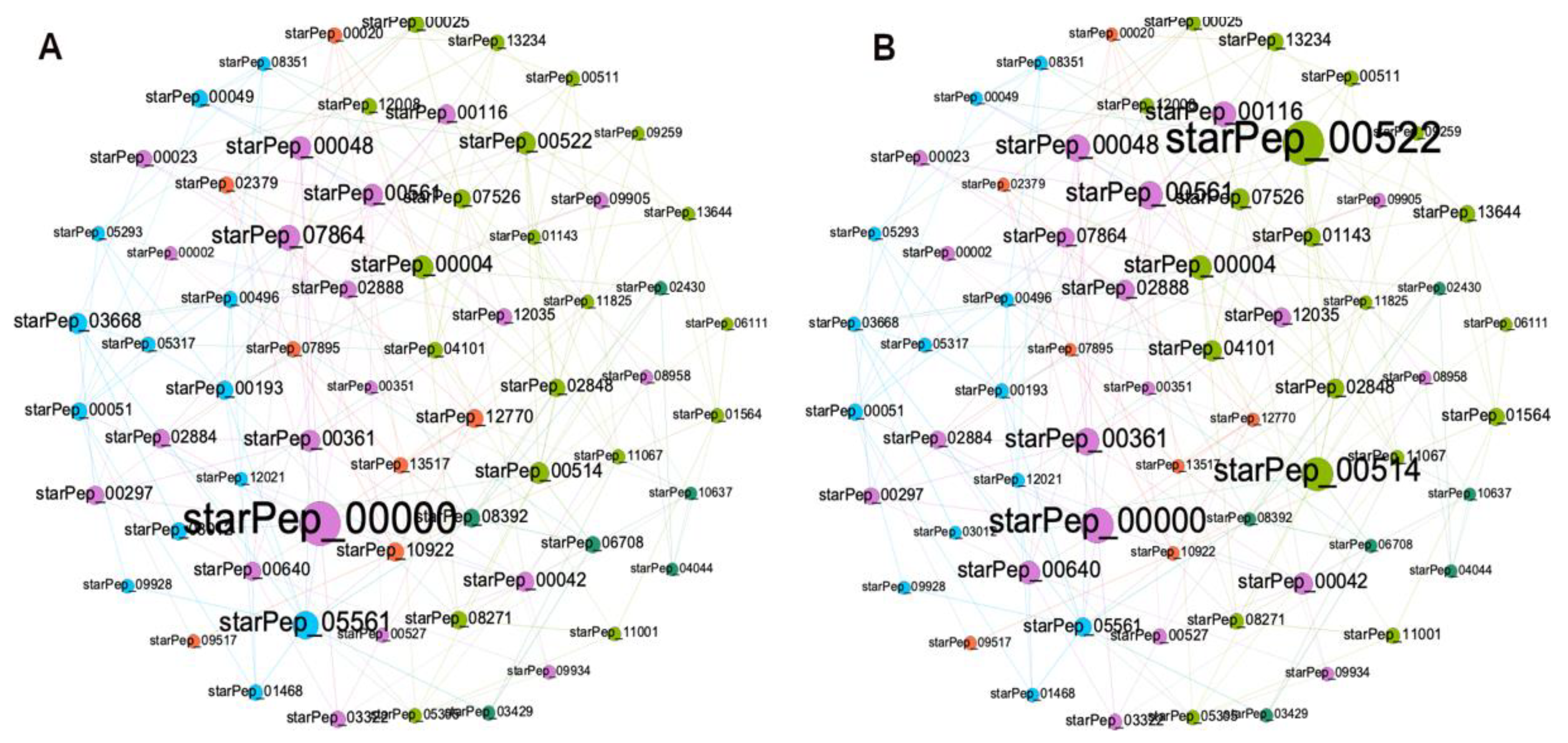
Figure 6 show the HSPN constructed from the reduced space made up of 66 ABFPs resulted from the HC ∪ HB of the subset 7 (cutoff 0.40). As can be seen in
Figure 6A and 6B, most of the main ABFPs identified in the HSPN model were transferred to the reduced space such the cases of starPep_00000, starPep_00042, starPep_00048, starPep_03668, starPep_05561 and starPep_00004, that additionally were among the 10-top relevant ABFPs by the HC and HB centralities (
Table 1). Its noteworthy that other ABFPs, not identified among the top-ranked in the HSPN model, brought up significantly when constructing the HSPN with the 66 representatives e.g. starPep_00522 and starPep_00514 (
Figure 6A and 6B; File 4SM).
On the other hand, 5 out of the 20 singlentons appeared in the representative subset. They are the following ones starPep_00002, starPep_04044, starPep_05305, starPep_09934, starPep_10637, being all reported as non-toxic except starPep_00002. It was not by chance that the priviliged scaffold of starPep_04044 was also selected, which indicate the scaffold extraction algorithm implemented in StarPep toolbox works.
The subset of 66 representative ABFPs extrated from the HSPN model is described in File 4SM that includes their IDs, sequences, lengths, the cluster where they belonged to in the HSPN model, their centralities measures both in the HSPN model and in the new HSPN constructed (Figure 6). This non-redundat subset of 66 peptides represents the chemical space of the ABFPs, and it is advisable to be used as reference to map new ABFP sequences, also in multi-references similarity searches against unlabel peptide datasets and for design purposes.
3.3.3. Visualizing Mining of the METNs
The reduced subset is also useful for unravelling information from METNs which are even more complex than similarity networks since they include other layers containing additional nodes with associated peptide metadata. Consequently, counting on a representative subset of the original space aids to analyse the huge amount of information associated to the ABFPs. As previosly-mentioned microbial biofilms are responsible for most of chornic and medical device-related infetions as well as for the microbial resistant to several antibiotic classes, thus, the identification of promising ABFP scaffolds is an urgent task.
As an illustrative example of the METNs contribution to the identification of promising ABFPs scaffolds, six relevant ABFPs according to the HC and HB centralities were selected from the representative subset (Figure 6) for visual analysis of their metadata. This time key metadata for the development/design of ABF agents from ABFPs were chosen, e.g. other associated activities of the ABFPs and the targets what they have been evaluated on (Figure 7). The study cases were starPep_00000 (blue), starPep_00193 (yellow), starPep_00004 (green), starPep_00025 (pink), starPep_00514 (black), starPep_00522 (cyan).
The
Figure 7A is organized in such way that desired activities were placed outside at the left part of the METN while the undesired ones at the right. The six candidates under study, in addition to the antibiofilm activity, have also the antifungal and the antibacterial, specifically against Gram-positive and Gram-negative strains which are very convinient for developing next-generation antimicrobial agents able to target both planktonic and biofilm microbial forms. However 4 of them were reported as hemolitycs and 5 were “toxic to mammals”. The starPep_00522 (in cyan) is the only one that was neither reported as “haemolytic” nor “toxic to mammals”and therefore its peptidic scaffold can be used for developing antibiofilm agents for clinical purposes. Addtionally, the
Figure 7B also shows that starPep_00522 has been evaluated in Escherichia. Coli, Pseudomonas aeruginosa, Candida albicans, Cryptococcus neoformans where the two first targets classified within the ESKAPEE pathogens, considered the most threatening antimicrobial resistant microbes [
63]. This visual analysis of the METNs can be extended to all 66 ABFPs of the representative set.
3.4. External Representative ABFPs on the representative Antibiofilm HSPN
In a recent work, Li at al. arrived to 14 representative ABFPs out of a total of 51 peptides with a reported antibiofilm activity either by inhibition of the biofilm formation or by the eradication of pre-formed biofilms. The selection was based on the identification of 14 ABFP classes according to their mechanisms of action. They also evaluated them against the biofilm and planktonic forms of Gram-positive bacterium Streptococcus mutans, the Gram-negative bacterium Pseudomonas aeruginosa, and the fungus Candida albicans. Those ABFPs with MBICs (minimal biofilm inhibitory concentrations) that are lower than their minimal inhibitory concentration (MICs) (minimal inhibitory concentrations) represented promising candidates against biofilm-related infections.
Table 4 shows the 14 representative candidates categorized by antibiofilm mechanisms of action (Information taken from
Table S2 published in [
64].
This set made up of 14 ABFP classes representing different mechanism of action to carry out the antibiofilm activity was mapped on the structural/chemical space of the 66 ABFPs, drawn by the HSPN model (Figure 8A). As the subset of 66 ABFPs showed the best representativeness of the antibiofilm chemical space was used along with the most suitable HSPN projection for the overlapping purpose. The HSPN that plots nodes coordinates from their 2 most relevant principal components, estimated from a non-redundant set of AF descriptors, is the most real approach to display the peptide’s location in the network, allowing a more accurate visual inspection of the similarity and distribution of the 14 representative ABFPs on the reported chemical space (Figure 8A).
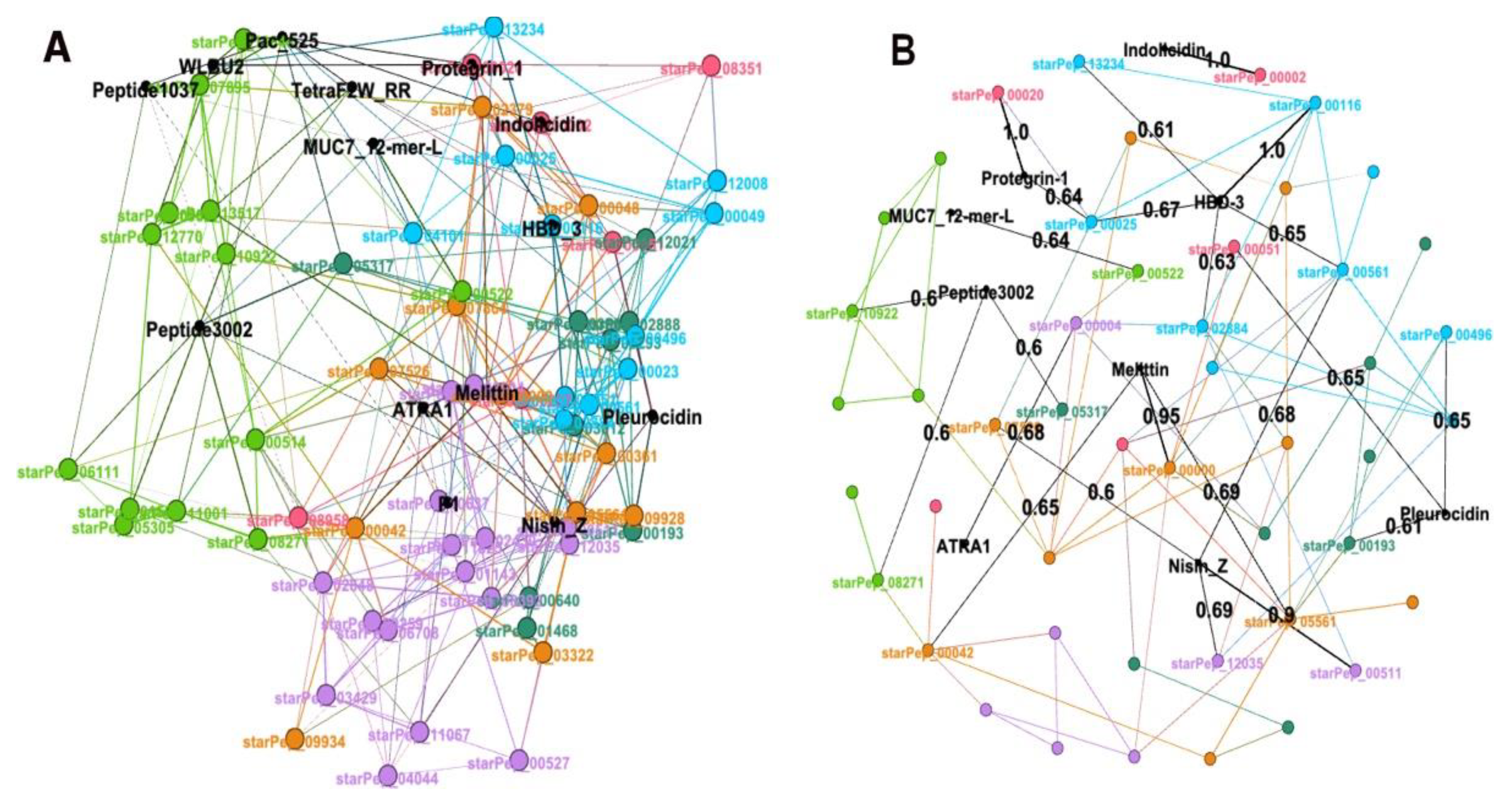
Figure 8A shows the node and names corresponding to 14 ABFPs in black colour while the other 66 from StarPepDB were labelled according to the colours assigned to each one of the 5 network communities. As can be observed in
Figure 8A, all 14 ABFPs were framed within the antibiofilm HSPN. In fact, the Indolicidin, Protegrin-1 and HBD-3 overlaped perfectly with starPep_00002, starPep_00020 and starPep_00116, respectively. The 14 mechanisms of action classes were distributed among all the 5 HSPN communities, which may indicate a connection between structural patterns (motifs) found within network communities with the antibiofilm mode of action. In order to illustrate this fact, 6 ABFPs that showed antibiofilm activity against both bacteria and fungi (pleurocidin, Pac-525, protegrin-1, TetraF2W-RR, WLBU2, and melittin) overlaped perfeclty or nearly over different commnunities. Pleurocidin, Pac-525, protegrin-1 and melittin are evidently overlaped on the communities coloured in blue, green, pink and light purple. WLBU2 was also placed in the green community as the Pac-525, probably because both act using similar mechanisms of action involving the interaction with the lipopolysaccharides to destroy or penetrate the bacterila membrane. Although, TetraF2W-RR is within the antibifilm space represented by the HSPN, it did not overlap on any specific community. However, it was placed between green and pink communities containing members such as Pac-525, WLBU2, Indolicidin and protegrin-1 which mode of action are closely related to the reported for TetraF2W-RR, bacterial membrane disruption. These 4 ABFPs are ariginine (R)-rich peptides containing repeated units of R allowing the interaction with negatively-charged bacterial membranes, the formation of transmembrane pores and cell penetration [
65].
The
Figure 8B complements the information extracted from the
Figure 8A. It shows those representative ABFPs sharing AF similarities > 0.60 with some of the 66 ABFPs extracted from StarPepDB. Black nodes represent the 9 out of the 14 representative ABFPs that fulfill this condition while the black edges display the similarity relationships from black nodes (origin) to coloured nodes (target). Target nodes labelled as starPep_XXXXX retained the same colour identifying them at the network communities in
Figure 8A. The
Figure 8B confirms that Indolicidin, Protegrin-1 and HBD-3 share the max. similarity (1.0) with those ABFPs that overlapped with starPep_00002, starPep_00020 and starPep_00116 in
Figure 8A. The location of pleurocidin and protegrin-1 was also supported by their highest similarities (0.65 and 1.0) with members of the communities blue (starPep_00496) and pink (starPep_00020), respectively. However, as the pleurocidin shows multiple actions such as membrane disturbance and permeabilization, binding to bacterial DNA and interference with several cellular functions, also display similiarities with members (starPep_00193, starPep_00051) from other 2 communities (
Figure 8B).
On the other hand,
Figure 8B also served to correct the location of melittin that is actually overlapped over the orange community showing 0.95 of similarity to starPep_00000 (a Melittin derivative), despite in
Figure 8A looked over the light purple community. Although the 14 ABFPs could be mapped within the HSPN, the
Figure 8B confirmed certain singularity of Pac-525, peptide 1037, TetraF2W-RR, P1 and WLBU2 within the representative ABFP space, at not sharing AF similarities higher than 60%. Such singularity was also confirmed by evaluating the pairwise identity of these last 5 ABFPs against the 66 representative ones (
Figue 9B). The 9 ABFPs that were clearly mapped at AF similarities > 0.60 were also compared by pairwise global alignments (
Figure 9A)
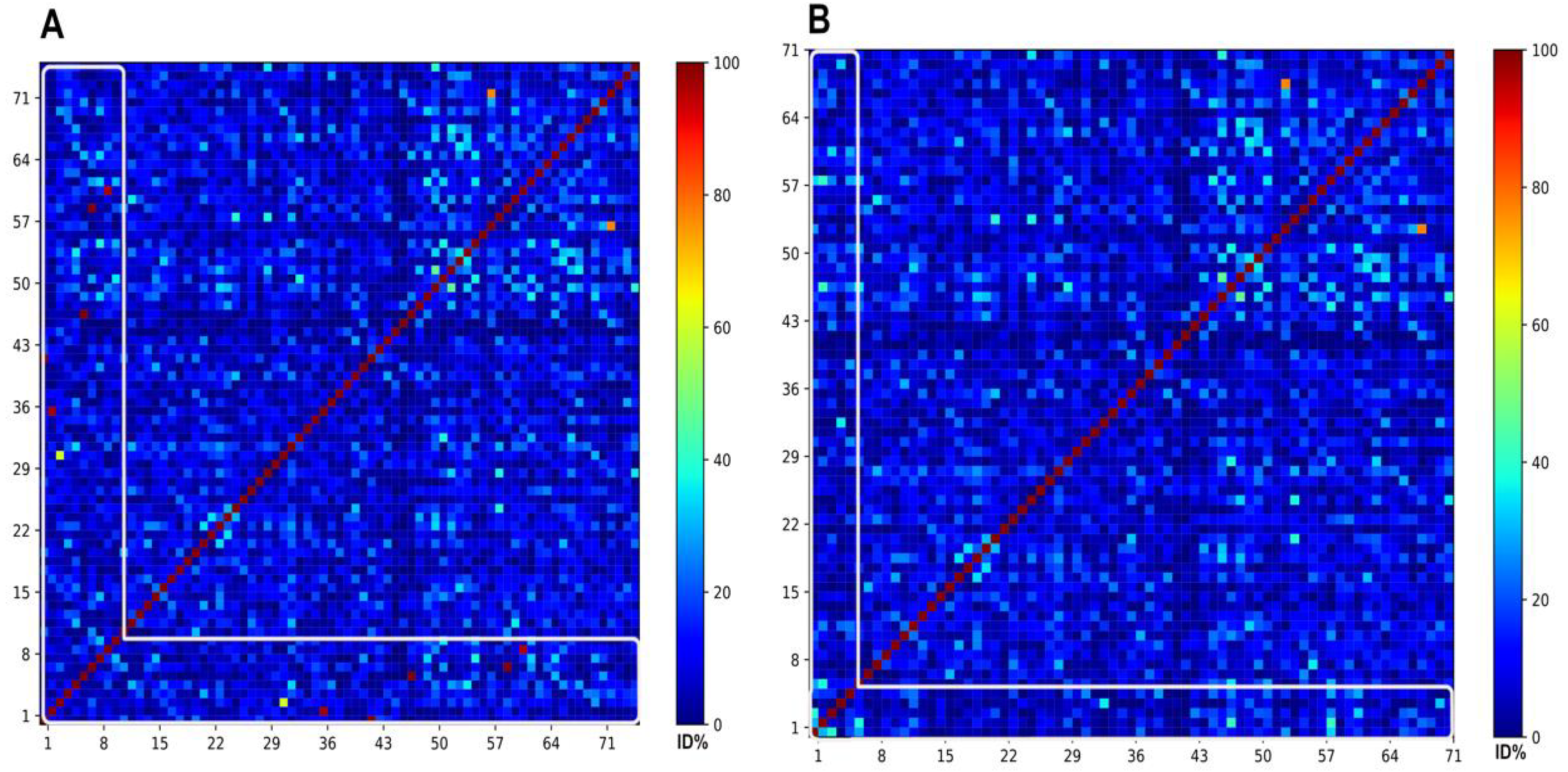
The inferior part of
Figure 9A, framed by the white line, display 5 red dots that corresponds to those ABFPs (Indolicidin, Protegrin-1, HBD-3, Melittin and Nisin) sharing network edges weighted with AF similarities higher than 0.90, the edge weighted with 0.69 is likely represented by the yellow dot and the remaining slightly superior to to 0.60 are depicted in cyan colours. While the
Figure 9B confirmed that the similarities shared by the all 5 unmapped ABFPs were actually below 0.60. All dots were moslty coloured in blue and some fews in cyan may represent the values close but below to 0.60.
It is important to say, as the AF and AB similarities are defined under different methodological frameworks, they may characterize the same pairwise relation with different values, despite they are correlated. The file 5SM shows the pariwise identities values of 9 and 5 ABFPs from the 14 mode of action classes against the 66 representing the antibiofilm chemical space.
3.5. Motif Discovery Assisted by Complex Networks
The identification of motifs accounting for the antibiofilm activity can be assisted by the exploration of ABFP similarity networks looking for sequence patterns within network communities. Although, the HSPN representing the ABFP chemical space was built using an AF distance metrics (Euclidean) and the network communities are estimated considering parameters from the nodes and edges properties [
37]; such clusters should contain peptides sharing similar features. Thus, the communities from the HSPN model of the 174 ABFPs resulted the source for the motif discovery. The sequence diversity at each community was evaluated by global alignments. The
Figure 2SM displays the heatmaps that mirrors the pairwise sequence identities for communities containing more than 2 peptides that correspond to the clusters 4, 7, 9, 11, 14, 15, 17, 22 including singletons clustered (
File 3SM). The
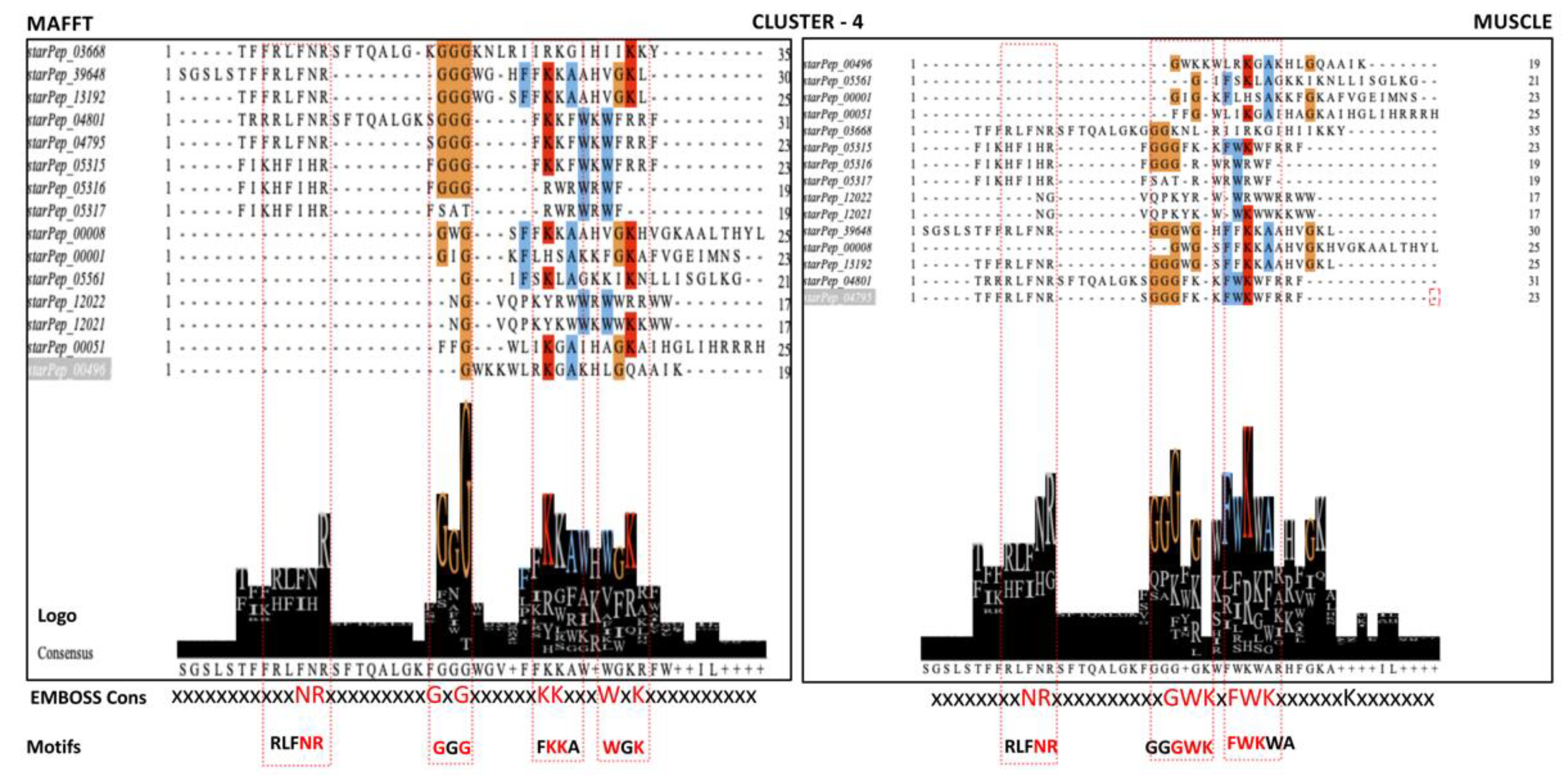
Figure 2SM evidenced a high sequence diversity within all communities. Consequently, iterative alignment algorithms like MAFFT and MUSCLE were applied to deal with the high sequence diversity. The multiple sequence alignments (MSAs) were visualized with the Jalview which allowed the estimation of their corresponding consensus sequences and Seq2Logos. The consensus sequences from the MSAs were also estimated by the EMBOSS Cons. The full exploration of the MSAs considering their corresponding consensus sequence and Seq2Logos allowed the identification of conserved regions considered as ABF motifs. The strategy carried out for the identification of the motifs in the MSAs performed on cluster 4 of the HSPN is illustrated in
Figure 9, while for all communities/clusters is displayed in
Figure 3SM.
Figure 9.
Motifs detection by the multiple sequence alignment (MSA) algorithms MAFFT and MUSCLE on the network cluster 4. The MSAs are visualized with the Jalview program which also estimates a Seq2Logo and the consensus from the alignment positions. Another consensus sequence that served as a guide for motif location was estimated by the EMBOSS Con.
Figure 9.
Motifs detection by the multiple sequence alignment (MSA) algorithms MAFFT and MUSCLE on the network cluster 4. The MSAs are visualized with the Jalview program which also estimates a Seq2Logo and the consensus from the alignment positions. Another consensus sequence that served as a guide for motif location was estimated by the EMBOSS Con.
Table 5 listed ABFP motifs identified by each MSA method at each network community or cluster. The consensus estimated by the EMBOSS Cons was the preferred template for motif identification because it gives a more legible output. High scored amino acids/positions are represented by capital letters, less scored but positive residues by lower-case letters while non-consensus positions by x (
Table 5).
As part of the motif discovery process, the alignment-based search was complemented by evaluating an AF approach. Based on its high performance and versatility to identify motifs in OMICs data, the STREME algorithm was applied to find unaligned patterns ranging from 3–5 aa length at each network community [
43]. STREME computes a score for the detected motifs meeting the statistical significance (
p-value threshold < 0.05); set also as a stopping search criterium.
Table 6 displays the discriminating motifs against control sequences at each ABFP cluster/community. Motifs appearing in more than 20% of the query peptides are listed according to their statistical significance (score).
Motifs highlighted in bold in
Table 5 and
Table 6 indicates they are closely related or included into each other. That’s means that both MSA algorithms and STREME showed some degree of agreement in the motif detection discovery. However, both approaches also identify singular motifs, not highlighted. This fact demonstrates that the application of AB and AF approaches was a right choice for a full motif exploration. Given that both methods identified a relative high number of motifs between 33 – 35, enrichment analyses were further performed in order to filter the discovery motifs shown in
Table 5 and
Table 6.
Motif enrichment analyses are used to determine if a group of sequences contains a statistically significant number of matches to a given motif. In this sense, we used the SEA algorithm [
44] to select what motifs from both tables significantly were enriched in two sets of ABFPs. The first set was the reported by Li
et al. consisting in 14 representative ABFPs of the antibiofilm modes of action [
64], and the second one encompassed 192 non-redundant ABFPs, extracted from the 214 ABFPs registered in BaAMP database [
24] (
File 3SM). Eight members from the representative subset were included among the 192 ABFPs, but all them have showed antibiofilm activity at different levels. As a screening criterium, ABFPs enriched in both the representative and the extended dataset were selected.
Table 7 list the discovered ABFP motifs by the AB and AF approaches at the network communities.
Our motif search approach assisted by the complex networks are not so far from the few findings reported in the literature. Recently, Anastasiu
et al. found that the following motifs “RIRV,” “RIVQRIK,” and “IGKEFKR” appeared with more frequency in 242 ABFPs collected from APD and BaAMP databases in respect to a curated negative set [
28], when using the “MERCI” software [
67]. In this sense, we agree with them in the detection of the “RIRV” which was fully integrated in the RIRVR motif detected in cluster 14 by the MAFFT algorithm, and also enriched in the BaAMP dataset. Although “RIVQRIK” and “IGKEFKR” were not detected as such, we could identify in clusters 15 and among the singletons by the MSA methods, the “RIV” and “FIK” patterns, which are part of them. These two last three-amino acid motifs were also enriched in the extended dataset.
In this previous report, authors also found that the dipeptides “IR/RI”, “WR/RW”, and “KK” were the most common among the selected ABFPs [
67]. Certainly, these dipeptides are present in the motifs discovered with the intervention of complex networks, at relatively high frequency. For example, the “IR/RI”, “WR/RW” and “KK” dipeptides appears in 9/7, 7/8 and 12 of the total motifs, respectively. In addition to them, also the “RR” and “KL” dipeptides displayed a similar representation among the motifs.
In a recent report arginine-rich motifs for the antibiofilm activity from peptides designed for sequestering the nucleotide second messenger c-di-GMP, involved in the formation of
P. aeruginosa and
K. pneumoniae biofilms were revisited [
68]. The key role of the DRR and [RK]RxxD motifs from these sequestering peptides (SP) to specifically bind to c-di-GMP was demonstrated by nuclear magnetic resonance (NMR)-based experiments [
69]. These motifs associated to SPs are difficult to discover by bioinformatics methods since their sourcing peptides are probably still not registered or underrepresented in databases. However, the peptide R4F4 (RRRRFFFF), with a proven antibiofilm activity on
P. aeruginosa through c-di-GMP sequestration [
68,
70] bears a more frequent motif (RRRR) among the arginine-rich ABFPs. In fact, “RRRR” was detected in our complex network-assisted motif search (
Table 5 and 6).
On the other hand, despite the role of WWW motif for disrupting preformed biofilms from methicillin-resistant
Staphylococcus aureus was unravelled by NMR and arginine scan experiments in 2017 [
71], the WWW motif hardly appears in ABFP databases, being only represented by the designed peptide TetraF2W-RR [
72].
Therefore, as the computational motif discovery is highly influenced by the peptide database composition and by the searching algorithm, here we provide new ABFP motifs discovered from combining network science with AB- and AF-based computational tools for motif detection. The motifs listed in 5, 6 and specially in
Table 7 are useful for the “
in silico” generation of peptide libraries addressed to the antibiofilm activity, as well as for the optimization of antibiofilm candidates. Finally, predicted motifs that actually account/improve the antibiofilm activity could be also used as motif-based descriptors for developing machine-learning models to screen peptide libraries and peptidomes as part of the discovery process.