Submitted:
28 March 2023
Posted:
29 March 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Subjects and data acquisition
2.2. Feature set and modelling
2.3. Confident Learning, Interpretation, and Evaluation
- Test performance on the given test labels using the Gaussian process classifier;
- Test performance on the corrected test labels using the Gaussian process classifier;
- Test performance on the given test labels using the Gaussian process classifier + confident learning on training data;
- Test performance on the corrected test labels using the Gaussian process classifier + confident learning on training data.
3. Results
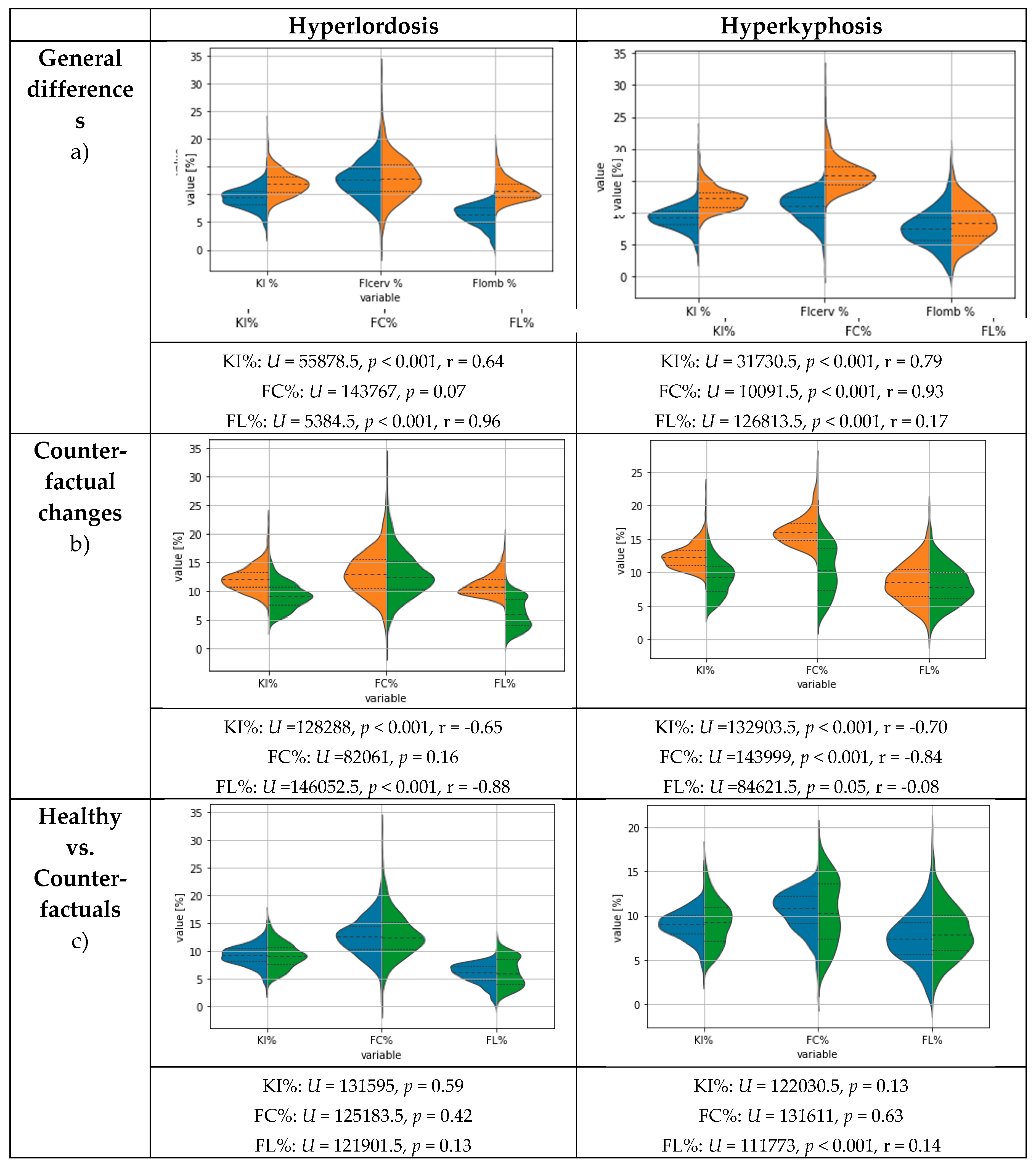
3.1. Re-evaluation results
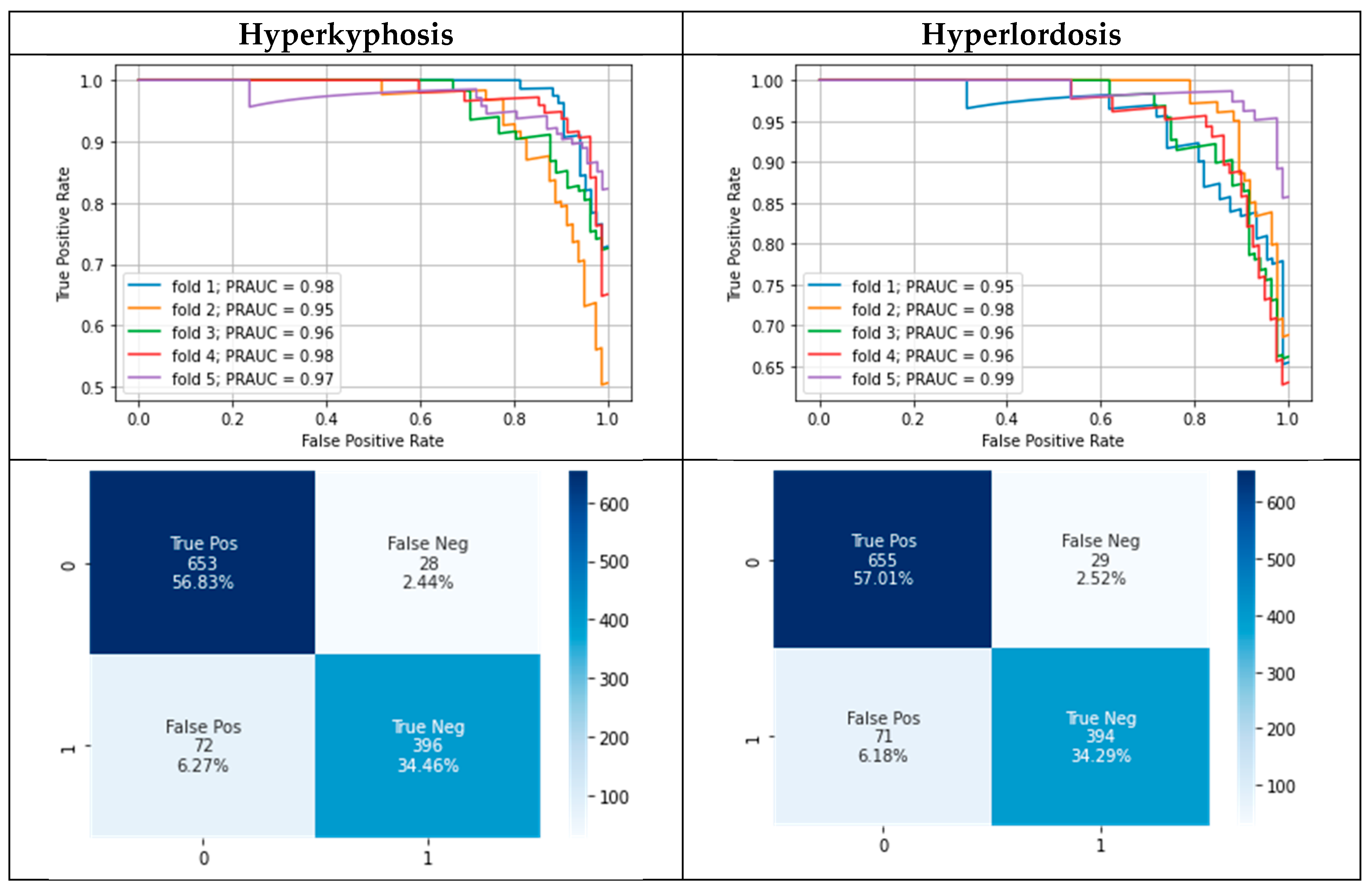
3.2. Modelling results
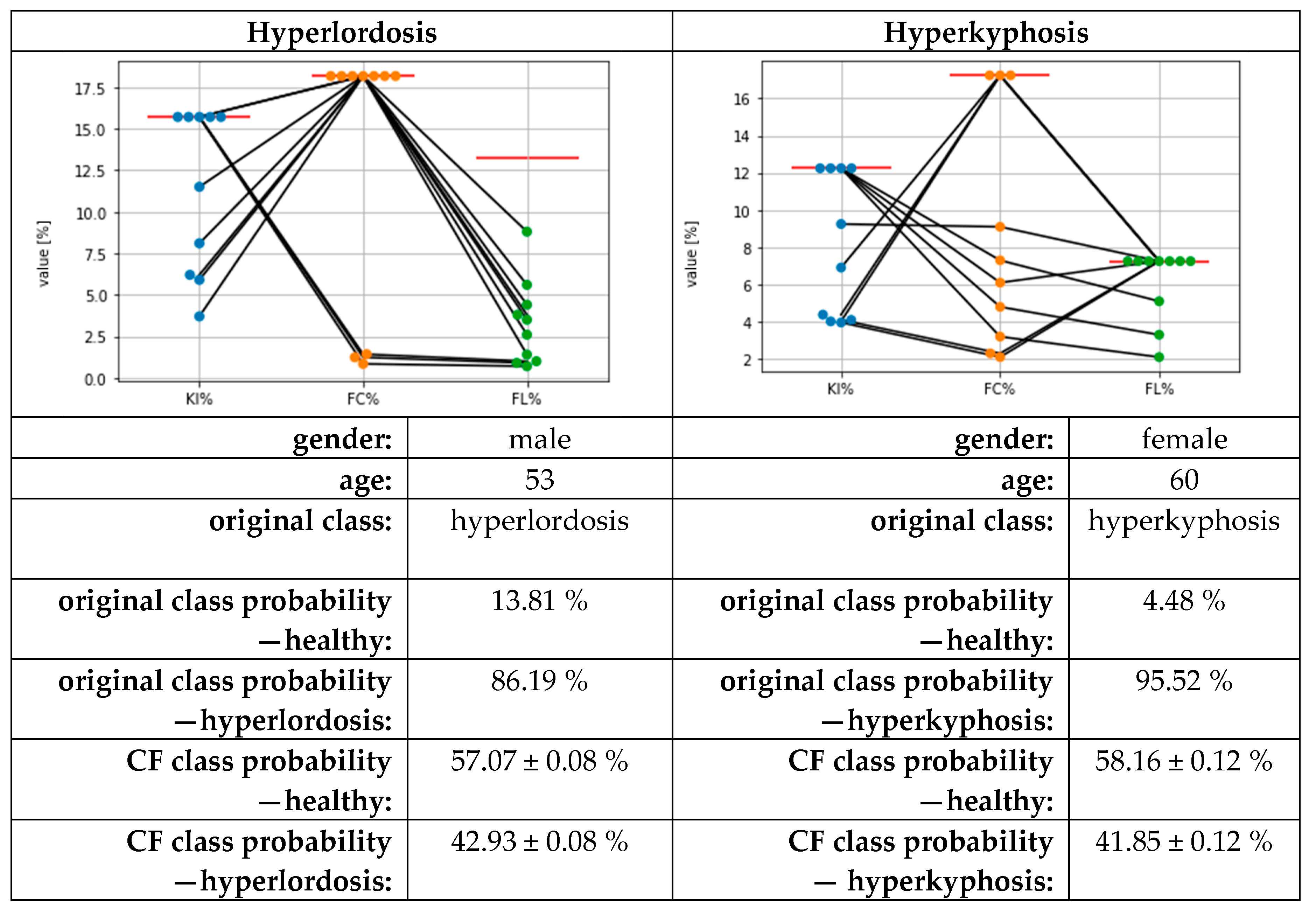
3.3. Results for Counterfactual Explanations
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dindorf, C.; Teufl, W.; Taetz, B.; Becker, S.; Bleser, G.; Fröhlich, M. Feature extraction and gait classification in hip replacement patients on the basis of kinematic waveform data. Biomedical Human Kinetics 2021, 13, 177–186. [Google Scholar] [CrossRef]
- Horst, F.; Lapuschkin, S.; Samek, W.; Müller, K.-R.; Schöllhorn, W.I. Explaining the unique nature of individual gait patterns with deep learning. Sci. Rep. 2019, 9, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Phinyomark, A.; Petri, G.; Ibáñez-Marcelo, E.; Osis, S.T.; Ferber, R. Analysis of Big Data in Gait Biomechanics: Current Trends and Future Directions. J. Med. Biol. Eng. 2018, 38, 244–260. [Google Scholar] [CrossRef] [PubMed]
- Halilaj, E.; Rajagopal, A.; Fiterau, M.; Hicks, J.L.; Hastie, T.J.; Delp, S.L. Machine learning in human movement biomechanics: Best practices, common pitfalls, and new opportunities. J. Biomech. 2018, 81, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Arnaout, R.; Curran, L.; Zhao, Y.; Levine, J.C.; Chinn, E.; Moon-Grady, A.J. An ensemble of neural networks provides expert-level prenatal detection of complex congenital heart disease. Nat. Med. 2021, 27, 882–891. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Bell, D.; Antani, S.; Xue, Z.; Yu, K.; Horning, M.P.; Gachuhi, N.; Wilson, B.; Jaiswal, M.S.; Befano, B.; et al. An Observational Study of Deep Learning and Automated Evaluation of Cervical Images for Cancer Screening. J. Natl. Cancer Inst. 2019, 111, 923–932. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.; Xu, G.; Li, C.; He, L.; Luo, L.; Wang, Z.; Jing, B.; Deng, Y.; Jin, Y.; Li, Y.; et al. Real-time artificial intelligence for detection of upper gastrointestinal cancer by endoscopy: a multicentre, case-control, diagnostic study. The Lancet Oncology 2019, 20, 1645–1654. [Google Scholar] [CrossRef]
- Lau, H.; Tong, K.; Zhu, H. Support vector machine for classification of walking conditions of persons after stroke with dropped foot. Hum. Mov. Sci. 2009, 28, 504–514. [Google Scholar] [CrossRef] [PubMed]
- Wahid, F.; Begg, R.K.; Hass, C.J.; Halgamuge, S.; Ackland, D.C. Classification of Parkinson’s Disease Gait Using Spatial-Temporal Gait Features. IEEE J. Biomed. Health Inform. 2015, 19, 1794–1802. [Google Scholar] [CrossRef]
- Mitchell, E.; Monaghan, D.; O’Connor, N.E. Classification of sporting activities using smartphone accelerometers. Sensors (Basel) 2013, 13, 5317–5337. [Google Scholar] [CrossRef]
- Begg, R.; Kamruzzaman, J. Neural networks for detection and classification of walking pattern changes due to ageing. Australas. Phys. Eng. Sci. Med. 2006, 29, 188–195. [Google Scholar] [CrossRef] [PubMed]
- Khodabandehloo, E.; Riboni, D.; Alimohammadi, A. HealthXAI: Collaborative and explainable AI for supporting early diagnosis of cognitive decline. Future Generation Computer Systems 2021, 116, 168–189. [Google Scholar] [CrossRef]
- Paulo, J.; Peixoto, P.; Nunes, U.J. ISR-AIWALKER: Robotic Walker for Intuitive and Safe Mobility Assistance and Gait Analysis. IEEE Trans. Human-Mach. Syst. 2017, 47, 1110–1122. [Google Scholar] [CrossRef]
- Laroche, D.; Tolambiya, A.; Morisset, C.; Maillefert, J.F.; French, R.M.; Ornetti, P.; Thomas, E. A classification study of kinematic gait trajectories in hip osteoarthritis. Comput. Biol. Med. 2014, 55, 42–48. [Google Scholar] [CrossRef] [PubMed]
- Teufl, W.; Taetz, B.; Miezal, M.; Lorenz, M.; Pietschmann, J.; Jöllenbeck, T.; Fröhlich, M.; Bleser, G. Towards an Inertial Sensor-Based Wearable Feedback System for Patients after Total Hip Arthroplasty: Validity and Applicability for Gait Classification with Gait Kinematics-Based Features. Sensors 2019, 19. [Google Scholar] [CrossRef]
- Dindorf, C.; Konradi, J.; Wolf, C.; Taetz, B.; Bleser, G.; Huthwelker, J.; Werthmann, F.; Bartaguiz, E.; Kniepert, J.; Drees, P.; et al. Classification and Automated Interpretation of Spinal Posture Data Using a Pathology-Independent Classifier and Explainable Artificial Intelligence (XAI). Sensors 2021, 21, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Fedorak, C.; Ashworth, N.; Marshall, J.; Paull, H. Reliability of the visual assessment of cervical and lumbar lordosis: how good are we? Spine (Phila Pa. 1976) 2003, 28, 1857–1859. [Google Scholar] [CrossRef] [PubMed]
- Moreira, R.; Teles, A.; Fialho, R.; Baluz, R.; Santos, T.C.; Goulart-Filho, R.; Rocha, L.; Silva, F.J.; Gupta, N.; Bastos, V.H.; et al. Mobile Applications for Assessing Human Posture: A Systematic Literature Review. Electronics 2020, 9, 1196. [Google Scholar] [CrossRef]
- Northcutt, C.G.; Athalye, A.; Mueller, J. Pervasive Label Errors in Test Sets Destabilize Machine Learning Benchmarks 2021.
- Northcutt, C.G.; Jiang, L.; Chuang, I.L. Confident Learning: Estimating Uncertainty in Dataset Labels 2021.
- Zhang, M.; Gao, J.; Lyu, Z.; Zhao, W.; Wang, Q.; Ding, W.; Wang, S.; Li, Z.; Cui, S. Characterizing Label Errors: Confident Learning for Noisy-Labeled Image Segmentation. In Medical image computing and computer assisted intervention -- MICCAI 2020: 23rd International Conference, Lima, Peru, October 4-8, 2020, Proceedings. Part I / Anne L. Martel, Purang Abolmaesumi, Danail Stoyanov, Diana Mateus, Maria A. Zuluaga, S. Kevin Zhou, Daniel Racoceanu, Leo Joskowicz (eds.); Martel, A., Abolmaesumi, P., Stoyanov, D., Mateus, D., Zuluaga, M.A., Zhou, S.K., Racoceanu, D., Joskowicz, L., Eds.; Springer: Cham, 2020; pp. 721–730. ISBN 978-3-030-59709-2. [Google Scholar]
- Northcutt, C.G.; Wu, T.; Chuang, I.L. Learning with Confident Examples: Rank Pruning for Robust Classification with Noisy Labels, 2017. Available online: https://arxiv.org/pdf/1705.01936.
- European Union. Regulation (EU) 2016/679 of the european parliament and of the council of 27 april 2016 on the protection of natural persons with regard to the processing of personal data and on the free movement of such data, and repealing directive 95/46/ec (General Data Protection Regulation). Official Journal of the European Union 2016, L 119, 1–88.
- Holzinger, A.; Biemann, C.; Pattichis, C.S.; Kell, D.B. What do we need to build explainable AI systems for the medical domain? Available online:. Available online: http://arxiv.org/pdf/1712.09923v1 (accessed on 20 February 2020).
- Dindorf, C.; Teufl, W.; Taetz, B.; Bleser, G.; Fröhlich, M. Interpretability of Input Representations for Gait Classification in Patients after Total Hip Arthroplasty. Sensors 2020, 20, 1–14. [Google Scholar] [CrossRef]
- Horst, F.; Slijepcevic, D.; Lapuschkin, S.; Raberger, A.-M.; Zeppelzauer, M.; Samek, W.; Breiteneder, C.; Schöllhorn, W.I.; Horsak, B. On the Understanding and Interpretation of Machine Learning Predictions in Clinical Gait Analysis Using Explainable Artificial Intelligence. Available online: http://arxiv.org/pdf/1912a.07737v1 (accessed on 10 March 2020).
- Adadi, A.; Berrada, M. Peeking Inside the Black-Box: A Survey on Explainable Artificial Intelligence (XAI). IEEE Access 2018, 6, 52138–52160. [Google Scholar] [CrossRef]
- Ribeiro, M.T.; Singh, S.; Guestrin, C. Why Should I Trust You?: Explaining the Predictions of Any Classifier. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. KDD ’16: The 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, 13-17.08. 2016; Krishnapuram, B., Shah, M., Smola, A., Aggarwal, C., Shen, D., Rastogi, R., Eds.; New York, 2016; pp 1135–1144.
- Lundberg, S.M.; Lee, S.-I. A Unified Approach to Interpreting Model Predictions. In 31st Conference on Neural Information Processing Systems (NIPS 2017); Curran Associates Inc.: Red Hook, NY, United States, 2017; pp. 1–10. [Google Scholar]
- Shrikumar, A.; Greenside, P.; Kundaje, A. Learning important features through propagating activation differences. In Proceedings of the 34th International Conference on Machine Learning; ICML: Sydney, Australia, 2017; pp. 3145–3153. [Google Scholar]
- Molnar, C. Interpretable Machine Learning: A Guide for Making Black Box Models Explainable; Leanpub: n.p., 2018. [Google Scholar]
- World Medical Association Declaration of Helsinki: ethical principles for medical research involving human subjects. JAMA 2013, 310, 2191–2194. [CrossRef] [PubMed]
- Kechagias, V.A.; Grivas, T.B.; Papagelopoulos, P.J.; Kontogeorgakos, V.A.; Vlasis, K. Truncal Changes in Patients Suffering Severe Hip or Knee Osteoarthritis: A Surface Topography Study. Clin Orthop Surg 2021, 13, 185. [Google Scholar] [CrossRef] [PubMed]
- Khallaf, M.E.; Fayed, E.E. Early postural changes in individuals with idiopathic Parkinson’s disease. Parkinsons. Dis. 2015, 2015, 369454. [Google Scholar] [CrossRef] [PubMed]
- Zytek, A.; Arnaldo, I.; Liu, D.; Berti-Equille, L.; Veeramachaneni, K. The Need for Interpretable Features: Motivation and Taxonomy, 2022. Available online: https://arxiv.org/pdf/2202.11748.
- Ludwig, O.; Dindorf, C.; Kelm, J.; Simon, S.; Nimmrichter, F.; Fröhlich, M. Reference Values for Sagittal Clinical Posture Assessment in People Aged 10 to 69 Years. Int. J. Environ. Res. Public Health 2023, 20. [Google Scholar] [CrossRef]
- Lemaitre, G.; Nogueira, F.; Aridas, C.K. Imbalanced-learn: A Python Toolbox to Tackle the Curse of Imbalanced Datasets in Machine Learning, 2016. Available online: https://arxiv.org/pdf/1609.06570.
- Buchanan, J.J.; Schneider, M.D.; Armstrong, R.E.; Muyskens, A.L.; Priest, B.W.; Dana, R.J. Gaussian Process Classification for Galaxy Blend Identification in LSST. ApJ 2022, 924, 94. [Google Scholar] [CrossRef]
- Desai, R.; Porob, P.; Rebelo, P.; Edla, D.R.; Bablani, A. EEG Data Classification for Mental State Analysis Using Wavelet Packet Transform and Gaussian Process Classifier. Wireless Pers Commun 2020, 115, 2149–2169. [Google Scholar] [CrossRef]
- Wang, B.; Wan, F.; Mak, P.U.; Mak, P.; Vai, M.I. EEG signals classification for brain computer interfaces based on Gaussian process classifier. In Information, Communications and Signal Processing, 2009. ICICS 2009. 7th International Conference on. Signal Processing (ICICS), Macau, China, 12/8/2009 - 12/10/2009; IEEE, 2009; pp 1–5, ISBN 978-1-4244-4656-8.
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Mothilal, R.K.; Sharma, A.; Tan, C. Diverse Counterfactual Explanations (DiCE) for ML: How to explain a machine learning model such that the explanation is truthful to the model and yet interpretable to people? Available online:. Available online: https://github.com/interpretml/DiCE (accessed on 2 June 2022).
- Hsieh, C.; Moreira, C.; Ouyang, C. DiCE4EL: Interpreting Process Predictions using a Milestone-Aware Counterfactual Approach. In 2021 3rd International Conference on Process Mining (ICPM). 2021 3rd International Conference on Process Mining (ICPM), Eindhoven, Netherlands, 10/31/2021 - 11/4/2021; IEEE, uuuu-uuuu; pp 88–95, ISBN 978-1-6654-3514-7.
- Jones, E.; Oliphant, T.; Peterson, P.; et al. SciPy: Open Source Scientific Tools for Python. Available online: http://www.scipy.org (accessed on 3 September 2019).
- Cohen, J. A power primer. Psychol. Bull. 1992, 112, 155–159. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Yu, L. Toward Integrating Feature Selection Algorithms for Classification and Clustering. IEEE Transactions on Knowledge and Data Engineering 2005, 17, 491–502. [Google Scholar] [CrossRef]
- Patias, P.; Grivas, T.B.; Kaspiris, A.; Aggouris, C.; Drakoutos, E. A review of the trunk surface metrics used as Scoliosis and other deformities evaluation indices. Scoliosis 2010, 5, 12. [Google Scholar] [CrossRef]
- Grath, R.M.; Costabello, L.; van Chan; Sweeney, P.; Kamiab, F.; Shen, Z.; Lecue, F. Interpretable Credit Application Predictions With Counterfactual Explanations, 2018. Available online: https://arxiv.org/pdf/1811.05245.
- Teufl, W.; Taetz, B.; Miezal, M.; Dindorf, C.; Fröhlich, M.; Trinler, U.; Hogam, A.; Bleser, G. Automated detection of pathological gait patterns using a one-class support vector machine trained on discrete parameters of IMU based gait data. Clinical Biomechanics 2021, 89, 1–7. [Google Scholar] [CrossRef] [PubMed]
| Hyperkyphosis | Hyperlordosis | |||
| n | % | n | % | |
| Highlighted labels | 130 | 11.31% | 110 | 9.57% |
| Agreement of the first two reviewers | 94 | 72.31% | 89 | 80.91% |
| Labels additionally assessed by a third expert | 36 | 27.69% | 21 | 19.09% |
| Highlighted labels corrected | 112 | 86.15% | 98 | 89.09% |
| Type | Feature | Description |
| subject characteristics | age | in years |
| gender | male/female | |
| height | body height in cm | |
| weight | body weight in kg | |
| BMI | weight/height² | |
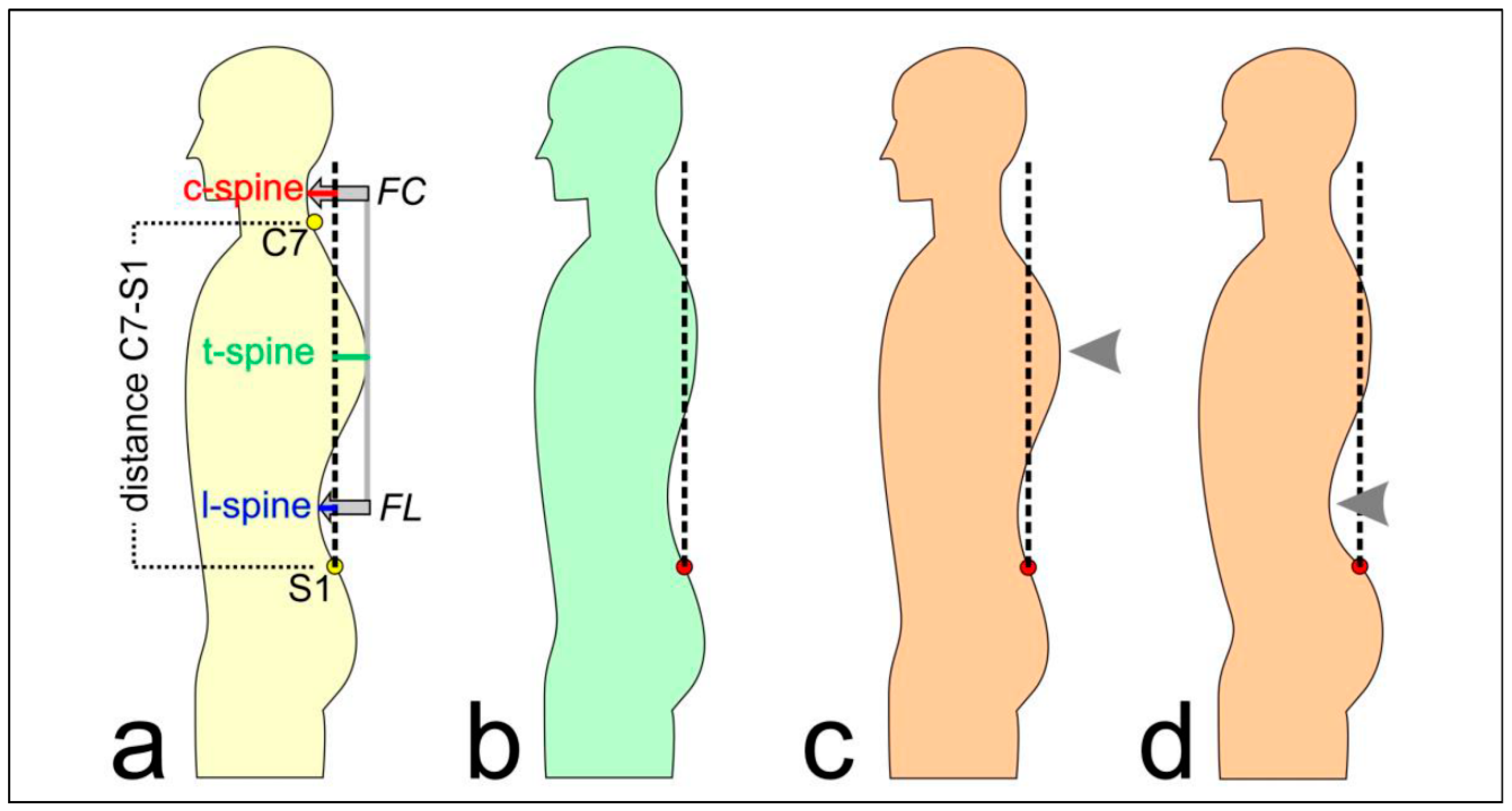
| directly measured by system | distance C7-S1 | vertical distance between the 7th cervical and the 1st sacral vertebrae in mm |
| c-spine | horizontal distance between the apex of the cervical lordosis and the perpendicular axis through the 1st sacral vertebrae in mm | |
| t-spine | horizontal distance between the apex of the thoracal kyphosis and the perpendicular axis through the 1st sacral vertebrae in mm | |
| l-spine | horizontal distance between the apex of the lumbar lordosis and the perpendicular axis through the 1st sacral vertebrae in mm | |
| calculated features | KI | (FC-FL)/2 |
| FC | abs(c-spine - t-spine) | |
| FL | abs(l-spine - t-spine) | |
| normalized features | KI% | KI*100/ distance C7-S1 |
| FC% | FC*100/ distance C7-S1 | |
| FL% | FL*100/ distance C7-S1 |
| Hyperkyphosis | Hyperlordosis | |||
| n | % | n | % | |
| Highlighted labels | 130 | 11.31% | 110 | 9.57% |
| Agreement of the first two reviewers | 94 | 72.31% | 89 | 80.91% |
| Labels additionally assessed by a third expert | 36 | 27.69% | 21 | 19.09% |
| Highlighted labels corrected | 112 | 86.15% | 98 | 89.09% |
| Hyperkyphosis | Hyperlordosis | ||
| Test performance (on given test labels) using Gaussian process classifier | MPRAUC | 0.80 ± 0.06 | 0.84 ± 0.05 |
| MF1 | 0.78 ± 0.03 | 0.77 ± 0.03 | |
| MMCC | 0.64 ± 0.05 | 0.63 ± 0.05 | |
| Test performance (on corrected test labels) using Gaussian process classifier | MPRAUC | 0.97 ± 0.01 | 0.97 ± 0.01 |
| MF1 | 0.90 ± 0.03 | 0.88 ± 0.04 | |
| MMCC | 0.85 ± 0.05 | 0.82 ± 0.06 | |
| Test performance (on given test labels) using Gaussian process classifier (+ confident learning on training data) |
MPRAUC | 0.78 ± 0.06 | 0.83 ± 0.04 |
| MF1 | 0.76 ± 0.04 | 0.77 ± 0.03 | |
| MMCC | 0.61 ± 0.05 | 0.64 ± 0.05 | |
| Test performance (on corrected test labels) using Gaussian process classifier (+ confident learning on training data) |
MPRAUC | 0.97 ± 0.01 | 0.97 ± 0.02 |
| MF1 | 0.89 ± 0.04 | 0.89 ± 0.03 | |
| MMCC | 0.82 ± 0.06 | 0.82 ± 0.06 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





