Submitted:
14 March 2023
Posted:
15 March 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
1.1. Related Work
2. Results
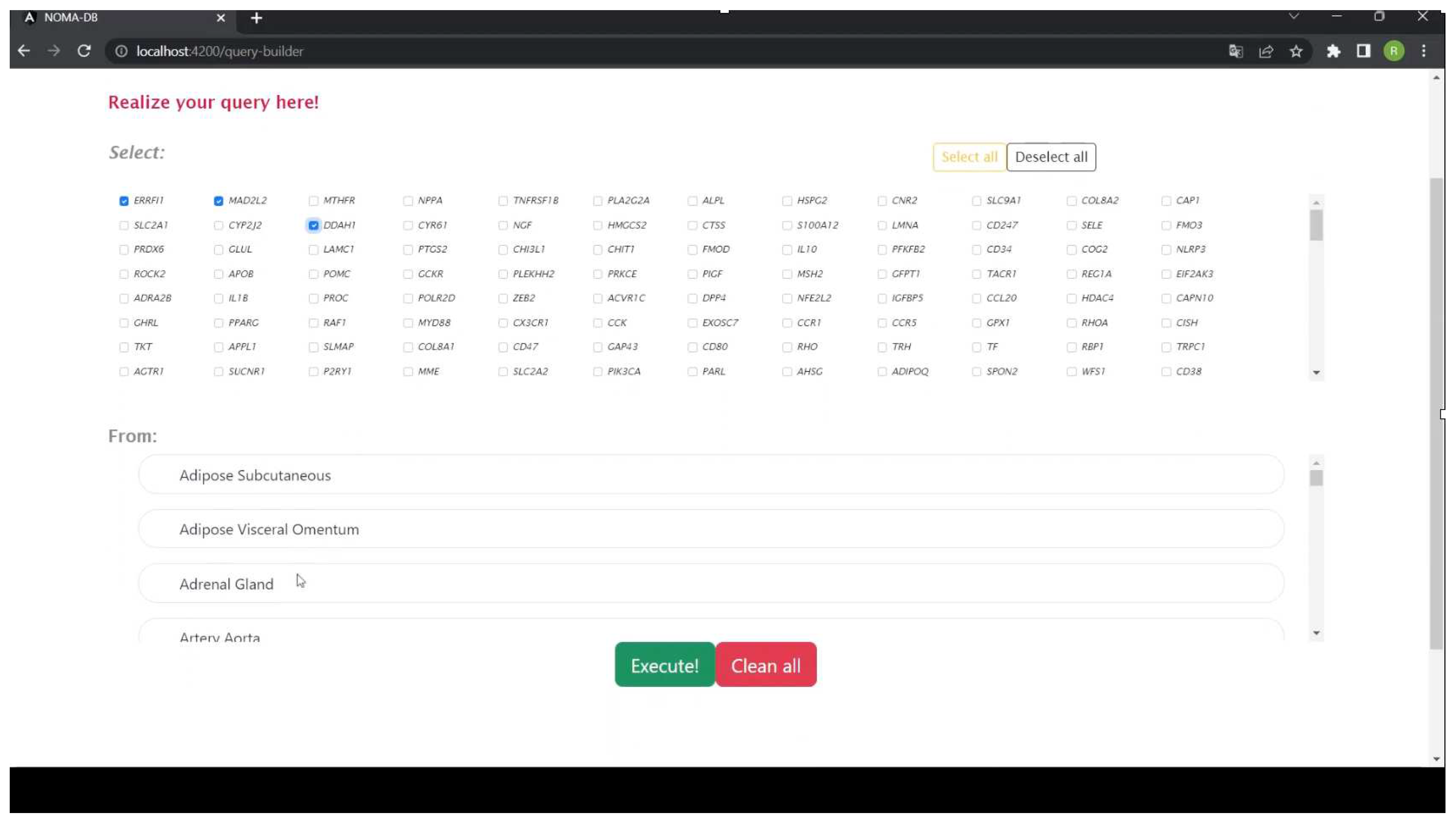
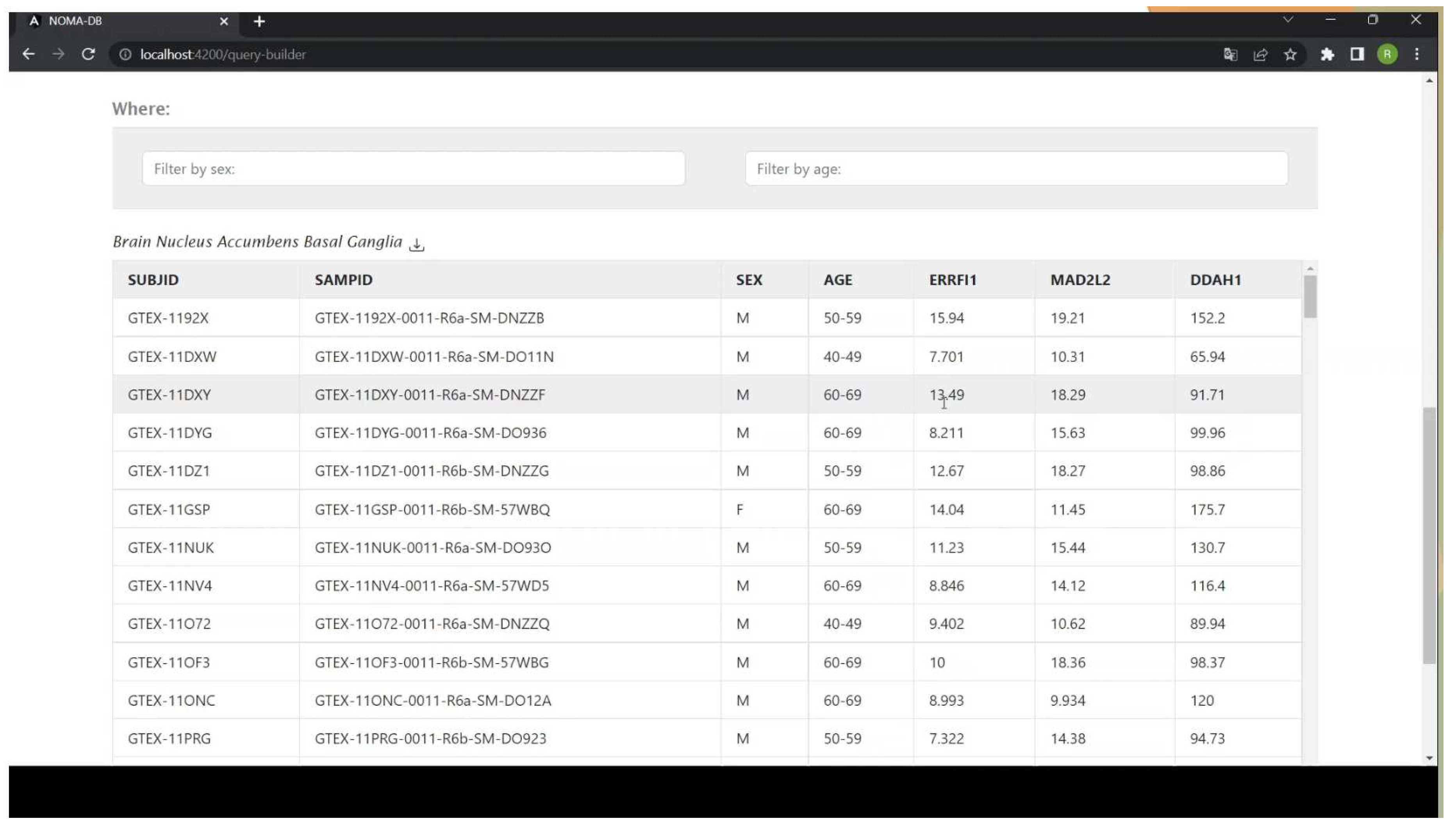
2.1. Using NOMA-DB for Studying Gene Changes with Age
3. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Acknowledgments
Conflicts of Interest
References
- Bond, K.M.; McCarthy, M.M.; Rubin, J.B.; Swanson, K.R. Molecular omics resources should require sex annotation: a call for action. Nature methods 2021, 18, 585–588. [Google Scholar] [CrossRef]
- Partridge, L. Intervening in ageing to prevent the diseases of ageing. Trends in Endocrinology & Metabolism 2014, 25, 555–557. [Google Scholar]
- Childs, B.G.; Gluscevic, M.; Baker, D.J.; Laberge, R.M.; Marquess, D.; Dananberg, J.; Van Deursen, J.M. Senescent cells: an emerging target for diseases of ageing. Nature reviews Drug discovery 2017, 16, 718–735. [Google Scholar] [CrossRef] [PubMed]
- Rockwood, K.; Howlett, S.E. Age-related deficit accumulation and the diseases of ageing. Mechanisms of ageing and development 2019, 180, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Leonardi, G.C.; Accardi, G.; Monastero, R.; Nicoletti, F.; Libra, M. Ageing: from inflammation to cancer. Immunity & Ageing 2018, 15, 1–7. [Google Scholar]
- De Magalhães, J.P. How ageing processes influence cancer. Nature Reviews Cancer 2013, 13, 357–365. [Google Scholar] [CrossRef] [PubMed]
- Bi, S.; Liu, Z.; Wu, Z.; Wang, Z.; Liu, X.; Wang, S.; Ren, J.; Yao, Y.; Zhang, W.; Song, M.; et al. SIRT7 antagonizes human stem cell aging as a heterochromatin stabilizer. Protein & cell 2020, 11, 483–504. [Google Scholar]
- Gallo Cantafio, M.E.; Grillone, K.; Caracciolo, D.; Scionti, F.; Arbitrio, M.; Barbieri, V.; Pensabene, L.; Guzzi, P.H.; Di Martino, M.T. From single level analysis to multi-omics integrative approaches: a powerful strategy towards the precision oncology. High-throughput 2018, 7, 33. [Google Scholar] [CrossRef]
- Bond, S.T.; Calkin, A.C.; Drew, B.G. Sex differences in white adipose tissue expansion: emerging molecular mechanisms. Clinical Science 2021, 135, 2691–2708. [Google Scholar] [CrossRef]
- Spinelli, R.; Parrillo, L.; Longo, M.; Florese, P.; Desiderio, A.; Zatterale, F.; Miele, C.; Raciti, G.A.; Beguinot, F. Molecular basis of ageing in chronic metabolic diseases. Journal of Endocrinological Investigation 2020, 43, 1373–1389. [Google Scholar] [CrossRef]
- Teruya, T.; Goga, H.; Yanagida, M. Human age-declined saliva metabolic markers determined by LC–MS. Scientific reports 2021, 11, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Nassa, G.; Tarallo, R.; Guzzi, P.H.; Ferraro, L.; Cirillo, F.; Ravo, M.; Nola, E.; Baumann, M.; Nyman, T.A.; Cannataro, M.; et al. Comparative analysis of nuclear estrogen receptor alpha and beta interactomes in breast cancer cells. Molecular BioSystems 2011, 7, 667–676. [Google Scholar] [CrossRef]
- Kerber, R.A.; O’Brien, E.; Cawthon, R.M. Gene expression profiles associated with aging and mortality in humans. Aging Cell 2009, 8, 239–250. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Memczak, S.; Qu, J.; Belmonte, J.C.I.; Liu, G.H. Single-cell omics in ageing: a young and growing field. Nature Metabolism 2020, 2, 293–302. [Google Scholar] [CrossRef] [PubMed]
- Guzzi, P.H.; Cannataro, M. μ-CS: An extension of the TM4 platform to manage Affymetrix binary data. BMC bioinformatics 2010, 11, 315. [Google Scholar] [CrossRef] [PubMed]
- Boheler, K.R.; Volkova, M.; Morrell, C.; Garg, R.; Zhu, Y.; Margulies, K.; Seymour, A.M.; Lakatta, E.G. Sex-and age-dependent human transcriptome variability: implications for chronic heart failure. Proceedings of the national academy of Sciences 2003, 100, 2754–2759. [Google Scholar] [CrossRef]
- Oosenbrug, E.; Marinho, R.P.; Zhang, J.; Marzolini, S.; Colella, T.J.; Pakosh, M.; Grace, S.L. Sex differences in cardiac rehabilitation adherence: a meta-analysis. Canadian Journal of Cardiology 2016, 32, 1316–1324. [Google Scholar] [CrossRef]
- Mercatelli, D.; Pedace, E.; Veltri, P.; Giorgi, F.M.; Guzzi, P.H. Exploiting the molecular basis of age and gender differences in outcomes of SARS-CoV-2 infections. Computational and Structural Biotechnology Journal 2021, 19, 4092–4100. [Google Scholar] [CrossRef]
- Valdes, A.M.; Glass, D.; Spector, T.D. Omics technologies and the study of human ageing. Nature Reviews Genetics 2013, 14, 601–607. [Google Scholar] [CrossRef]
- Zierer, J.; Menni, C.; Kastenmüller, G.; Spector, T.D. Integration of ‘omics’ data in aging research: from biomarkers to systems biology. Aging cell 2015, 14, 933–944. [Google Scholar] [CrossRef]
- Lorusso, J.S.; Sviderskiy, O.A.; Labunskyy, V.M. Emerging omics approaches in aging research. Antioxidants & Redox Signaling 2018, 29, 985–1002. [Google Scholar]
- Hühne, R.; Thalheim, T.; Sühnel, J. AgeFactDB—the JenAge Ageing Factor Database—towards data integration in ageing research. Nucleic acids research 2014, 42, D892–D896. [Google Scholar] [CrossRef] [PubMed]
- Fabris, F.; Magalhães, J.P.d.; Freitas, A.A. A review of supervised machine learning applied to ageing research. Biogerontology 2017, 18, 171–188. [Google Scholar] [CrossRef]
- Ai, R.; Jin, X.; Tang, B.; Yang, G.; Niu, Z.; Fang, E.F. Ageing and Alzheimer’s Disease: Application of Artificial Intelligence in Mechanistic Studies, Diagnosis, and Drug Development. In Artificial Intelligence in Medicine; Springer, 2021; pp. 1–16. [Google Scholar]
- Miloulis, S.T.; Kakkos, I.; Anastasiou, A.; Matsopoulos, G.K.; Koutsouris, D. Application of Artificial Intelligence Towards Successful Ageing: A Holistic Approach. In Modern Challenges and Approaches to Humanitarian Engineering; IGI Global, 2022; pp. 172–193. [Google Scholar]
- Lonsdale, J.; Thomas, J.; Salvatore, M.; Phillips, R.; Lo, E.; Shad, S.; Hasz, R.; Walters, G.; Garcia, F.; Young, N.; et al. The genotype-tissue expression (GTEx) project. Nature genetics 2013, 45, 580–585. [Google Scholar] [CrossRef] [PubMed]
- Shi, M.W.; Zhang, N.A.; Shi, C.P.; Liu, C.J.; Luo, Z.H.; Wang, D.Y.; Guo, A.Y.; Chen, Z.X. SAGD: a comprehensive sex-associated gene database from transcriptomes. Nucleic Acids Research 2019, 47, D835–D840. [Google Scholar] [CrossRef] [PubMed]
- Stanfill, A.G.; Cao, X. Enhancing Research Through the Use of the Genotype-Tissue Expression (GTEx) Database. Biological research for nursing 2021, 23, 533–540. [Google Scholar] [CrossRef]
- Pressler, M.P.; Horvath, A.; Entcheva, E. Sex-dependent transcription of cardiac electrophysiology and links to acetylation modifiers based on the GTEx database. Frontiers in Cardiovascular Medicine 2022, 9. [Google Scholar] [CrossRef]
- Ortuso, F.; Mercatelli, D.; Guzzi, P.H.; Giorgi, F.M. Structural genetics of circulating variants affecting the SARS-CoV-2 spike/human ACE2 complex. Journal of Biomolecular Structure and Dynamics 2021, 1–11. [Google Scholar] [CrossRef]
- de Magalhaes, J.P.; Toussaint, O. GenAge: a genomic and proteomic network map of human ageing. FEBS letters 2004, 571, 243–247. [Google Scholar] [CrossRef]
- Gu, S.; Jiang, M.; Guzzi, P.H.; Milenković, T. Modeling multi-scale data via a network of networks. Bioinformatics 2022, 38, 2544–2553. [Google Scholar] [CrossRef]
- Li, Q.; Newaz, K.; Milenković, T. Improved supervised prediction of aging-related genes via weighted dynamic network analysis. BMC bioinformatics 2021, 22, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Agapito, G.; Guzzi, P.H.; Cannataro, M. Parallel extraction of association rules from genomics data. Applied Mathematics and Computation 2019, 350, 434–446. [Google Scholar] [CrossRef]
- Guzzi, P.H.; Lomoio, U.; Scicchitano, R.; Veltri, P. NOMA-DB: a framework for management and analysis of ageing-related gene-expression data. In Proceedings of the 2022 IEEE International Conference on Bioinformatics and Biomedicine (BIBM) IEEE; IEEE, 2022; pp. 1905–1910. [Google Scholar]
- Aging Atlas: a multi-omics database for aging biology. Nucleic Acids Research 2021, 49, D825–D830. [CrossRef] [PubMed]
- Tacutu, R.; Thornton, D.; Johnson, E.; Budovsky, A.; Barardo, D.; Craig, T.; Diana, E.; Lehmann, G.; Toren, D.; Wang, J.; et al. Human Ageing Genomic Resources: new and updated databases. Nucleic Acids Research 2017, 46, D1083–D1090. [Google Scholar] [CrossRef] [PubMed]
- Rani, J.; Mittal, I.; Pramanik, A.; Singh, N.; Dube, N.; Sharma, S.; Puniya, B.L.; Raghunandanan, M.V.; Mobeen, A.; Ramachandran, S. T2DiACoD: a gene atlas of type 2 diabetes mellitus associated complex disorders. Scientific Reports 2017, 7, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Nowakowska, M.; Zghebi, S.S.; Ashcroft, D.M.; Buchan, I.; Chew-Graham, C.; Holt, T.; Mallen, C.; Van Marwijk, H.; Peek, N.; Perera-Salazar, R.; et al. The comorbidity burden of type 2 diabetes mellitus: patterns, clusters and predictions from a large English primary care cohort. BMC medicine 2019, 17, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Iglay, K.; Hannachi, H.; Joseph Howie, P.; Xu, J.; Li, X.; Engel, S.S.; Moore, L.M.; Rajpathak, S. Prevalence and co-prevalence of comorbidities among patients with type 2 diabetes mellitus. Current medical research and opinion 2016, 32, 1243–1252. [Google Scholar] [CrossRef]
- Succurro, E.; Marini, M.A.; Fiorentino, T.V.; Perticone, M.; Sciacqua, A.; Andreozzi, F.; Sesti, G. Sex-specific differences in prevalence of nonalcoholic fatty liver disease in subjects with prediabetes and type 2 diabetes. Diabetes Research and Clinical Practice 2022, 190, 110027. [Google Scholar] [CrossRef]
- Bellary, S.; Kyrou, I.; Brown, J.E.; Bailey, C.J. Type 2 diabetes mellitus in older adults: clinical considerations and management. Nature Reviews Endocrinology 2021, 17, 534–548. [Google Scholar] [CrossRef]
- Pirillo, A.; Casula, M.; Olmastroni, E.; Norata, G.D.; Catapano, A.L. Global epidemiology of dyslipidaemias. Nature Reviews Cardiology 2021, 18, 689–700. [Google Scholar] [CrossRef]
- Pearson-Stuttard, J.; Holloway, S.; Polya, R.; Sloan, R.; Zhang, L.; Gregg, E.W.; Harrison, K.; Elvidge, J.; Jonsson, P.; Porter, T. Variations in comorbidity burden in people with type 2 diabetes over disease duration: A population-based analysis of real world evidence. EClinicalMedicine 2022, 52, 101584. [Google Scholar] [CrossRef] [PubMed]
- Guerrero-Fernández de Alba, I.; Orlando, V.; Monetti, V.M.; Mucherino, S.; Gimeno-Miguel, A.; Vaccaro, O.; Forjaz, M.J.; Poblador Plou, B.; Prados-Torres, A.; Riccardi, G.; et al. Comorbidity in an older population with type-2 diabetes mellitus: identification of the characteristics and healthcare utilization of high-cost Patients. Frontiers in pharmacology 2020, 11, 586187. [Google Scholar] [CrossRef] [PubMed]
- Dworzynski, P.; Aasbrenn, M.; Rostgaard, K.; Melbye, M.; Gerds, T.A.; Hjalgrim, H.; Pers, T.H. Nationwide prediction of type 2 diabetes comorbidities. Scientific reports 2020, 10, 1–13. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




