Submitted:
08 March 2023
Posted:
13 March 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
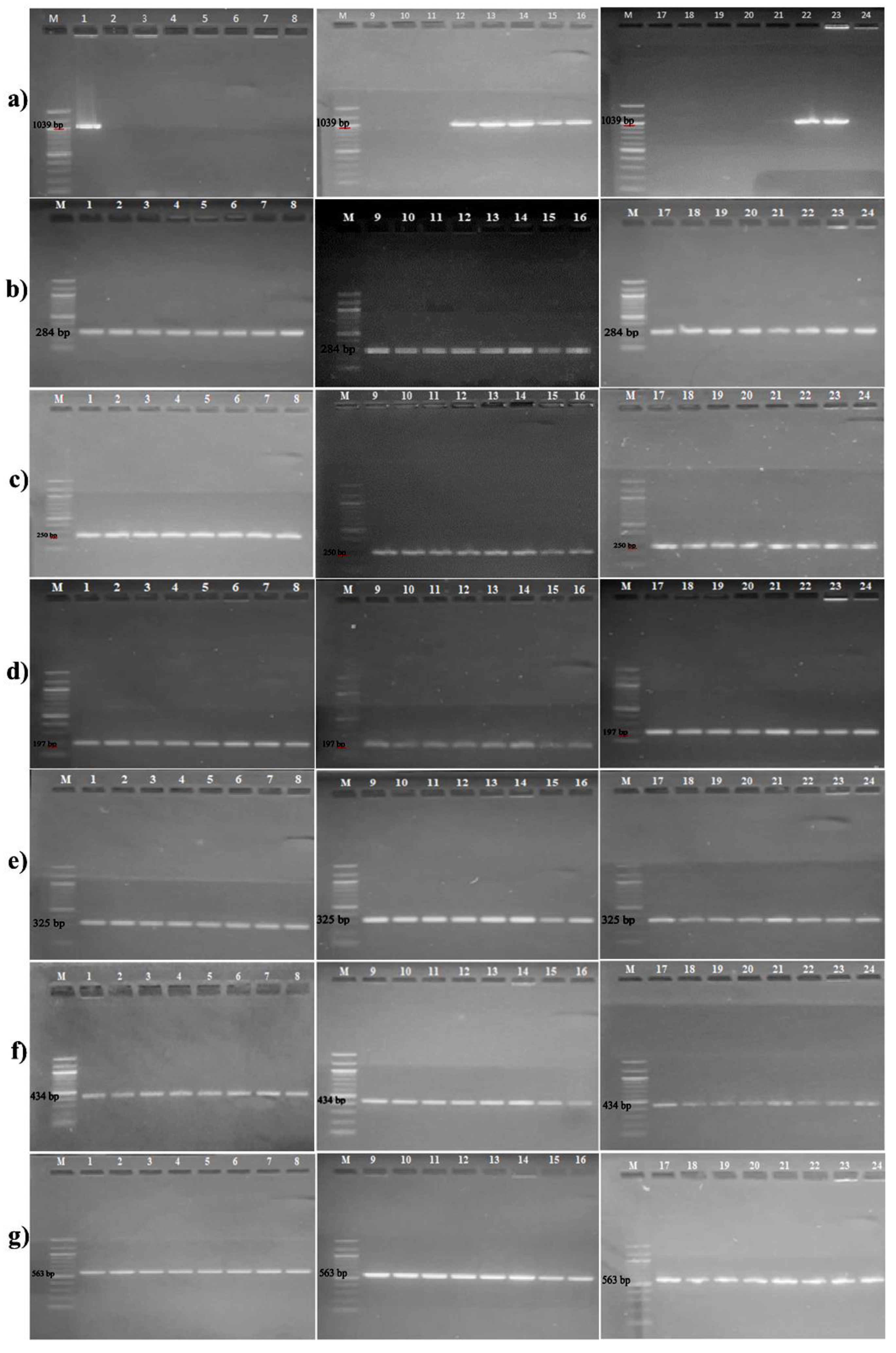
2.2. Detection of Antibiotic Resistance Genes
2.3. Determination of Minimum Inhibitory Concentration Values
2.4. Aluminum Chlorohydrate Exposure and Isolation of Resistant Bacteria Against Test Antibiotics
2.5. Quantitative Reverse Transcriptase PCR Analysis of Target Genes
3. Results and Discussion
3.1. Detection of Antibiotic Resistance Genes
3.2. MIC Values of Aluminum Chlorohydrate
3.3. Antibiotic Resistance Development After Aluminum Chlorohydrate Exposure
3.4. Increase in the Minimum Inhibitory Concentration Values of the Test Bacteria
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Ethical Statements
References
- Landers, T.F.; Cohen, B.; Wittum, T.E.; Larson, E.L. A review of antibiotic use in food animals: perspective, policy, and potential. Public Health Rep 2012, 127, 4–22. [Google Scholar] [CrossRef] [PubMed]
- Jin, M.; Lu, J.; Chen, Z.; Nguyen, S.H.; Mao, L.; Li, J.; Yuan, Z.; Guo, J. Antidepressant fluoxetine induces multiple antibiotics resistance in Escherichia coli via ROS-mediated mutagenesis. Environ Int 2018, 120, 421–430. [Google Scholar] [CrossRef] [PubMed]
- Morehead, M.S.; Scarbrough, C. Emergence of global antibiotic resistance. Prim Care. 2018, 45, 467–484. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention. Antibiotic resistance threats in the United States 2019. Available online: https://www.cdc.gov/ drugresistance/pdf/threats-report/2019-ar-threats-report508.pdf (accessed on 15 October 2021).
- World Health Organization. Antibiotic resistance. Available online: https:// www.who.int/news-room/factsheets/detail/antibiotic-resistance (accessed on 23 August 2021).
- Gurpinar, S.S.; Kart, D.; Eryilmaz, M. The effects of antidepressants fluoxetine, sertraline, and amitriptyline on the development of antibiotic resistance in Acinetobacter baumannii. Arch Microbiol 2022, 204, 230. [Google Scholar] [CrossRef] [PubMed]
- Haran, J.P.; McCormick, B.A. Aging, frailty, and the microbiome-how dysbiosis influences human aging and disease. Gastroenterology 2021, 160, 507–523. [Google Scholar] [CrossRef] [PubMed]
- Shanahan, F.; Ghosh, T.S.; O'toole, P.W. The healthy microbiome-What is the definition of a healthy gut microbiome? Gastroenterology 2021, 160, 483–494. [Google Scholar] [CrossRef]
- Otto, M. Staphylococcus epidermidis--the 'accidental' pathogen. Nat Rev Microbiol 2009, 7, 555–567. [Google Scholar] [CrossRef]
- Brooks, G.F.; Carroll, K.C.; Butel, J.S.; Morse, S.A.; Mietzner, T.A. Jawetz, Melnickk ve Adelberg Tıbbi Mikrobiyoloji. Ç. Dr. Osman Şadi Yenen: Nobel Kitapevleri, Ankara, 2014; p. 167. [Google Scholar]
- Fournière, M.; Latire, T.; Souak, D.; Feuilloley, M.G.J.; Bedoux, G. Staphylococcus epidermidis and Cutibacterium acnes: Two major sentinels of skin microbiota and the influence of cosmetics. Microorganisms 2020, 8, 1752. [Google Scholar] [CrossRef] [PubMed]
- Vuong, C.; Otto, M. Staphylococcus epidermidis infections. Microbes Infect 2002, 4, 481–489. [Google Scholar] [CrossRef]
- Eryilmaz, M.; Gurpinar, S.S. Investigation of the antibacterial efficacy of some commonly used antiseptics in hospitals against biofilm forming and non-biofilm forming Staphylococcus epidermidis strains. J Fac Pharm Ankara 2017, 41, 1–8. [Google Scholar]
- Cerca, N.; Martins, S.; Cerca, F.; Jefferson, K.K.; Pier, G.B.; Oliveira, R.; Azeredo, J. Comparative assessment of antibiotic susceptibility of coagulase-negative Staphylococci in biofilm versus planktonic culture as assessed by bacterial enumeration or rapid XTT colorimetry. J Antimicrob Chemother 2005, 56, 331–336. [Google Scholar] [CrossRef]
- Morgenstern, M.; Erichsen, C.; von Rüden, C.; Metsemakers, W.J.; Kates, S.L.; Moriarty, T.F.; Hungerer, S. Staphylococcal orthopaedic device-related infections in older patients. Injury 2016, 47, 1427–1434. [Google Scholar] [CrossRef] [PubMed]
- Sabaté Brescó, M.; Harris, L.G.; Thompson, K.; Stanic, B.; Morgenstern, M.; O'Mahony, L.; Richards, R.G.; Moriarty, T.F. Pathogenic mechanisms and host interactions in Staphylococcus epidermidis device-related infection. Front Microbiol 2017, 8, 1401. [Google Scholar] [CrossRef] [PubMed]
- Bouiller, K.; Ilic, D.; Wicky, P.H.; Cholley, P.; Chirouze, C.; Bertrand, X. Spread of clonal linezolid-resistant Staphylococcus epidermidis in an intensive care unit associated with linezolid exposure. Eur J Clin Microbiol Infect Dis 2020, 39, 1271–1277. [Google Scholar] [CrossRef] [PubMed]
- Callewaert, C.; Hutapea, P.; Van de Wiele, T.; Boon, N. Deodorants and antiperspirants affect the axillary bacterial community. Arch Dermatol Res 2014, 306, 701–710. [Google Scholar] [CrossRef] [PubMed]
- Glatz, M.; Jo, J.H.; Kennedy, E.A.; Polley, E.C.; Segre, J.A.; Simpson, E.L.; Kong, H.H. Emollient use alters skin barrier and microbes in infants at risk for developing atopic dermatitis. PLoS One 2018, 13, e0192443. [Google Scholar] [CrossRef] [PubMed]
- Aras, A.; Eryilmaz, M. Microbiological contamination of cosmetic products. J Fac Pharm Ankara 2022, 46, 262–276. [Google Scholar]
- Holzle, E. Antiperspirants; Dermatopharmacology of Topical Preparations, Springer-Verlag: Berlin Heidelberg, 2000; pp. 401–402. [Google Scholar]
- Martini, M.C. Déodorants et antitranspirants [Deodorants and antiperspirants]. Ann Dermatol Venereol 2020, 147, 387–395. [Google Scholar] [CrossRef] [PubMed]
- Clinical and Laboratory Standards Institute (CLSI). Performance Standartds for Antimicrobial Susceptibility Testing, CLSI supplement M100, 30th ed.; Clinical and Laboratory Standards Institute: Wayne, PA, 2020. [Google Scholar]
- Vandecasteele, S.J.; Peetermans, W.E.; Merckx, R.; Van Eldere, J. Quantification of expression of Staphylococcus epidermidis housekeeping genes with Taqman quantitative PCR during in vitro growth and under different conditions. J Bacteriol 2001, 183, 7094–7101. [Google Scholar] [CrossRef]
- Yamada, M.; Yoshida, J.; Hatou, S.; Yoshida, T.; Minagawa, Y. Mutations in the quinolone resistance determining region in Staphylococcus epidermidis recovered from conjunctiva and their association with susceptibility to various fluoroquinolones. Br J Ophthalmol 2008, 92, 848–851. [Google Scholar] [CrossRef]
- Betanzos-Cabrera, G.; Juárez-Verdayes, M.A.; González-González, G.; Cancino-Díaz, M.E.; Cancino-Díaz, J.C. Gatifloxacin, moxifloxacin, and balofloxacin resistance due to mutations in the gyrA and parC genes of Staphylococcus epidermidis strains isolated from patients with endophthalmitis, corneal ulcers and conjunctivitis. Ophthalmic Res 2009, 42, 43–48. [Google Scholar] [CrossRef] [PubMed]
- Juárez-Verdayes, M.A.; Parra-Ortega, B.; Hernández-Rodríguez, C.; Betanzos-Cabrera, G.; Rodríguez-Martínez, S.; Cancino-Diaz, M.E.; Cancino-Diaz, J.C. Identification and expression of nor efflux family genes in Staphylococcus epidermidis that act against gatifloxacin. Microb Pathog 2012, 52, 318–325. [Google Scholar] [CrossRef] [PubMed]
- Sully, E.K.; Malachowa, N.; Elmore, B.O.; Alexander, S.M.; Femling, J.K.; Gray, B.M.; DeLeo, F.R.; Otto, M.; Cheung, A.L.; Edwards, B.S.; Sklar, L.A.; Horswill, A.R.; Hall, P.R.; Gresham, H.D. Selective chemical inhibition of agr quorum sensing in Staphylococcus aureus promotes host defense with minimal impact on resistance. PLoS Pathog 2014, 10, e1004174. [Google Scholar] [CrossRef] [PubMed]
- Eladli, M.G.; Alharbi, N.S.; Khaled, J.M.; Kadaikunnan, S.; Alobaidi, A.S.; Alyahya, S.A. Antibiotic-resistant Staphylococcus epidermidis isolated from patients and healthy students comparing with antibiotic-resistant bacteria isolated from pasteurized milk. Saudi J Biol Sci 2019, 26, 1285–1290. [Google Scholar] [CrossRef] [PubMed]
- Zen, J.M.; Yang, T.H.; Kumar, A.S.; Chen, Y.J.; Hsu, J.C.; Shih, Y. Detection of aluminum chlorohydrate content in antiperspirant deodorants using screen-printed silver electrodes by one drop analysis. Electroanalysis 2009, 21, 2272–2276. [Google Scholar] [CrossRef]
- Scientific Committee on Consumer Safety (SCCS). The SCCS Notes of Guidance for the Testing of Cosmetic Ingredients and Their Safety Evaluation 10th Revision. SCCS/1628/18: 24-25, 2018.
- Kart, D.; Gurpinar, S.S.; Eryilmaz, M. Assessment of the anti-quorum sensing effect of Lactobacillus sp. metabolites on expression levels of QS-related genes in Pseudomonas aeruginosa PAO1. Turk Bull Hyg Exp Biol 2020, 77, 311–318. [Google Scholar] [CrossRef]
- Yilmaz, E. Quinolones. Turkiye Klinikleri J Inf Dis-Special Topics 2017, 10, 99–105. [Google Scholar]
- Thai, T.; Salisbury, B.H.; Zito, P.M. Ciprofloxacin. [Updated 2022 Sep 2]. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2022 Jan. Available online: https://www.ncbi.nlm.nih.gov/books/NBK535454/.
- Shariati, A.; Arshadi, M.; Khosrojerdi, M.A.; Abedinzadeh, M.; Ganjalishahi, M.; Maleki, A.; Heidary, M.; Khoshnood, S. The resistance mechanisms of bacteria against ciprofloxacin and new approaches for enhancing the efficacy of this antibiotic. Front Public Health 2022, 10, 1025633. [Google Scholar] [CrossRef] [PubMed]
- DeMarco, C.E.; Cushing, L.A.; Frempong-Manso, E.; Seo, S.M.; Jaravaza, T.A.; Kaatz, G.W. Efflux-related resistance to norfloxacin, dyes, and biocides in bloodstream isolates of Staphylococcus aureus. Antimicrob Agents Chemother 2007, 51, 3235–3239. [Google Scholar] [CrossRef]
- Qin, R.; Xiao, K.; Li, B.; Jiang, W.; Peng, W.; Zheng, J.; Zhou, H. The combination of catechin and epicatechin callate from Fructus Crataegi potentiates beta-lactam antibiotics against methicillin-resistant Staphylococcus aureus (MRSA) in vitro and in vivo. Int J Mol Sci 2013, 14, 1802–1821. [Google Scholar] [CrossRef]
- Bostanmaneshrad, A.; Nowroozi, J.; Eslami, G.; Hashemi, A. The expression of efflux pump genes in methicillin-resistant Staphylococcus aureus (MRSA) strains isolated from blood cultures in Iran. Arch Clin Infect Dis 2020, 15, e99804. [Google Scholar] [CrossRef]
- Sancak, B. Resistance mechanisms of MRSA: Epidemiology in the World and Turkey. Ankem Derg 2012, 26, 38–47. [Google Scholar]
- Çiftci, İ.H.; Altindis, M.; Cetinkaya, Z.; Asik, G.; Aktepe, O.C. Investigation of mecA genes in Staphylococcus strains isolated from clinical samples. Kocatepe Med J 2009, 10, 17–20. [Google Scholar]
- Xu, Z.; Shah, H.N.; Misra, R.; Chen, J.; Zhang, W.; Liu, Y.; Cutler, R.R.; Mkrtchyan, H.V. The prevalence, antibiotic resistance and mecA characterization of coagulase negative Staphylococci recovered from non-healthcare settings in London, UK. Antimicrob Resist Infect Control 2018, 7, 73. [Google Scholar] [CrossRef]
- Tan, Y.S.; Zhang, R.K.; Liu, Z.H.; Li, B.Z.; Yuan, Y.J. Microbial adaptation to enhance stress tolerance. Front Microbiol 2022, 13, 888746. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Lu, J.; Zhang, S.; Li, J.; Mao, L.; Yuan, Z.; Bond, P.L.; Guo, J. Non-antibiotic pharmaceuticals promote the transmission of multidrug resistance plasmids through intra- and intergenera conjugation. ISME J 2021, 15, 2493–2508. [Google Scholar] [CrossRef]
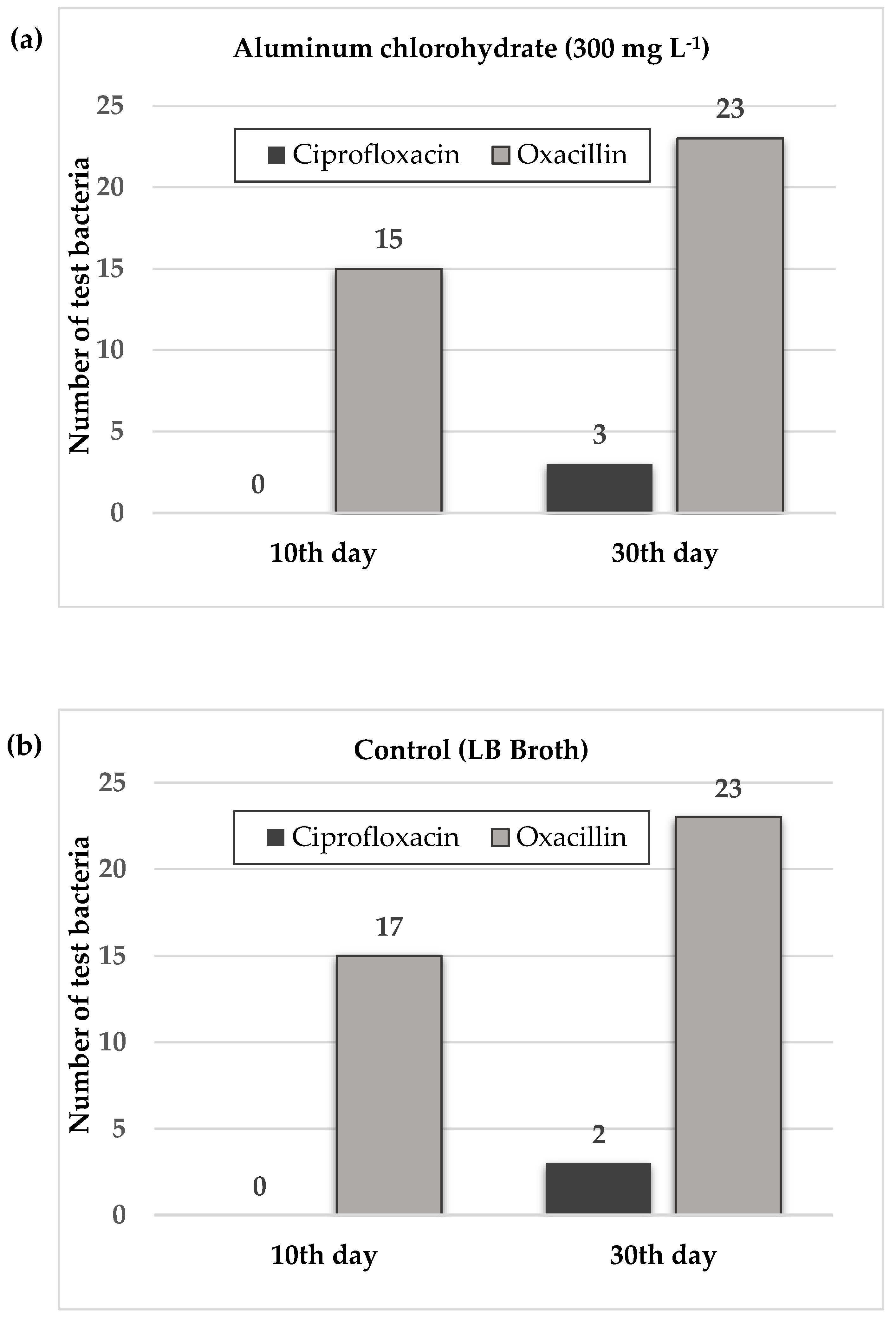
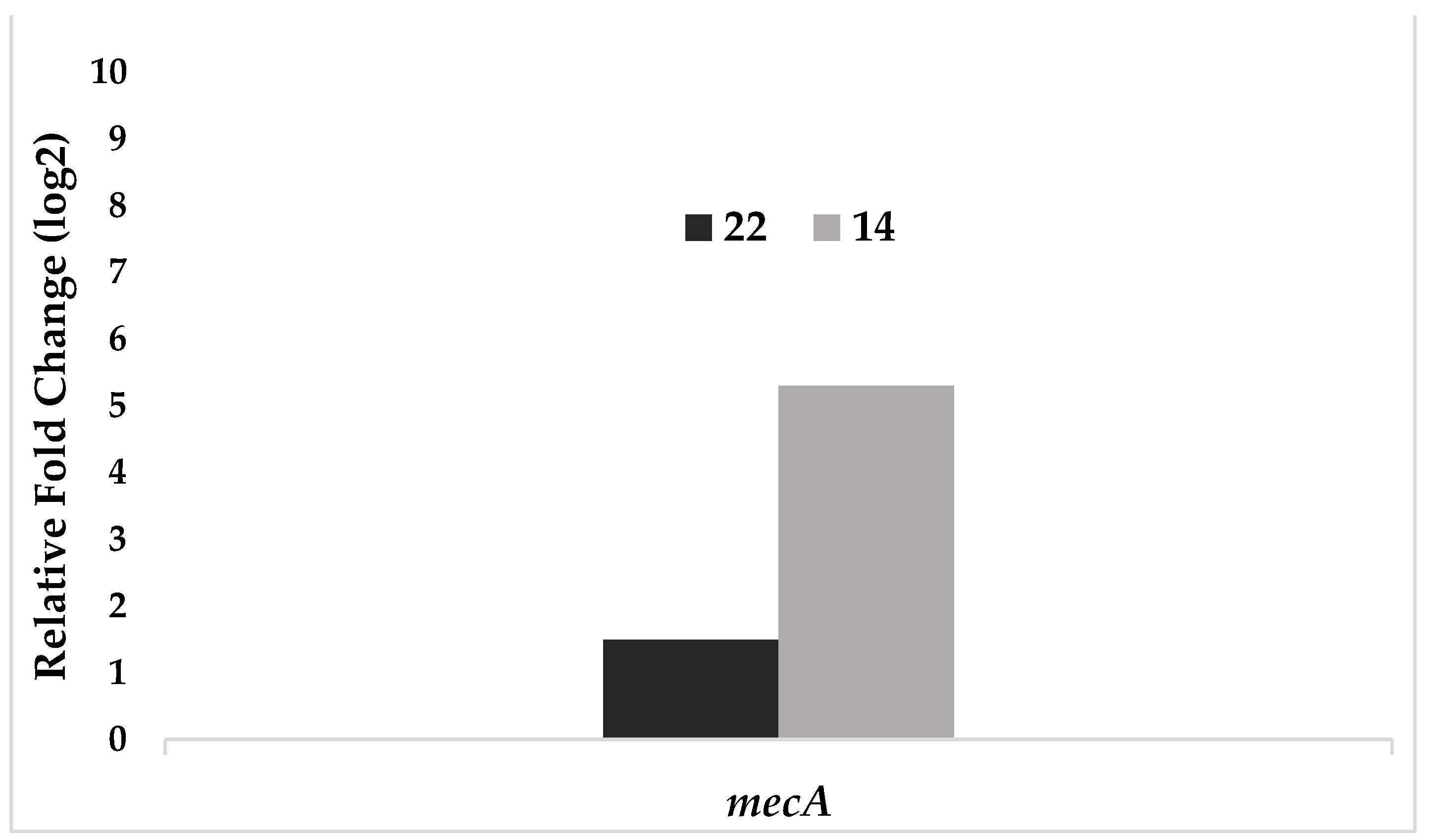
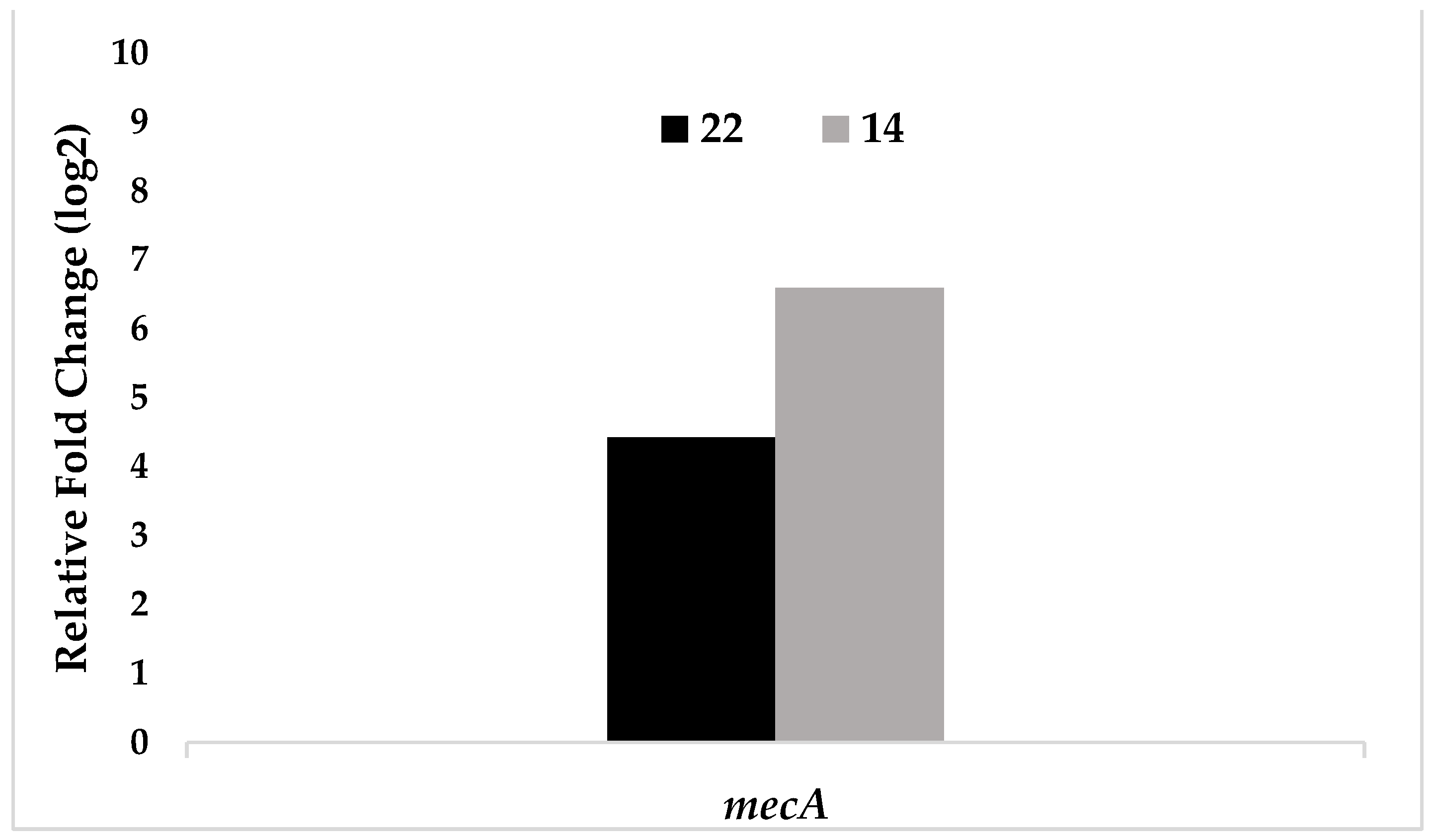
| Exposure day |
Test antibiotics |
Aluminum chlorohydrate (300 mg L-1) |
Control* |
|---|---|---|---|
| 10th | CIP | - | - |
| OXA | 1, 2, 3, 5, 12, 13, 14, 15, 16, 17, 18, 20, 21, 22, 24 | 1, 2, 3, 4, 6, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 22 | |
| 30th | CIP | 18, 22, 24 | 14, 24 |
| OXA | 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 24 | 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 24 |
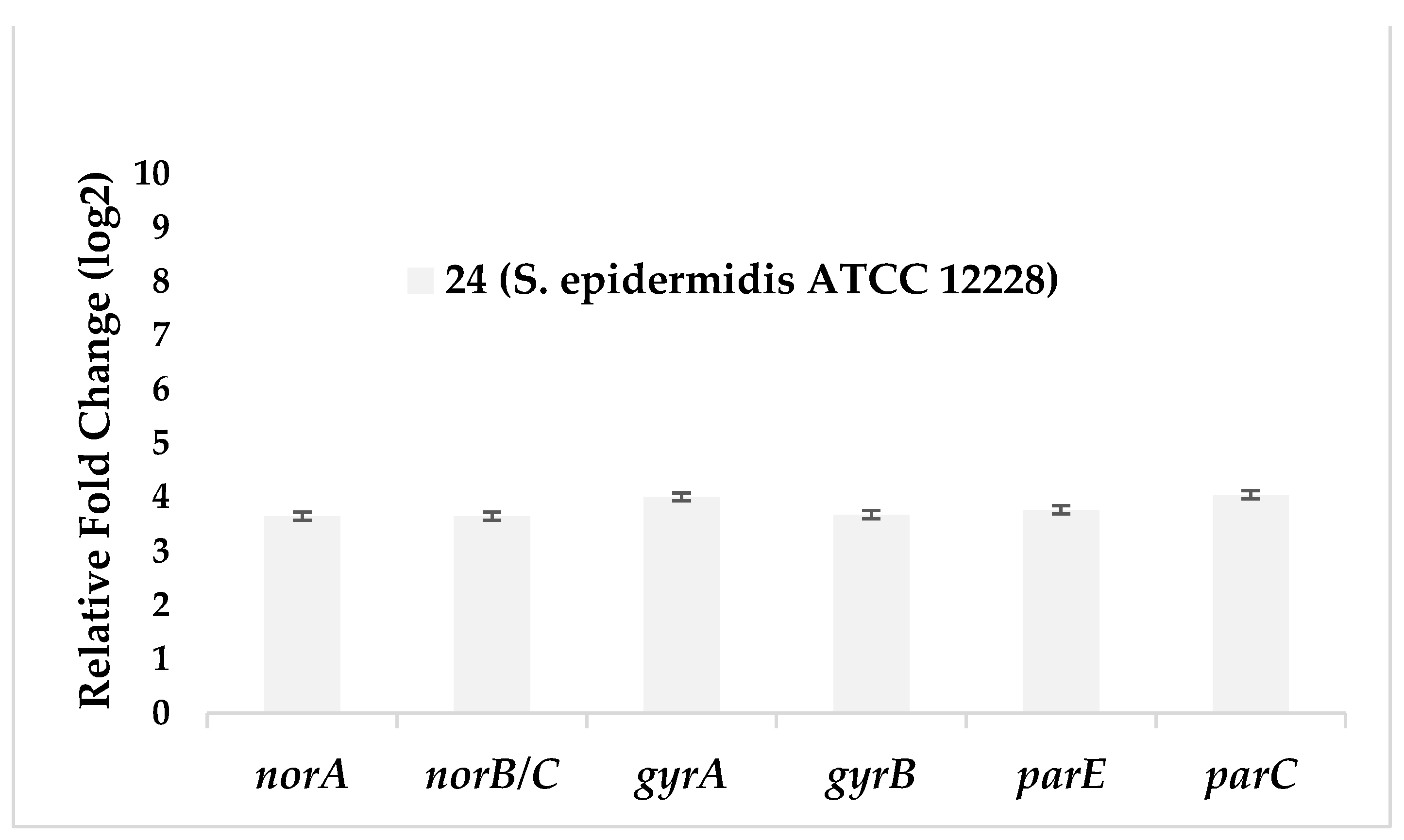
| Test bacteria | norA | norB/C | gyrA | gyrB | parE | parC |
|---|---|---|---|---|---|---|
| 24 (S. epidermidis ATCC 12228) |
3,65 | 3,65 | 4,01 | 3,68 | 3,77 | 4,05 |
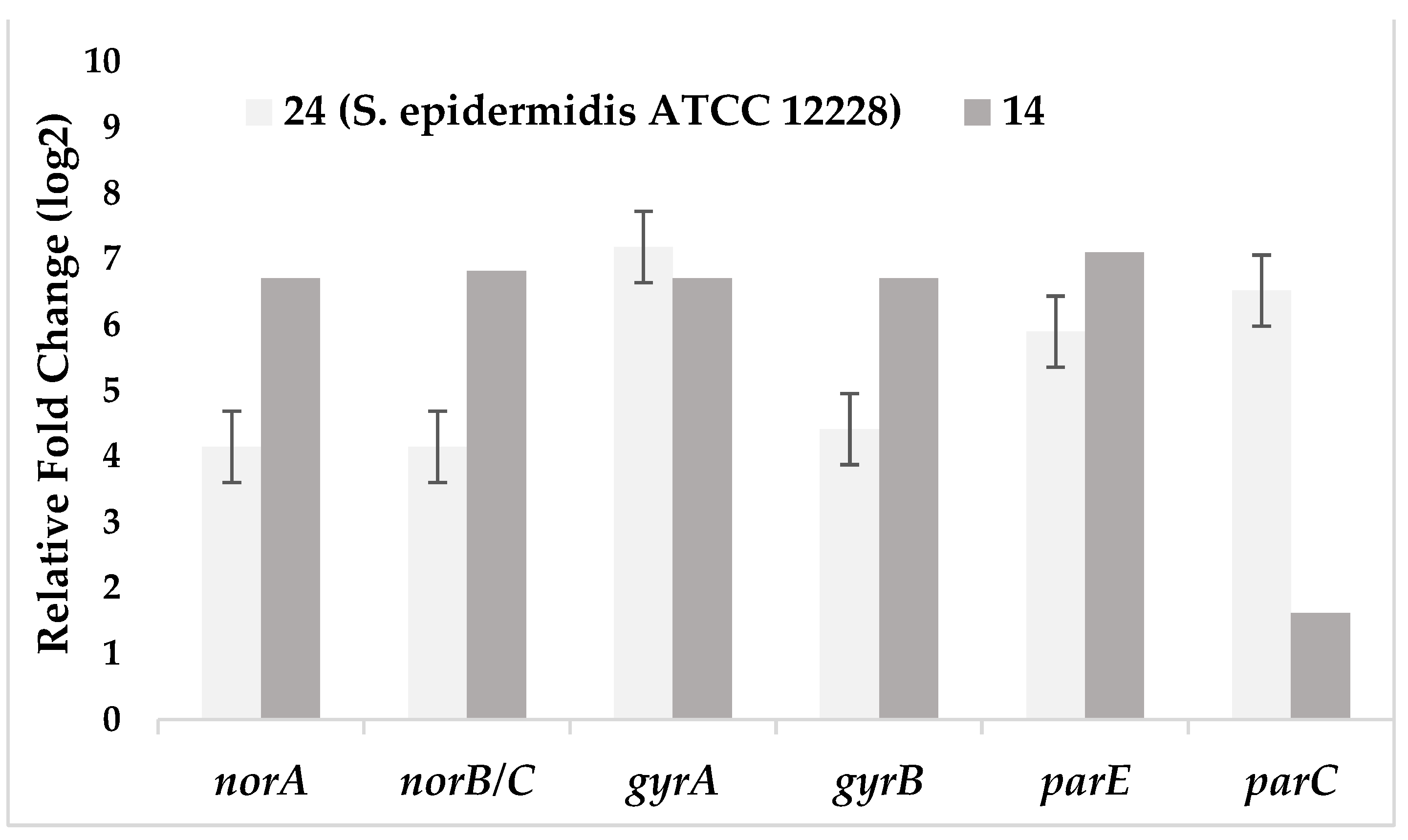
| Test bacteria | norA | norB/C | gyrA | gyrB | parE | parC |
|---|---|---|---|---|---|---|
| 24 (S. epidermidis ATCC 12228) |
4,14 | 4,14 | 7,18 | 4,41 | 5,89 | 6,52 |
| 14 | 6,71 | 6,82 | 6,71 | 6,71 | 7,10 | 1,61 |
| Test bacteria | mecA |
|---|---|
| 22 | 1,50 |
| 14 | 5,30 |
| Test bacteria | mecA |
|---|---|
| 22 | 4,42 |
| 14 | 6,59 |
| Test bacteria | Test antibiotics | Pre-exposure | Post-exposure (30th day) |
|---|---|---|---|
| 1 | CIP | 0,125 | 4 |
| 2 | OXA | 2 | 16 |
| 3 | OXA | 16 | >64 |
| 4 | OXA | 16 | >64 |
| 5 | OXA | 0,25 | 32 |
| 6 | OXA | 8 | 32 |
| 7 | OXA | 0,25 | 16 |
| 8 | OXA | 0,25 | 1 |
| 11 | OXA | 1 | 16 |
| CIP | 0,25 | 1 | |
| 12 | OXA | 8 | 32 |
| 13 | CIP | 0,25 | 8 |
| 14 | OXA | 64 | >64 |
| 16 | CIP | 0,125 | 0,5 |
| 17 | OXA | 0,5 | 16 |
| 18 | OXA | 0,25 | 64 |
| CIP | 0,25 | 8 | |
| 19 | OXA | 0,25 | 1 |
| 20 | OXA | 0,25 | 1 |
| CIP | <0,03125 | 0,125 | |
| 21 | OXA | 0,25 | 64 |
| CIP | 0,125 | 4 | |
| 22 | OXA | 2 | 4 |
| CIP | 0,25 | 4 | |
| 23 | OXA | 8 | 64 |
| 24 | OXA | 0,125 | 0,5 |
| CIP | 0,125 | 0,5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





