Submitted:
28 March 2023
Posted:
29 March 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Sample Collection, DNA Extraction, and Sequencing
2.2. Mitochondrial Genome Assembly
2.3. Phylogenetic Analyses
3. Results
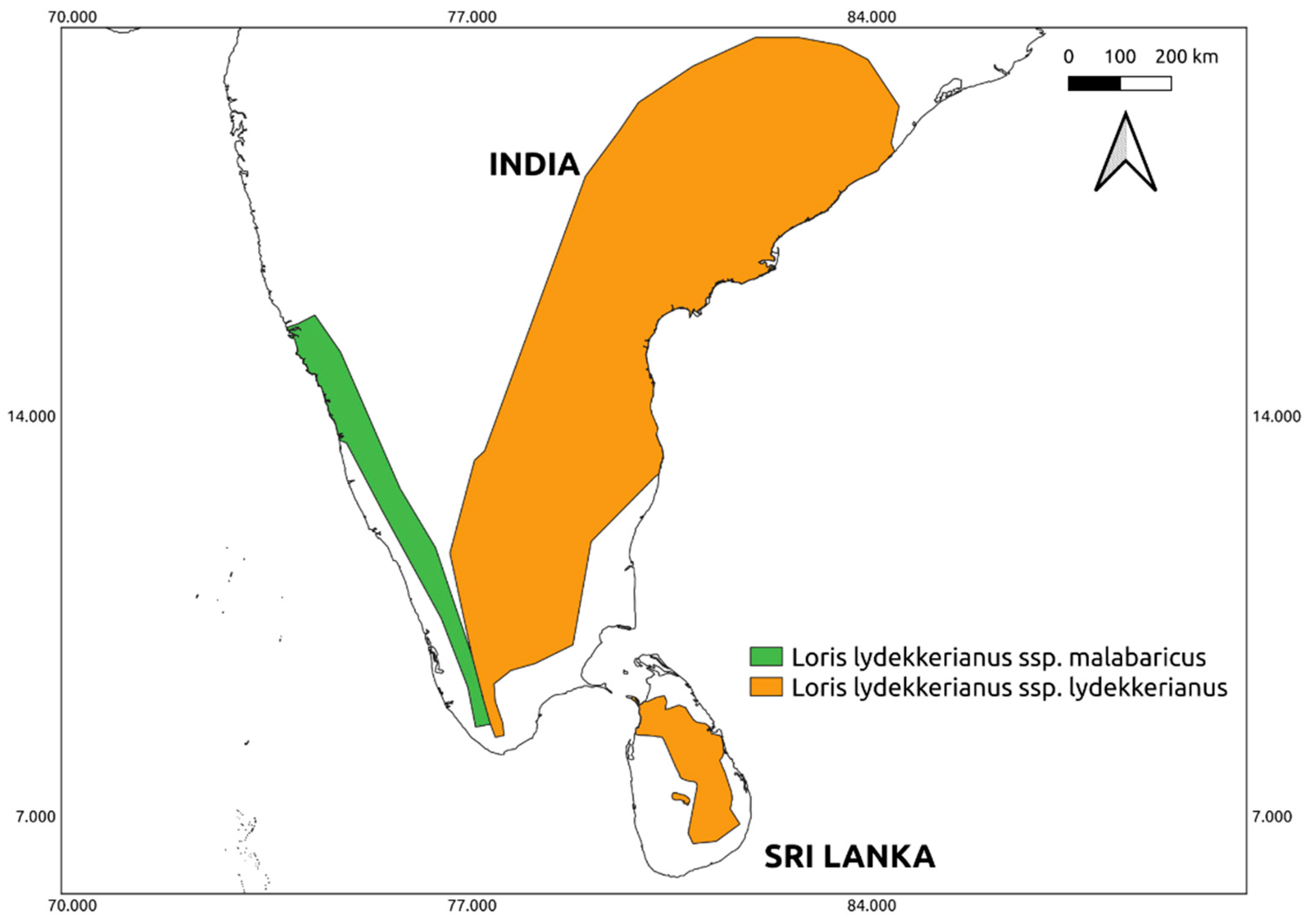
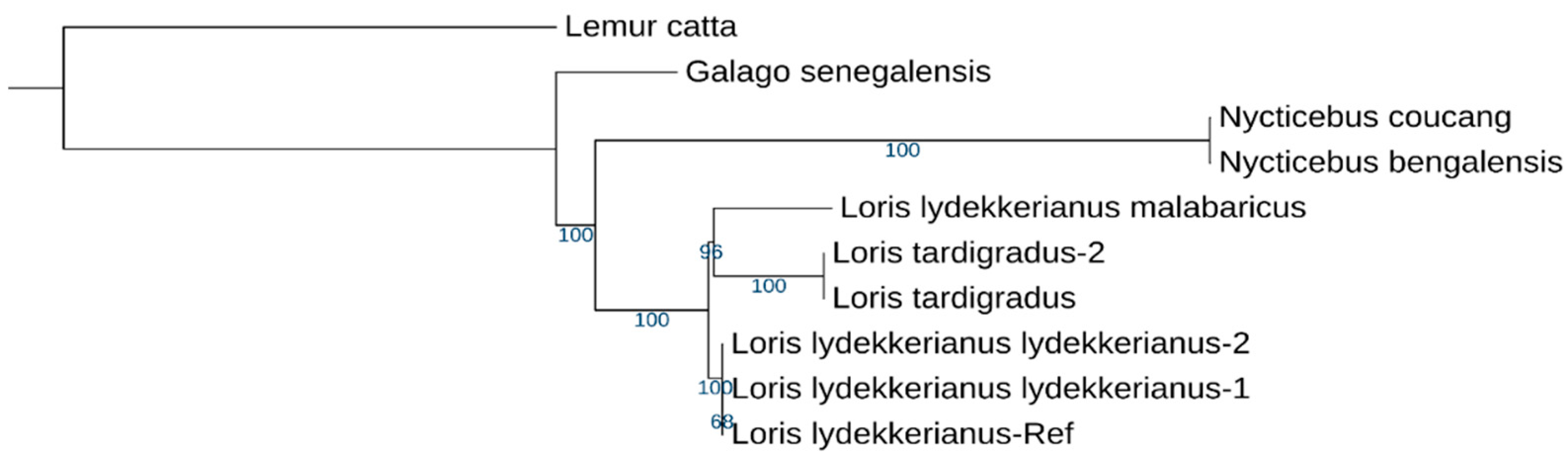
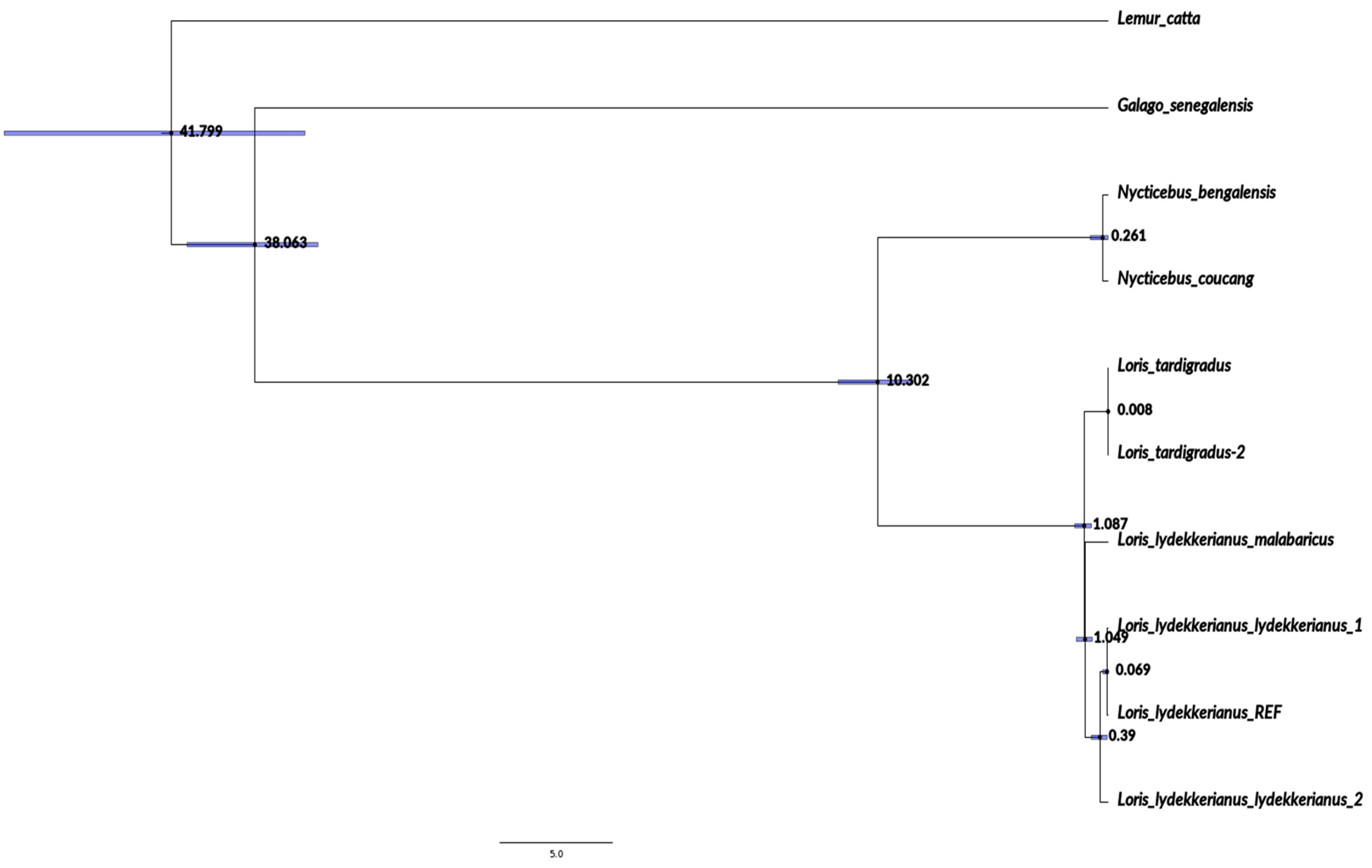
3.1. Phylogenetics Supports the Morphological Delineation of Mysore and Malabar Slender Lorises
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nekaris, K. a. I. Extreme Primates: Ecology and Evolution of Asian Lorises. Evol. Anthropol. Issues News Rev. 2014, 23, 177–187. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.; Singh, M.; Kumara, H.N.; Kumar, S.; Gnanaoliu, S.D.; Sasi, R. A Review of Research on the Distribution, Ecology, Behaviour, and Conservation of the Slender Loris Loris Lydekkerianus (Mammalia: Primates: Lorisidae) in India. J. Threat. Taxa 2021, 13, 19540–19552. [Google Scholar] [CrossRef]
- Nekaris, K.A.I.; Rasmussen, D.T. Diet and Feeding Behavior of Mysore Slender Lorises. Int. J. Primatol. 2003, 24, 33–46. [Google Scholar] [CrossRef]
- Radhakrishna, S.; Kumara, H.N. Behavioural Variation in the Mysore Slender Loris Loris Lydekkerianus Lydekkerianus. Curr. Sci. 2010, 99, 1226–1232. [Google Scholar]
- Radhakrishna, S.; Singh, M. Social Behaviour of the Slender Loris (Loris Tardigradus Lydekkerianus). Folia Primatol. (Basel) 2002, 73, 181–196. [Google Scholar] [CrossRef]
- Groves, C.P. Primate Taxonomy; Smithsonian Institution Press, 2001.
- Nijman, V.; Robbins, T.; Maddock, A.J.; Ang, A. Molecular Phylogeny, Taxonomy and Conservation of Slender Lorises. Primate Conserv. 2020, 34, 143–151. [Google Scholar]
- Kumara, H.N.; Singh, M.; Irfan-Ullah, M.; Kumar, S. Status, Distribution, and Conservation of Slender Lorises in India. In Leaping Ahead: Advances in Prosimian Biology; Masters, J., Gamba, M., Génin, F., Eds.; Developments in Primatology: Progress and Prospects; Springer: New York, NY, 2013; pp. 343–352. ISBN 978-1-4614-4511-1. [Google Scholar]
- Dittus, W.; Singh, M.; Kumara, H.N.; Kumar, A. IUCN Red List of Threatened Species: Loris Lydekkerianus. IUCN Red List Threat. Species 2022. [Google Scholar]
- Kumara, H.N.; Nekaris, A.; Singh, M. IUCN Red List of Threatened Species: Loris Lydekkerianus Ssp. Lydekkerianus. IUCN Red List Threat. Species 2022. [Google Scholar]
- Kumara, H.N.; Nekaris, A.; Singh, M. IUCN Red List of Threatened Species: Loris Lydekkerianus Ssp. Malabaricus. IUCN Red List Threat. Species 2022. [Google Scholar]
- Kumara, H.N.; Singh, M.; Kumar, S. Distribution, Habitat Correlates, and Conservation of Loris Lydekkerianus in Karnataka, India. Int. J. Primatol. 2006, 27, 941–969. [Google Scholar] [CrossRef]
- Radhakrishna, S.; Singh, M. Reproductive Biology of the Slender Loris (Loris Lydekkerianus Lydekkerianus). Folia Primatol. (Basel) 2004, 75, 1–13. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An Ultra-Fast All-in-One FASTQ Preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Jin, J.-J.; Yu, W.-B.; Yang, J.-B.; Song, Y.; dePamphilis, C.W.; Yi, T.-S.; Li, D.-Z. GetOrganelle: A Fast and Versatile Toolkit for Accurate de Novo Assembly of Organelle Genomes. Genome Biol. 2020, 21, 241. [Google Scholar] [CrossRef]
- Bernt, M.; Donath, A.; Jühling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Pütz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de Novo Metazoan Mitochondrial Genome Annotation. Mol. Phylogenet. Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef]
- Sievers, F.; Higgins, D.G. The Clustal Omega Multiple Alignment Package. In Multiple Sequence Alignment: Methods and Protocols; Katoh, K., Ed.; Methods in Molecular Biology; Springer US: New York, NY, 2021; pp. 3–16. ISBN 978-1-07-161036-7. [Google Scholar]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. PartitionFinder 2: New Methods for Selecting Partitioned Models of Evolution for Molecular and Morphological Phylogenetic Analyses. Mol. Biol. Evol. 2017, 34, 772–773. [Google Scholar] [CrossRef] [PubMed]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life (ITOL) v5: An Online Tool for Phylogenetic Tree Display and Annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Bouckaert, R.; Heled, J.; Kühnert, D.; Vaughan, T.; Wu, C.-H.; Xie, D.; Suchard, M.A.; Rambaut, A.; Drummond, A.J. BEAST 2: A Software Platform for Bayesian Evolutionary Analysis. PLOS Comput. Biol. 2014, 10, e1003537. [Google Scholar] [CrossRef] [PubMed]
- Seiffert, E.R.; Simons, E.L.; Attia, Y. Fossil Evidence for an Ancient Divergence of Lorises and Galagos. Nature 2003, 422, 421–424. [Google Scholar] [CrossRef] [PubMed]
- Harrison, T. Later Tertiary Lorisiformes. In Cenozoic Mammals of Africa; Werdelin, L., Ed.; University of California Press, 2010; p. 0 ISBN 978-0-520-25721-4.
- Helfrich, P.; Rieb, E.; Abrami, G.; Lücking, A.; Mehler, A. TreeAnnotator: Versatile Visual Annotation of Hierarchical Text Relations. In Proceedings of the Proceedings of the Eleventh International Conference on Language Resources and Evaluation (LREC 2018); European Language Resources Association (ELRA): Miyazaki, Japan, May 2018. [Google Scholar]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior Summarization in Bayesian Phylogenetics Using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A. FigTree v1. 4.2, a Graphical Viewer of Phylogenetic Trees 2014.
- Finstermeier, K.; Zinner, D.; Brameier, M.; Meyer, M.; Kreuz, E.; Hofreiter, M.; Roos, C. A Mitogenomic Phylogeny of Living Primates. PLOS ONE 2013, 8, e69504. [Google Scholar] [CrossRef] [PubMed]
- Kumara, H.N.; Sasi, R.; Chandran, S.; Radhakrishna, S. Distribution of the Grey Slender Loris (Loris Lyddekerianus Cabrera, 1908) in Tamil Nadu, Southern India. Folia Primatol. (Basel) 2016, 87, 291–302. [Google Scholar] [CrossRef] [PubMed]
| Sample |
Loris tardigradus |
Loris tardigradus-2 |
Loris lydekkerianus malabaricus |
Loris lydekkerianus lydekkerianus-1 |
Loris lydekkerianus lydekkerianus-2 |
Loris lydekkerianus-Ref |
|---|---|---|---|---|---|---|
| Loris tardigradus | - | 100 | 97.42 | 97.18 | 97.05 | 97.10 |
| Loris tardigradus-2 | 100 | - | 97.42 | 97.18 | 97.03 | 97.10 |
| Loris lydekkerianus malabaricus | 97.42 | 97.42 | - | 97.91 | 97.91 | 97.91 |
| Loris lydekkerianus lydekkerianus-1 | 97.18 | 97.18 | 97.91 | - | 99.84 | 99.92 |
| Loris lydekkerianus lydekkerianus-2 | 97.03 | 97.03 | 97.91 | 99.84 | - | 99.84 |
| Loris lydekkerianus-Ref | 97.10 | 97.10 | 97.91 | 99.92 | 99.84 | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





