1. Introduction
Since the beginning of the COVID-19 pandemic, tremendous efforts have been made by the scientific community to find therapeutic approaches for the treatment of SARS-CoV-2-induced respiratory disease. In this ongoing research, the repurposing of approved drugs has been considered the most rapid, affordable and efficient strategy [
1], and numerous available drugs have thus been tested in repositioning studies over the last two years.
Among the tested drugs, methotrexate (MTX), a widely used chemotherapy and immunosuppressant drug [
2,
3,
4,
5,
6,
7,
8,
9], has shown anti-viral effects against SARS-CoV-2 [
10,
11]. As other anti-folates, MTX exerts its anti-cancer function mainly by inhibiting the dihydrofolate reductase (DHFR), an enzyme involved in the de novo synthesis of the nucleosides required for nucleic acid production. This effect results in an anti-metabolic but also anti-inflammatory effects due to the subsequent direct or indirect inhibition of several cellular mechanisms, such as lymphocytes replication, polyamine production, redox cellular activities and cytokines release [
12] and [
13,
14,
15,
16,
17,
18,
19]. The activity on inflammation as well as nucleic acid metabolism and purine synthesis were suggested to be at the basis of the possible use of MTX in COVID-19 [
20]. However, additional evidences showed a role of this drug in regulating also the activity of the Angiotensin Converting Enzyme 2 (ACE2) and the interaction of a human helicase with Spike (S) and Transmembrane Serine Protease 2 (TMPRSS2) (
https://opendata.ncats.nih.gov/covid19 [
20,
21,
22]. All these data indicate that MTX could also interfere with viral entry and replication by targeting host proteins.
Belonging to the same drug class of MTX, pralatrexate (PTX) is another DHFR inhibitor used for the treatment of relapsed or refractory peripheral T-cell lymphoma (PTCL). Interestingly, an effect in counteracting COVID-19 disease has been observed also for this compound [
23], thus suggesting that also other DHFR inhibitors may have anti-viral effects against SARS-CoV-2.
Starting from these data, in this study we evaluated the anti-viral activity of several DHFR inhibitors, specifically PTX, trimetrexate (TMX), aminopterin, as well as MTX, pemetrexed and raltitrexed, against SARS-CoV-2. Then, we assessed the effects of these compounds in inhibiting virus entry and, taking advantage of our EXSCALATE platform for molecular docking simulations, we also examined other targets and mechanisms potentially involved in the inhibition of SARS-CoV-2 infection mediated by DHFR inhibitors that could explained the different anti-viral activities of these compounds.
2. Materials and Methods
Cells
Vero E6 cells were grown and treated as previously described [
24]. A549 ACE2+ cells, a kind gift of Prof. Steven J. Elledge (Harvard Medical School, Boston, MA USA), were maintained in Dulbecco's Modified Eagle Medium (DMEM; Gibco, Thermo-Fisher) supplemented with 10% fetal bovine serum (FBS; Gibco, Thermo-Fisher) at 37°C in a humidified atmosphere of 5% CO2.
Virus
Infections were carried out using the SARS-CoV-2 B.1 lineage [
11,
65] at a multiplicity of infection (MOI) of 0.05. All the infection experiments were performed in a biosafety level-3 (BLS-3) laboratory.
Cell viability studies of compounds
Cells were seeded into 24-well plates (2.5 x 104 cells/well) in DMEM supplemented with 10% FBS, and treated with the indicated doses of each compound at 37°C for 48 h. Cell viability was evaluated by measuring the ATP levels using CellTiter-Glo (Promega, Madison, WI, USA).
Evaluation of antiviral efficacy of compounds
Cells were infected at 37°C for 1 h with the SARS-CoV-2 at a MOI of 0.05. Infections were carried out in DMEM without FBS. Then, the virus was removed and cells washed with warm phosphate buffered saline (PBS) and cultured with medium containing 2% FBS in the presence or in the absence of different concentrations of each compound. Cells and supernatants were collected for further analysis at 48 h post infection (p.i).
Plaque Assay
Cells were seeded at a density of 5 x 105 cells/well in a 12-well plate and incubated at 37°C for 24 h. Supernatants from infected cells were serially diluted in DMEM without FBS and added to the cells. After 1 h incubation, media were removed and cells washed with warm PBS. Then cells were covered with an overlay consisting of DMEM with 0.4% SeaPlaque (Lonza, Basel, Switzerland). The plates were further incubated at 37°C for 48 h. Cells were fixed with 10% formaldehyde at room temperature for 3 h. Formaldehyde was aspirated and the agarose overlay was removed. Cells were then stained with crystal violet (1% CV w/v in a 20% ethanol solution), and viral titer (PFU/mL) of SARS-CoV-2 was determined by counting the number of plaques.
Viral RNA extraction and quantitative real-time RT-PCR (qRT-PCR)
RNA was extracted from clarified cell culture supernatants (16,000 g x 10 min) and from infected cells using QIAamp Viral RNA Mini Kit and RNeasy Plus mini kit (Qiagen, Hilden, Germany), respectively, according to the manufacturer’s instructions.
RNA was eluted in 30 μl of RNase-free water and stored at -80°C till use. The qRT-PCR was carried-out following previously described procedures [
66]. Briefly, reverse transcription and amplification of the S gene were performed using the one-step QuantiFast Sybr Green RT-PCR mix (Qiagen) as follows: 50°C for 10 min, 95oC for 5 min; 95oC for 10 sec, 60oC for 30 sec (40 cycles) (primers: RBD-qF1: 5’-CAATGGTTTAACAGGCACAGG-3’ and RBD-qR1: 5’-CTCAAGTGTCTGTGGATCACG-3). A standard curve was generated by determination of copy numbers derived from serial dilutions (103-109 copies) of a pGEM T-easy vector (Promega, Madison, WI, USA) containing the receptor binding domain of the S gene (primers: RBD-F: 5’-GCTGGATCCCCTAATATTACAAACTTGTGCC-3’; RBD-R: 5’-TGCCTCGAGCTCAAGTGTCTGTGGATCAC-3’).
Western blot analysis
Western blot was carried-out following previously described procedures with minor modifications [
67]. Protein samples (30 µg) obtained from lysis in RIPA buffer (Cell Signaling Technology, Danvers, MA, USA) of infected cells were separated on 10% SDS-PAGE and then transferred onto polyvinylidene difluoride (PVDF) membranes (Millipore, Sigma, Burlington, MA, USA). After being blocked with 3% BSA in TBS buffer containing 0.05% Tween20, the blot was probed with a human serum (1:1000 dilution) containing IgG to the SARS-CoV-2 nucleoprotein (NP) and with mouse anti-human GAPDH monoclonal antibody (G-9: Santa Cruz Biotechnology, Dallas, TX, USA). The antigen-antibody complexes were detected using peroxidase-conjugated goat anti-human or goat anti-mouse IgG (Sigma) and revealed using the enhanced chemiluminescence (ECL) system (Santa Cruz Biotechnology).
Data analysis
The half-cytotoxic concentration (CC50) and the half-maximal inhibitory concentration (IC50) for each compound were calculated from concentration-effect-curves after non-linear regression analysis using Prism8 Software (GraphPad Software, La Jolla, CA, USA). The selectivity index (SI) was calculated as the ratio of CC50 over IC50 [
36].
Statistical analysis
Data were analyzed for statistical significance using the 1-way ANOVA, and the Bonferroni post-test was used to compare data. Differences were considered significant when p < 0.05. Statistical tests were performed using Prism8 Software (GraphPad Software).
Pseudovirus entry assay
Cell-free lentiviral particles were generated as described previously[
68] and production protocols are available at the LeGO website (
http://www.LentiGO-Vectors. de). Lentiviral particles (2.9x105 TU/ml) used for this study contain the D614G variant of SPIKE without the last 19 amino acids to remove the ER-retention signal. Caco-2 cells, obtained from Cell Lines Service (CLS, #300137), were grown in DMEM High Glucose (4.5 g/L) (Capricorn, #DMEM-HXRXA) with 10% fetal bovine serum (Capricorn, #FBS-12A), L-Glutamine (Capricorn, #GLN-B), streptomycin (100 μg/ml), and 100 U/ml penicillin (Capricorn, #PS-B). Cells were seeded in 20 µl at a density of 8000 cells per well into white 384-well microtiter plates (Greiner Bio-One, #781073) and incubated at 37°C in the presence of 5% CO2 for 24 h. Compounds were added using Echo550 (Labcyte Inc.) directly prior virus addition. Virus addition was done in 10 µl/well and incubated for 48 h at 37°C. Detection was done by addition of 30 µl of 0,5 mM Luciferin (Biosynth Carbosynth; #FL08608) solution in PBS and incubation at 10 min in the dark at RT and measurement of Luminescence using the EnSight multimode plate reader with 100 ms detection time per well. Data analysis was performed using Microsoft Excel and GraphPad Prism 8. Test compound results were normalized relative to the DMSO control that represents 0% inhibition of lentiviral pseudovirus entry. Dose response curves were fitted to 4-parameter logistic functions in Prism8 Software (GraphPad Software) with no constrains.
Nsp13 enzymatic assay
SARS-CoV-2 nsp13 was expressed from pNIC-ZB vector (Addgene plasmid #159614;
http://n2t.net/addgene:159614; RRID:Addgene_159614) in Rosetta cells, using TB medium for culture and purified according to Newman et al. [
69]. SARS-CoV-2 nsp13 enzymatic activity was evaluated as reported[
37]. Briefly, nsp13 unwinding-associated activity was measured in black 384 well plates (PerkinElmer), in 40 μl reaction volume containing 20 mM Tris–HCl, pH 7.2, 50 mM NaCl, 2 μM Hel Capture oligo (5'- TGG TGC TCG AAC AGT GAC -3’) from Biomers, 5 mM MgCl2, 5% DMSO or inhibitor and 1 nM of purified nsp13. The reaction mixture containing the enzyme was pre-incubated for 10 min with inhibitor at room temperature (RT). The reaction was started adding 1 mM ATP and 750 nM annealed DNA substrate (5'- AGT CTT CTC CTG GTG CTC GAA CAG TGA C-Cy3-3', 5'- BHQ-2-GTC ACT GTT CGA GCA CCA CCT CTT CTG A-3’) from Biomers. After 15 min of incubation at 37°C, products were measured with Victor Nivo (Perkin) at 530/580 nm.
SARS-CoV-2 nsp12 RNA polymerase RNA dependent (RdRp) activity.
SARS-CoV-2 nsp12 was expressed from pET28a vector in BL21 DE3 cells, as previously described[
70]. Briefly, the protein was firstly purified with Ni-Sepharose column and eluted in a buffer contained 20 mM Tris pH8, 150 mM NaCl, 1M Imidazole and 4mM MgCl2. The fraction contained the protein was loaded in a HiTrapQ-HP column and eluted in a buffer containing 20 mM Tris pH8, 1M NaCl and 4mM MgCl2. The quality of the protein was analyzed through SDS-PAGE and the purified protein was stored at -80°C. The SARS-CoV-2 nsp12 RdRp activity was measured in black 96 well plates (PerkinElmer), in 25 µl reaction volume containing 50 mM Tris-HCl pH 8.0, 1 mM DTT, 2.5 mM MgCl2, 50 mM NaCl, 10% Glicerolo, 20 µM UTP, RNAse inhibitor 10 U/µl (Thermo Scientific), 12.5 nM polyA RNA template, 4% DMSO or inhibitor and 400 nM of purified nsp12. The reaction mixture containing the enzyme and the template RNA was incubated for 60 min with the inhibitor at 37°C. After the incubation, the reaction was stopped with the addition of 2 µl of 200 mM nuclease-free EDTA. Then, 170 µl of 1X PicoGreen (Invitrogen) in 1X TE were added to the mixture and the reaction was incubated for 5 min at RT, protected from light. Products were measured with Victor Nivo (Perkin) at 502/523 (em/ex) nm. Experiments were performed in triplicate, the results report average and standard deviation of two independent replicates.
3. Results
3.1. Inhibitors of DHFR exert antiviral activity against SARS-CoV-2 in two cell lines
Previous studies have indicated an MTX-mediated antiviral activity against SARS-CoV-2 infection, possibly due to MTX effects on host cellular processes [
11,
20,
21]. To analyze the potential anti-viral activity of other DHFR inhibitors, we first performed a classical cellular phenotypic assay on Vero E6 cells, as previously described [
24]. This cell line has been extensively used for SARS-CoV-like virus studies and is highly susceptible to cell death after infection [
25,
26,
27,
28,
29,
30]. We used cells that constitutively expressed EGFP fluorescent protein, which thus allow for monitoring the effects of drug treatment in modulating the virus-induced cytopathic effect (CPE) by measuring EGFP fluorescence. In parallel, the compounds cytotoxicity was determined in the absence of virus to establish the half-maximal cytotoxic concentration (CC50). Although cells did not reach 100% of confluence after treatment due to the anti-metabolic activity of this class of compounds, the observed half-maximal inhibitory concentrations (IC50) were much lower than CC50 values, indicating good therapeutic windows (
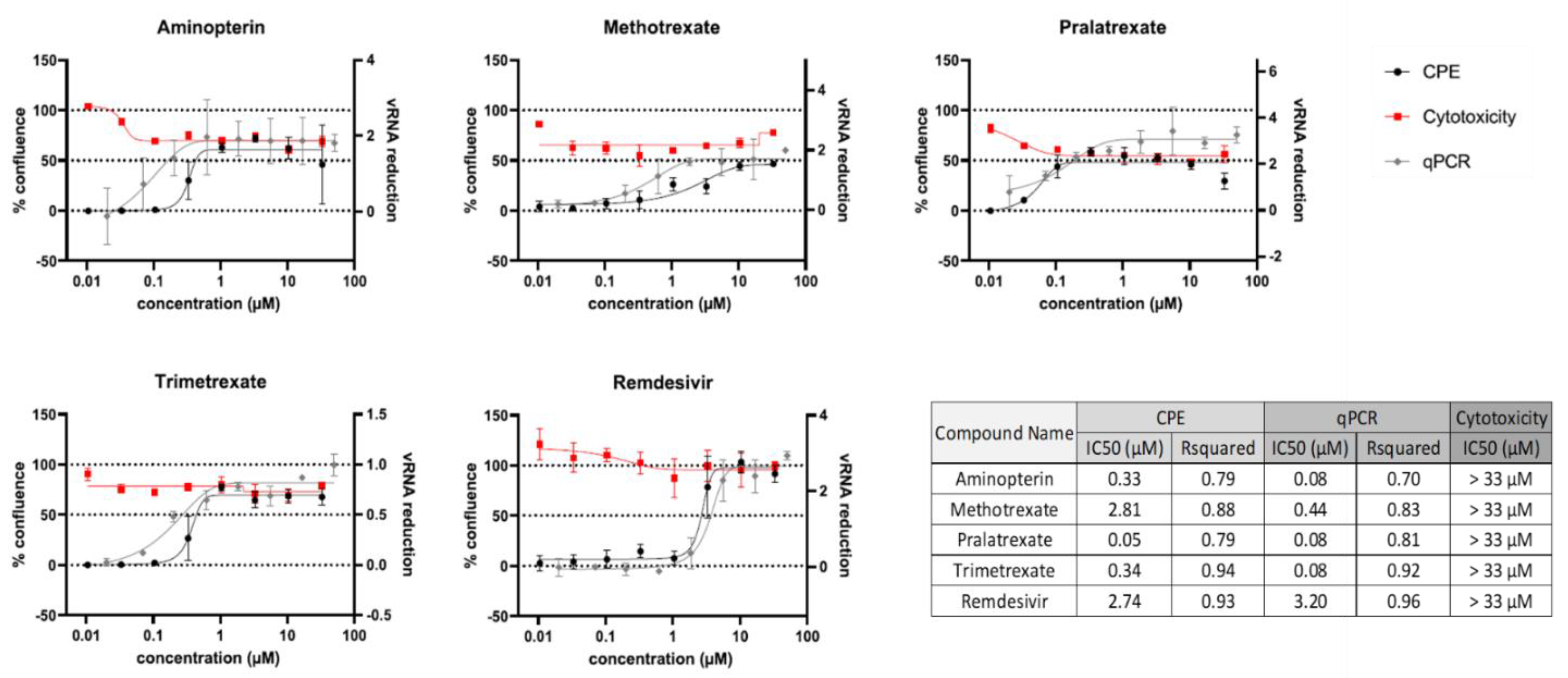
Figure 1). CPE analysis showed that PTX, TMX and aminopterin have the higher anti-viral activity in these cells, and quantitative PCR (qPCR) readouts evaluating the effects of compounds on viral replication were in line with these results (
Figure 1 and [
24]).
To further support our evidence, we tested this class of compounds on A549 ACE2+ cells, a human lung epithelial cell line engineered to stably overexpress the ACE2 receptor [
31,
32]. A549 ACE2+ cells are commonly used for in vitro screening and characterization of drug candidates against SARS-CoV-2 and have been already used to study MTX cellular pathways [
31,
33,
34,
35], using different approaches.
At first, a standard assay was carried out to measure the activity of each DHFR inhibitor on A549 ACE2+ cell metabolism and thus their selectivity index (SI) (Table1) [
11,
36]. To this end, cells were cultured for 48 h in the absence or presence of different drug concentrations. The cells treated with these compounds displayed a normal surface-adherent phenotype at all concentrations tested (
Figure 2A,
Figure 3A and
Figure 4A), and the CC50 value for PTX and TMX was found to be 0.008 µM and 0.01 µM, respectively (
Figure 2B,
Figure 3B and
Table 1). On the other hand, MTX and raltitrexed displayed a lower tolerability with CC50 values of 1.18 and 0.89 µM, respectively (
Figure 4B and
Table 1). Lastly, pemetrexed and aminopterin had the lowest tolerability with a CC50 higher than 2 µM (
Table 1).
To assess the anti-viral activity of the compounds, A549 ACE2+ cells were then infected with SARS-CoV-2 for 1 h at a multiplicity of infection (MOI) of 0.05 [
11] and, after infection, cells were cultured in the absence or presence of different concentrations of DHFR inhibitors. Supernatants were then collected at 48 h post infection (p.i.) and tested for viral genome copy numbers by quantitative real-time PCR (qRT-PCR). Three out of the six DHFR inhibitors tested showed a strong antiviral effect displaying a IC50 < 1 µM (
Table 1). The most active compounds were PTX, TMX and MTX, which significantly reduced the virus yield displaying a dose-dependent inhibition of viral replication (
Figure 2C,
Figure 3C and
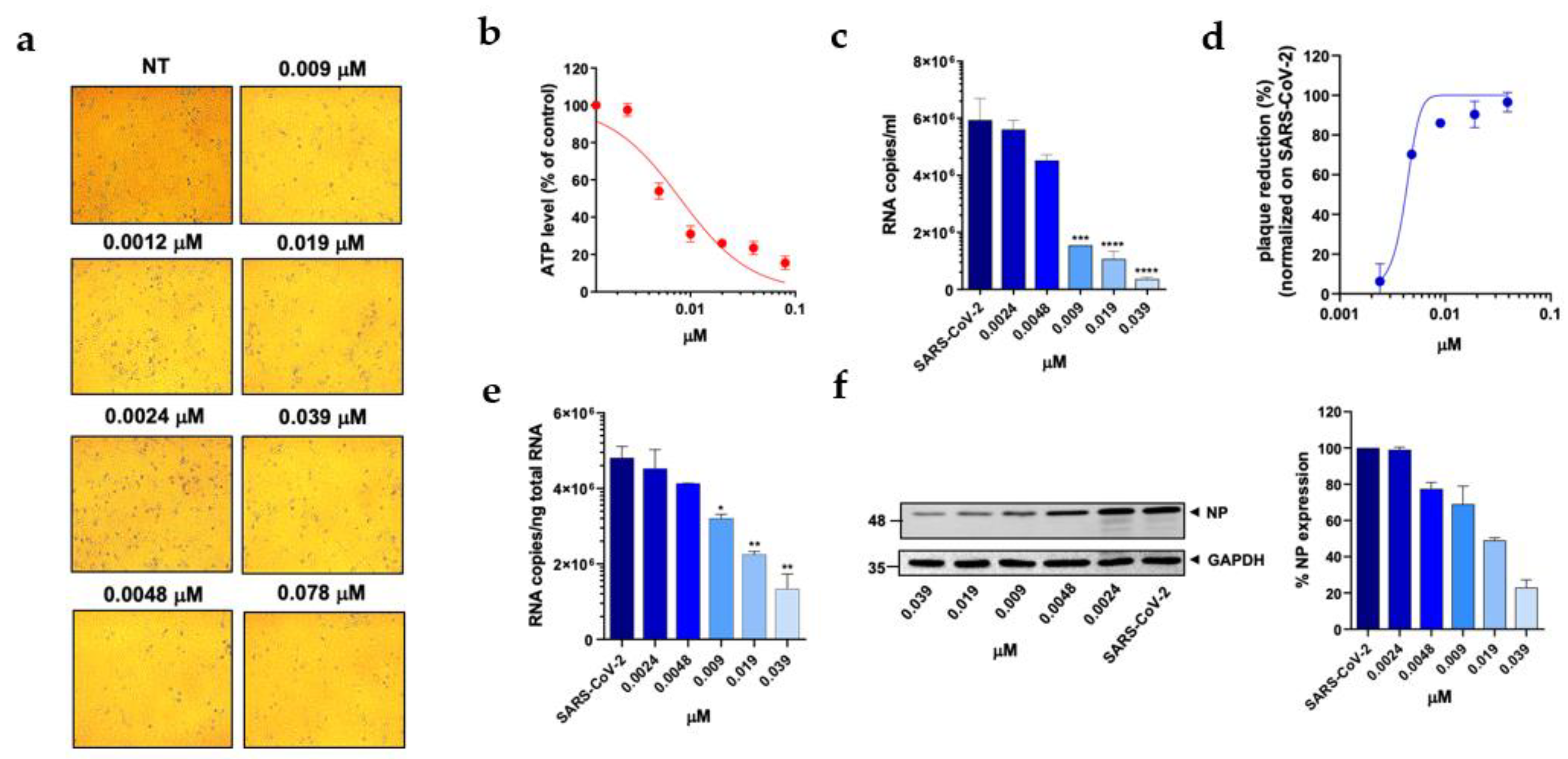
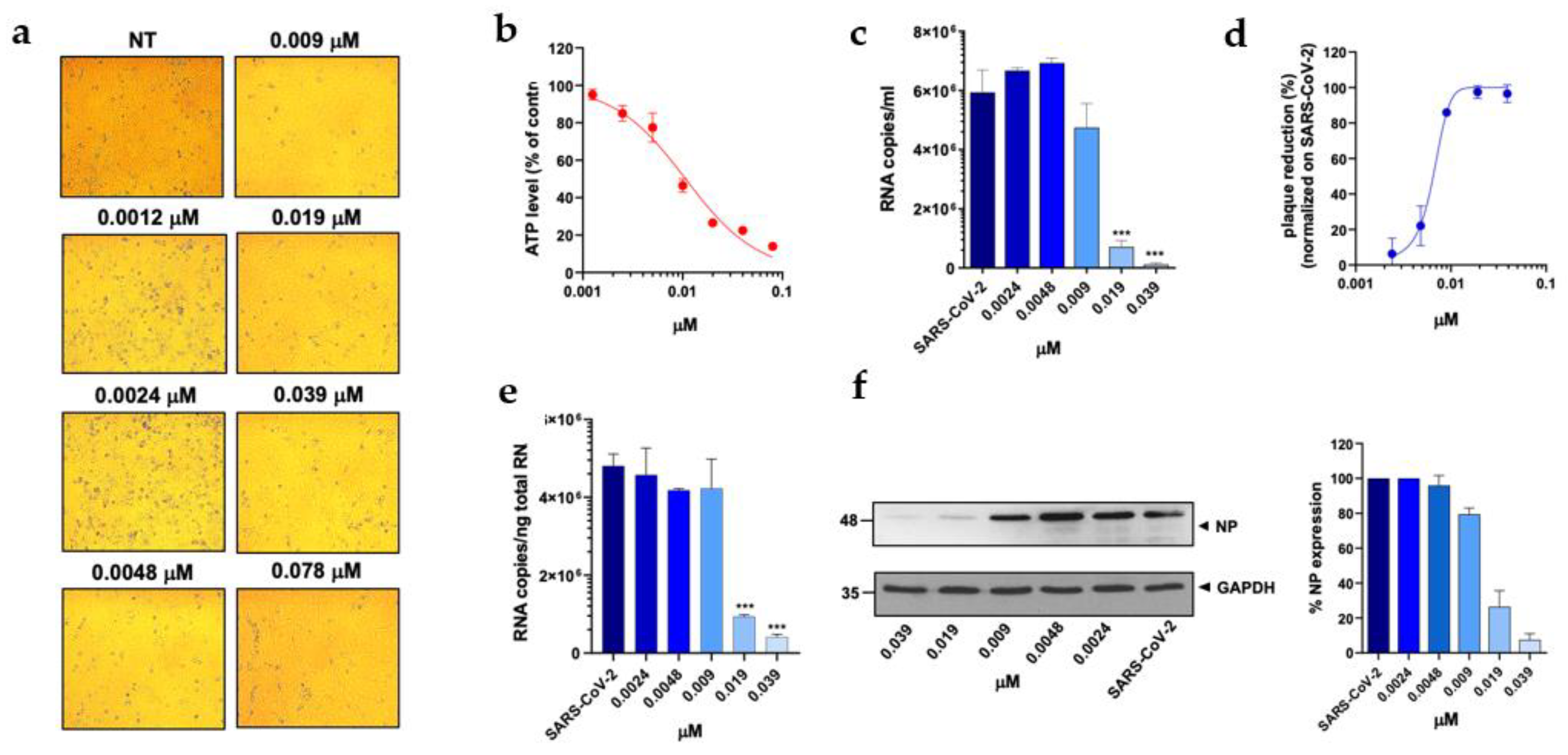
Figure 4C). In particular, PTX exhibited a 90.4% to 96.5% inhibition of viral titer at 0.019 µM and 0.039 µM, respectively (
Figure 2D). The efficacy of the treatment was also confirmed at the intracellular level by qRT-PCR and western blot (WB) on SARS-CoV-2 nucleoprotein (NP) (
Figure 2E and F), with the IC50 value calculated to be 0.004 µM. At the same time, TMX significantly reduced the SARS-CoV-2 virus yield, with an 86% reduction at a concentration of 0.009 µM and a 97.5% and 96.5% inhibition at 0.019 µM and 0.039 µM, respectively (
Figure 3D). The efficacy of the treatment was confirmed at the intracellular level by qRT-PCR and WB on NP (
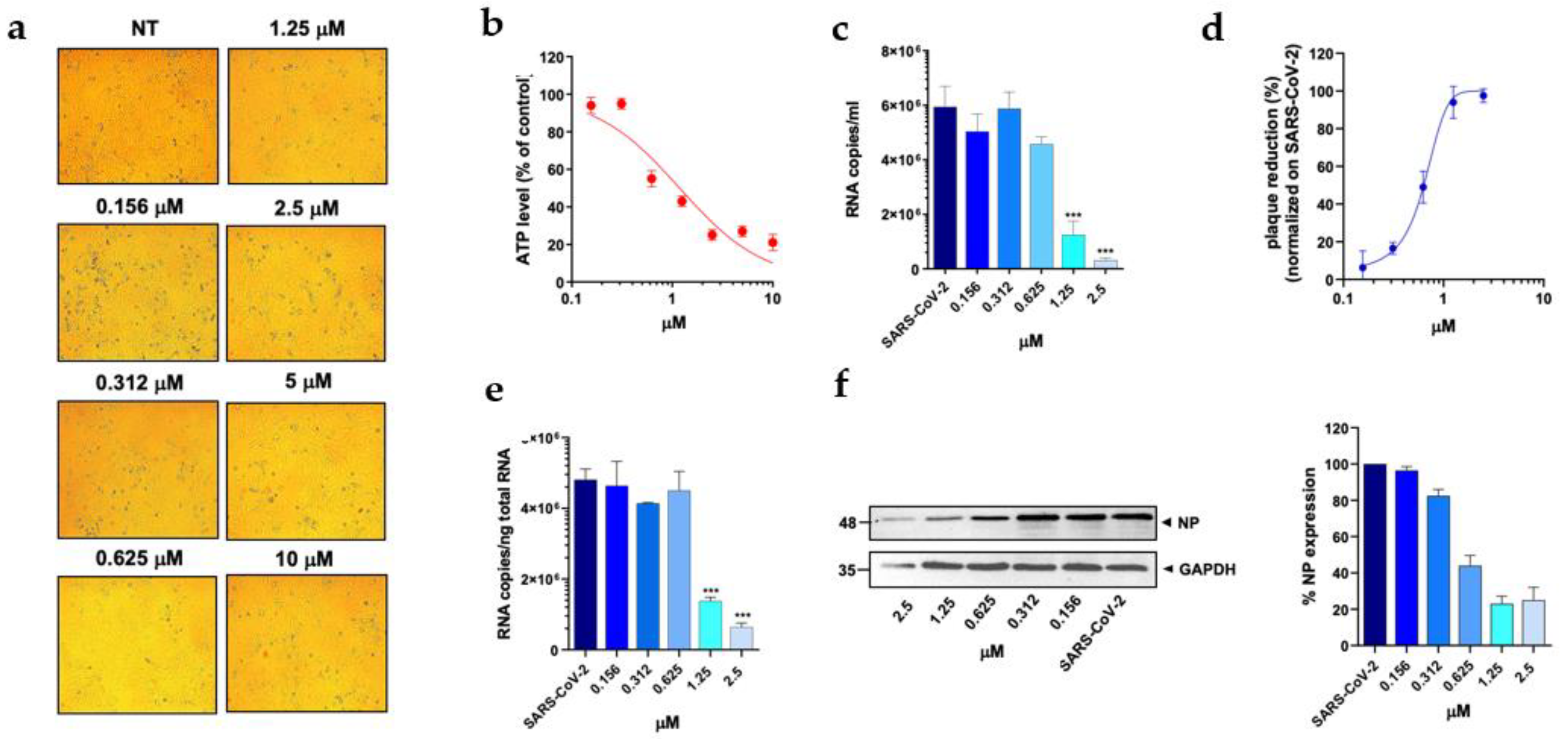
Figure 3E and F), with the IC50 value calculated to be 0.007 µM. Among the 3 compounds, MTX was the least efficient, with an IC50 value calculated to be 0.63 µM (Table1). In particular, MTX exhibited a 94% to 98% inhibition of viral titer at 1.25 µM and 2.5 µM, respectively (
Figure 4D). The efficacy of the treatment was also confirmed at the intracellular level by qRT-PCR and WB on SARS-CoV-2 NP (
Figure 4E and F). Collectively, these data indicate that PTX, TMX, and MTX, possess a high antiviral activity in the low nanomolar range. On the contrary, the remaining 3 tested compounds, aminopterin, pemetrexed and raltitrexed, showed low antiviral activities with IC50 values > 1 µM, and specifically of 1.3, 5 and 5 µM, respectively (
Table 1). Additionally, taken together these data demonstrate that the anti-cytopathic effect of DHFR inhibitors is due not only to the antimetabolic action of these compounds, but also to a specific antiviral activity.
3.2. PTX, TMX and MTX Inhibit the Activity of SARS-CoV-2 Viral Key Enzymes
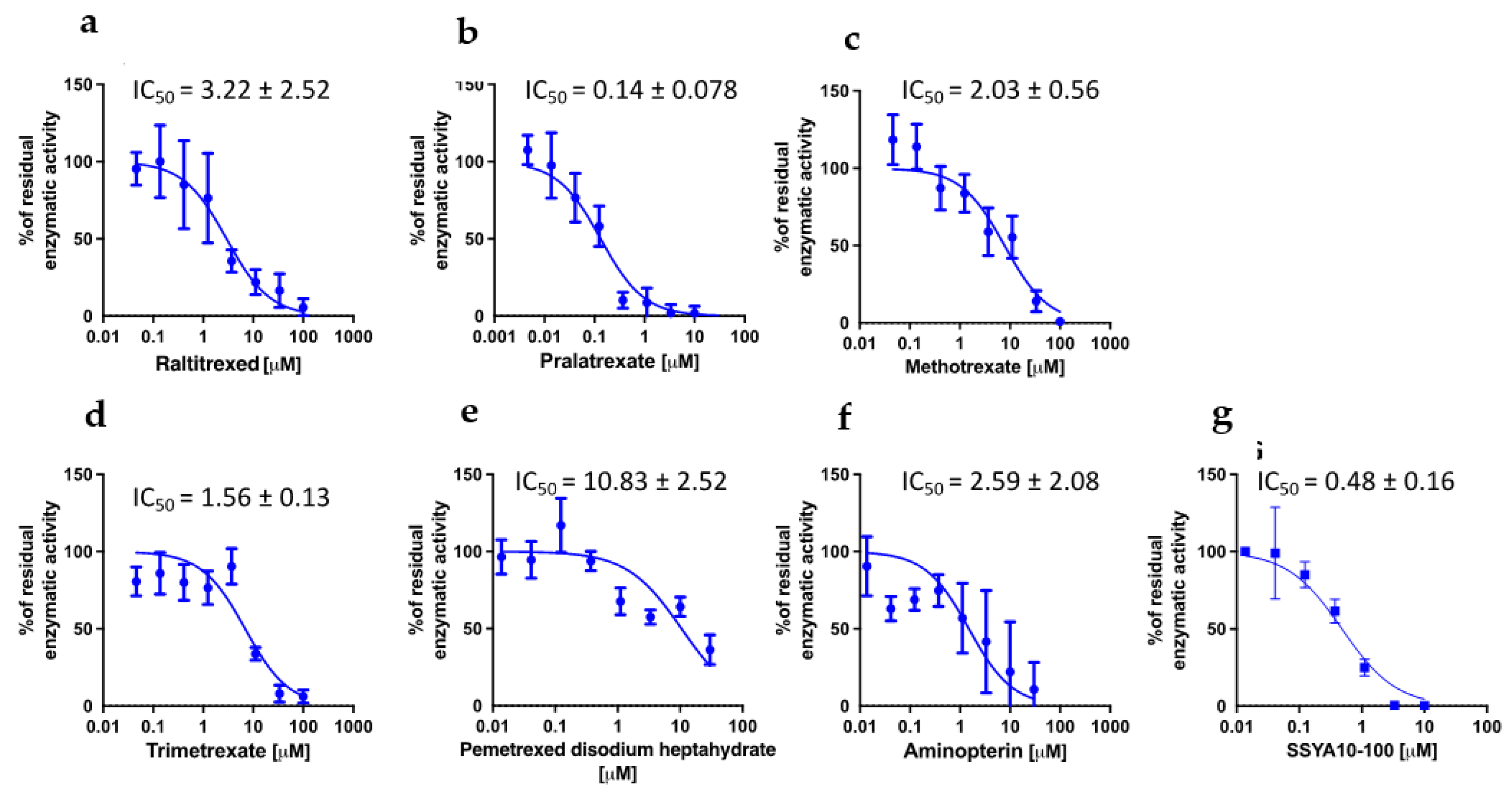
We then sought to understand whether the different antiviral activities of the compounds could be associated to different effects on viral proteins. To identify the potential targets of DHFR inhibitors among the viral proteins, we took advantage of the high-throughput screening campaign run in the context of the E4C consortium (
https://www.exscalate4cov.eu/). We performed a virtual screening using our EXSCALATE platform for molecular docking simulations, as already described. The simulation was performed using LiGen™ (Ligand Generator), the de novo structure-based virtual screening software, designed and developed to run on HPC architectures, which represents the most relevant tool of the EXSCALATE platform (
https://ieeexplore.ieee.org/document/9817028). From this analysis, we obtained the docking score values that predict the binding affinity of the molecules in the protein binding site and based on this information we thus tested the 6 compounds for their potential inhibitory effects against 2 important enzymes in mediating SARS-CoV-2 replication: nsp13 helicase, which is essential for viral replication [
37], and RNA-dependent RNA polymerase (RdRp), which is a highly versatile enzyme that is involve in RNA viral genome replication process. In line with data on anti-viral activity, PTX and TMX were the most effective in inhibiting the unwinding activity of nsp13 helicase, displaying IC50 values of 0.14 and 1.56 µM, respectively, while MTX showed and IC50 of 2.03 µM. Compared to PTX, TMX, and MTX, the other tested compounds were less effective in this assay, showing progressively higher IC50 values: 2.59 µM for aminopterin, 3.22 µM for raltitrexed and 10.83 µM for pemetrexed (
Figure 5). Finally, none of the tested compounds was able to inhibit RdRp (IC50 > 30 µM for all the tested compounds).
Altogether these data demonstrate that PTX, TMX and MTX have the highest anti-viral effect against SARS-CoV-2 due to a dual mechanism of action. Positive strand RNA viruses remodel cell metabolism to create a suitable microenvironment to survive and replicate in host cells. Indeed, the CPE observed in infected cells is ascribed to viral hijacking of cellular resources to fulfil viral needs. Anti-metabolite drugs, aimed to subtract nucleotides required for the synthesis of viral RNA or impair protein translation, act as broad-spectrum anti-viral drugs. Thus, DHFR inhibitors, on one side, inhibit cell metabolism, and on the other side, they can inhibit the function of key viral enzymes, thus exhibiting a pleiotropic effect. On the contrary, pemetrexed, raltitrexed and aminopterin that have shown a lower antiviral activity (IC50 > 1.5 µM), do not affect viral replication mechanisms, suggesting that their activity is only due to the anti-metabolic effect of this drug class.
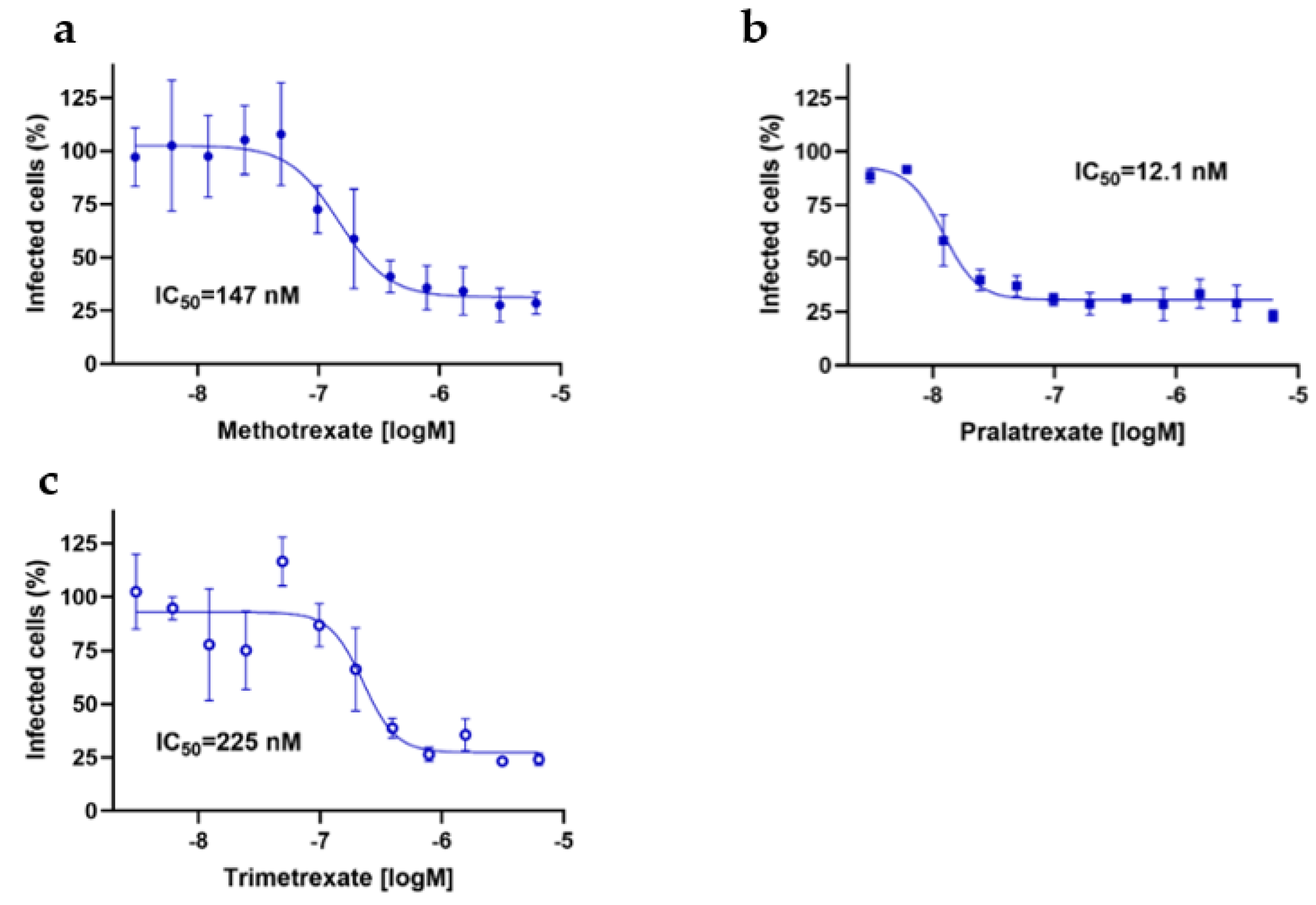
3.3. DHFR Inhibitors Inhibit Also SARS-CoV-2 Viral Entry
Since previous studies indicated that MTX can inhibit also viral entry by acting on virus-host interactions [
21], we finally investigated whether these mechanisms could be controlled also by PTX and TMX. Thus, we used the pseudovirus entry assay to investigate the potential effects of PTX and TMX on viral entry. To this end, permissive Caco-2 cells naturally expressing ACE2 were infected with SARS-CoV-2 pseudovirus. Notably, the pseudovirus used was a Lentiviral vector pseudotyped expressing SARS-CoV-2 spike protein (D614G) on the surface and carrying a luciferase (Luc2) reporter, thus allowing for following the infection.
Again, in line with the high anti-viral activity of these compounds, PTX, TMX and, confirming previous studies, also MTX significantly inhibited luciferase output with IC50 values in the nanomolar range, thus further demonstrating that these drugs exert their anti-viral activity through a polypharmacological effect as they can strongly interfere also with viral entry (
Figure 6). Furthermore, the potential inhibitory activity of DHFR inhibitors on serine protease TMPRSS2, which is important for spike protein priming, was assessed. Only PTX displayed a significant inhibition of TMPRSS2, with an IC50 of 0.45 µM, and this activity was not shared with TMX and MTX (IC50 > 30 µM; data not shown). Notably, PTX was the compound displaying the highest anti-viral activity, and also the one showing the highest and widest ability to target both viral proteins and mechanisms of viral entry, thus further demonstrating that the different anti-viral activities that we observed using different DHFR inhibitors depend on the pleiotropy of their mechanism of action.
4. Discussion
By combining in silico studies with experiments using human COVID-19 in vitro models, in this study we demonstrated that, in addition to MTX, also PTX and TMX possess strong anti-viral effects against SARS-CoV-2, and that their superior effects compared to other DHFR inhibitors in counteracting the viral infection are due to a polypharmacological activity, targeting not only the metabolism but also virus entry and other mechanisms involved in virus replication.
Although different studies have sought to explain the effects of MTX using different in vitro models of COVID-19, also suggesting a dual activity of the drug on both viral entry and virus replication in the host, to date the underlying mechanisms of these effects have not been completely explained and targets have not been clearly identified. Taking advantage of our EXSCALATE platform for molecular docking simulations, we identified potential targets of MTX and also of PTX and TMX, and then experimentally demonstrated the inhibition of NSP13 helicase as a new target for the mediation of the superior anti-viral activity of all three these DHFR inhibitors and the influence on viral entry as a common mechanism underlying the anti-viral effects of these drugs. Moreover, we identified TMPRSS2 as an additional target protein inhibited by the most effective PTX.
To fight SARS-CoV-2-induced respiratory disease is still a top priority for the scientific community, and investigating and explaining the potential anti-viral effects of drugs, such as DHFR inhibitors, that are used by patients that would be highly exposed to severe complications in case of infection, is of utmost importance. MTX is widely and successfully used for the treatment of cancer and autoimmune diseases, but its toxicity, poor pharmacokinetic and narrow safety range are certainly the major issues associated with its prolonged use [
38,
39] and often lead to dose reduction or withdrawal of treatment [
8,
40,
41]. MTX bioavailability is relatively high, but it is highly bound to plasma proteins and shows a very low volume of distribution, suggesting that its biodistribution may not be sufficient to reach the primary target tissues of the lung, while accumulating instead in liver and intestine [
13,
42]). PTX and TMX are known, respectively, as anti-cancer and anti-opportunistic infection agents [
43,
44,
45]). PDX has a more effective biodistribution ([
46] and
https://www.drugbank.com/), showed greater in vitro and in vivo anti-tumor efficacy [
47,
48]) and gave promising results in in vivo studies and clinical trials about toxicity, indicating a safer profile compared to MTX [
46,
47,
49,
50]. In addition, PTX is under study for treatment of Non-Small Cell Lung Cancer (NSCLC), suggesting that it can reach the lung more efficiently compared to MTX. On the other hand, TMX is the less studied drug among DHFR inhibitors. Differently from the above-mentioned drugs, it targets specifically DHFR but not the thymidylate synthase, suggesting that toxicity concerns typical of the other members of the same family could be overcome in this case. Interestingly, TMX has also an indication for lung fungal infections [
51] and was reported to be distributed into the respiratory tract [
52].
Our studies demonstrated that PTX and TMX have strong anti-viral efficacy against SARS-CoV-2 and that their higher anti-viral activity (nanomolar range) compared to the other compounds belonging to this drug class is due to their polypharmacological profile and pleiotropic effects. The anti-metabolic activity observed for the most effective DHFR inhibitors only partially explain the antiviral activity of these compounds, and a direct role on viral entry and replication mechanisms significantly contribute to the anti-CPE activity of these compounds, resulting in an additive antiviral effect. Thus, considering also their pharmacokinetic features, PTX and TMX could be even more effective compared to MTX in the management of SARS-CoV-2 infection-associated complications in patients affected by chronic disease who are already using these drugs.
First clinical evidence on the potential beneficial effects of the treatment with a DHFR inhibitor in COVID-19 disease was observed in patient receiving MTX for treating psoriasis or rheumatoid arthritis [
53,
54,
55,
56,
57,
58,
59]. These studies suggested that MTX treatment did not worsen COVID-19 outcomes and rates of hospitalization in these patients, probably acting on inflammation associated to SARS-CoV-2 infection [
60,
61,
62]; however, as the cohort of patients was heterogeneous in terms of period of MTX treatments and clinical manifestations of COVID-19, no clear conclusion on the benefits of MTX therapies could be extrapolated. On the other hand, further supporting the potential beneficial effect of MTX in COVID-19 patients treatment, the Global Rheumatology Alliance physician-reported registry recently reviewed the COVID-19 situation in rheumatic patients, suggesting that odds of death were higher in patients receiving different Disease-Modifying Anti-Rheumatic Drug (DMARD) or not receiving any DMARD compared with patients treated with MTX alone [
63].
Knowing of the strong, pleiotropic anti-viral activity of PTX and TMX can thus be extremely useful for physicians managing SARS-CoV-2 infections in patients with cancers or autoimmune diseases treated with these drugs. These patients are in fact often immunocompromised, cannot undergo vaccination or are at higher risk of insufficient immune response after vaccines and of developing severe COVID-19 disease [
64]. In this context, treatment with DHFR inhibitors with strong antiviral activity and better safety profile as PTX and TMX can thus be advantageous, allowing for treating the primary disease and, eventually, controlling COVID-19 disease.
5. Conclusions
With this study, we confirm the importance of repurposing studies and of in silico/experimental synergy as very powerful methods to generate effective responses to diseases that are still untreatable. Using this approach, we demonstrated that PTX, TMX and MTX have stronger anti-viral effects against SARS-CoV-2 compared to other DHFR inhibitors, and such higher efficacy is due to their pleiotropic inhibitory activity on cell metabolism as well as both viral entry and replication mechanisms. These compounds can thus potentially give a clinical advantage in the management of SARS-CoV-2 infection-associated complications in patients affected by chronic disease who are already treated with this class of drugs.
Author Contributions
Conceptualization and supervision: DI, FC, ARB, AC and MA; writing DI, RN, FC; Methodology: CT and CM generated the in silico data; MK,BE, KR performed the antiviral characterization on Vero E6 cells; FC, AZ, AB, FF performed the antiviral characterization on ACE2+ cells; FE, AC, ET generated the data on viral helicase and RdRp; BE contributed to study viral entry; Editing: MA, RN and DI ; DI, FC, PG, ARB, AC and MA provided strategic input. All authors have read and agreed to the published version of the manuscript.
Funding
This research was conducted under the project “EXaSCale smArt pLatform Against paThogEns for Corona Virus—Exscalate4CoV” founded by the EU’s H2020-SC1-PHE-CORONAVIRUS-2020 call, grant N. 101003551.
Data Availability Statement
Conflicts of Interest
The authors declare no conflict of interest.
References
- Rameshrad, M.; Ghafoori, M.; Mohammadpour, A.H.; Nayeri, M.J.D.; Hosseinzadeh, H. A comprehensive review on drug repositioning against coronavirus disease 2019 (COVID19). Naunyn Schmiedebergs Arch Pharmacol 2020, 393, 1137-1152. [CrossRef]
- Yu, W.J.; Huang, D.X.; Liu, S.; Sha, Y.L.; Gao, F.H.; Liu, H. Polymeric Nanoscale Drug Carriers Mediate the Delivery of Methotrexate for Developing Therapeutic Interventions Against Cancer and Rheumatoid Arthritis. Front Oncol 2020, 10, 1734. [CrossRef]
- Amjad, M.T.; Chidharla, A.; Kasi, A. Cancer Chemotherapy. In StatPearls, Treasure Island (FL), 2022.
- Theti, D.S.; Jackman, A.L. The role of alpha-folate receptor-mediated transport in the antitumor activity of antifolate drugs. Clin Cancer Res 2004, 10, 1080-1089. [CrossRef]
- Friedman, B.; Cronstein, B. Methotrexate mechanism in treatment of rheumatoid arthritis. Joint Bone Spine 2019, 86, 301-307. [CrossRef]
- Adegbola, S.O.; Sahnan, K.; Warusavitarne, J.; Hart, A.; Tozer, P. Anti-TNF Therapy in Crohn's Disease. Int J Mol Sci 2018, 19. [CrossRef]
- Elmamoun, M.; Chandran, V. Role of Methotrexate in the Management of Psoriatic Arthritis. Drugs 2018, 78, 611-619. [CrossRef]
- Shah, R.A.; Nwannunu, C.E.; Limmer, A.L.; Patel, R.R.; Mui, U.N.; Tyring, S.K. Brief Update on Dermatologic Uses of Methotrexate. Skin Therapy Lett 2019, 24, 5-8.
- Kozminski, P.; Halik, P.K.; Chesori, R.; Gniazdowska, E. Overview of Dual-Acting Drug Methotrexate in Different Neurological Diseases, Autoimmune Pathologies and Cancers. Int J Mol Sci 2020, 21. [CrossRef]
- Xing, J.; Shankar, R.; Drelich, A.; Paithankar, S.; Chekalin, E.; Dexheimer, T.; Chua, M.S.; Rajasekaran, S.; Tseng, C.K.; Chen, B. Analysis of Infected Host Gene Expression Reveals Repurposed Drug Candidates and Time-Dependent Host Response Dynamics for COVID-19. bioRxiv 2020, 10.1101/2020.04.07.030734. [CrossRef]
- Caruso, A.; Caccuri, F.; Bugatti, A.; Zani, A.; Vanoni, M.; Bonfanti, P.; Cazzaniga, M.E.; Perno, C.F.; Messa, C.; Alberghina, L. Methotrexate inhibits SARS-CoV-2 virus replication "in vitro". J Med Virol 2021, 93, 1780-1785. [CrossRef]
- Chan, E.S.; Cronstein, B.N. Methotrexate--how does it really work? Nat Rev Rheumatol 2010, 6, 175-178. [CrossRef]
- Bedoui, Y.; Guillot, X.; Selambarom, J.; Guiraud, P.; Giry, C.; Jaffar-Bandjee, M.C.; Ralandison, S.; Gasque, P. Methotrexate an Old Drug with New Tricks. Int J Mol Sci 2019, 20. [CrossRef]
- Cronstein, B.N. Low-dose methotrexate: a mainstay in the treatment of rheumatoid arthritis. Pharmacol Rev 2005, 57, 163-172. [CrossRef]
- Cronstein, B.N.; Eberle, M.A.; Gruber, H.E.; Levin, R.I. Methotrexate inhibits neutrophil function by stimulating adenosine release from connective tissue cells. Proc Natl Acad Sci U S A 1991, 88, 2441-2445. [CrossRef]
- Cronstein, B.N.; Naime, D.; Ostad, E. The antiinflammatory mechanism of methotrexate. Increased adenosine release at inflamed sites diminishes leukocyte accumulation in an in vivo model of inflammation. J Clin Invest 1993, 92, 2675-2682. [CrossRef]
- Montesinos, M.C.; Desai, A.; Delano, D.; Chen, J.F.; Fink, J.S.; Jacobson, M.A.; Cronstein, B.N. Adenosine A2A or A3 receptors are required for inhibition of inflammation by methotrexate and its analog MX-68. Arthritis Rheum 2003, 48, 240-247. [CrossRef]
- Herman, S.; Zurgil, N.; Deutsch, M. Low dose methotrexate induces apoptosis with reactive oxygen species involvement in T lymphocytic cell lines to a greater extent than in monocytic lines. Inflamm Res 2005, 54, 273-280. [CrossRef]
- Phillips, D.C.; Woollard, K.J.; Griffiths, H.R. The anti-inflammatory actions of methotrexate are critically dependent upon the production of reactive oxygen species. Br J Pharmacol 2003, 138, 501-511. [CrossRef]
- Stegmann, K.M.; Dickmanns, A.; Gerber, S.; Nikolova, V.; Klemke, L.; Manzini, V.; Schlosser, D.; Bierwirth, C.; Freund, J.; Sitte, M., et al. The folate antagonist methotrexate diminishes replication of the coronavirus SARS-CoV-2 and enhances the antiviral efficacy of remdesivir in cell culture models. Virus Res 2021, 302, 198469. [CrossRef]
- Liu, X.; Huuskonen, S.; Laitinen, T.; Redchuk, T.; Bogacheva, M.; Salokas, K.; Pohner, I.; Ohman, T.; Tonduru, A.K.; Hassinen, A., et al. SARS-CoV-2-host proteome interactions for antiviral drug discovery. Mol Syst Biol 2021, 17, e10396. [CrossRef]
- Chen, Z.; Wang, C.; Feng, X.; Nie, L.; Tang, M.; Zhang, H.; Xiong, Y.; Swisher, S.K.; Srivastava, M.; Chen, J. Interactomes of SARS-CoV-2 and human coronaviruses reveal host factors potentially affecting pathogenesis. EMBO J 2021, 40, e107776. [CrossRef]
- Bae, J.Y.; Lee, G.E.; Park, H.; Cho, J.; Kim, J.; Lee, J.; Kim, K.; Kim, J.I.; Park, M.S. Antiviral Efficacy of Pralatrexate against SARS-CoV-2. Biomol Ther (Seoul) 2021, 29, 268-272. [CrossRef]
- Zaliani, A.; Vangeel, L.; Reinshagen, J.; Iaconis, D.; Kuzikov, M.; Keminer, O.; Wolf, M.; Ellinger, B.; Esposito, F.; Corona, A., et al. Cytopathic SARS-CoV-2 screening on VERO-E6 cells in a large-scale repurposing effort. Sci Data 2022, 9, 405. [CrossRef]
- Kistner, O.; Barrett, P.N.; Mundt, W.; Reiter, M.; Schober-Bendixen, S.; Dorner, F. Development of a mammalian cell (Vero) derived candidate influenza virus vaccine. Vaccine 1998, 16, 960-968. [CrossRef]
- Kaye, M. SARS-associated coronavirus replication in cell lines. Emerg Infect Dis 2006, 12, 128-133. [CrossRef]
- Barrett, P.N.; Portsmouth, D.; Ehrlich, H.J. Vero cell culture-derived pandemic influenza vaccines: preclinical and clinical development. Expert Rev Vaccines 2013, 12, 395-413. [CrossRef]
- Li, W.; Moore, M.J.; Vasilieva, N.; Sui, J.; Wong, S.K.; Berne, M.A.; Somasundaran, M.; Sullivan, J.L.; Luzuriaga, K.; Greenough, T.C., et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 2003, 426, 450-454. [CrossRef]
- Ogando, N.S.; Dalebout, T.J.; Zevenhoven-Dobbe, J.C.; Limpens, R.; van der Meer, Y.; Caly, L.; Druce, J.; de Vries, J.J.C.; Kikkert, M.; Barcena, M., et al. SARS-coronavirus-2 replication in Vero E6 cells: replication kinetics, rapid adaptation and cytopathology. J Gen Virol 2020, 101, 925-940. [CrossRef]
- Chu, H.; Chan, J.F.; Yuen, T.T.; Shuai, H.; Yuan, S.; Wang, Y.; Hu, B.; Yip, C.C.; Tsang, J.O.; Huang, X., et al. Comparative tropism, replication kinetics, and cell damage profiling of SARS-CoV-2 and SARS-CoV with implications for clinical manifestations, transmissibility, and laboratory studies of COVID-19: an observational study. Lancet Microbe 2020, 1, e14-e23. [CrossRef]
- Zhang, Y.; Guo, R.; Kim, S.H.; Shah, H.; Zhang, S.; Liang, J.H.; Fang, Y.; Gentili, M.; Leary, C.N.O.; Elledge, S.J., et al. SARS-CoV-2 hijacks folate and one-carbon metabolism for viral replication. Nat Commun 2021, 12, 1676. [CrossRef]
- Bugatti, A.; Filippini, F.; Bardelli, M.; Zani, A.; Chiodelli, P.; Messali, S.; Caruso, A.; Caccuri, F. SARS-CoV-2 Infects Human ACE2-Negative Endothelial Cells through an alpha(v)beta(3) Integrin-Mediated Endocytosis Even in the Presence of Vaccine-Elicited Neutralizing Antibodies. Viruses 2022, 14. [CrossRef]
- Yamagami, Y.; Kawami, M.; Ojima, T.; Futatsugi, S.; Yumoto, R.; Takano, M. Role of plasminogen activator inhibitor-1 in methotrexate-induced epithelial-mesenchymal transition in alveolar epithelial A549 cells. Biochem Biophys Res Commun 2020, 525, 543-548. [CrossRef]
- Kawami, M.; Miyamoto, M.; Yumoto, R.; Takano, M. Methotrexate influx via folate transporters into alveolar epithelial cell line A549. Drug Metab Pharmacokinet 2015, 30, 276-281. [CrossRef]
- Ojima, T.; Kawami, M.; Yumoto, R.; Takano, M. Differential mechanisms underlying methotrexate-induced cell death and epithelial-mesenchymal transition in A549 cells. Toxicol Res 2021, 37, 293-300. [CrossRef]
- Iaconis, D.; Bordi, L.; Matusali, G.; Talarico, C.; Manelfi, C.; Cesta, M.C.; Zippoli, M.; Caccuri, F.; Bugatti, A.; Zani, A., et al. Characterization of raloxifene as a potential pharmacological agent against SARS-CoV-2 and its variants. Cell Death Dis 2022, 13, 498. [CrossRef]
- Corona, A.; Wycisk, K.; Talarico, C.; Manelfi, C.; Milia, J.; Cannalire, R.; Esposito, F.; Gribbon, P.; Zaliani, A.; Iaconis, D., et al. Natural Compounds Inhibit SARS-CoV-2 nsp13 Unwinding and ATPase Enzyme Activities. ACS Pharmacol Transl Sci 2022, 5, 226-239. [CrossRef]
- Abolmaali, S.S.; Tamaddon, A.M.; Dinarvand, R. A review of therapeutic challenges and achievements of methotrexate delivery systems for treatment of cancer and rheumatoid arthritis. Cancer Chemother Pharmacol 2013, 71, 1115-1130. [CrossRef]
- Khan, S.; Durairaj, S. JAK Inhibition with Methotrexate as Treatment for COVID-19 Is a Double-Edged Sword. Int Arch Allergy Immunol 2020, 181, 563-564. [CrossRef]
- Hannoodee, M.; Mittal, M. Methotrexate. In StatPearls, Treasure Island (FL), 2022.
- Wang, W.; Zhou, H.; Liu, L. Side effects of methotrexate therapy for rheumatoid arthritis: A systematic review. Eur J Med Chem 2018, 158, 502-516. [CrossRef]
- Yousefi, G.; Shafaati, A.; Zarghi, A.; Foroutan, S.M. Pharmacokinetics and Biodistribution of Pegylated Methotrexate after IV Administration to Mice. Iran J Pharm Res 2018, 17, 111-123.
- Molina, J.R. Pralatrexate, a dihydrofolate reductase inhibitor for the potential treatment of several malignancies. IDrugs 2008, 11, 508-521.
- Purcell, W.T.; Ettinger, D.S. Novel antifolate drugs. Curr Oncol Rep 2003, 5, 114-125. [CrossRef]
- Sattler, F.R.; Allegra, C.J.; Verdegem, T.D.; Akil, B.; Tuazon, C.U.; Hughlett, C.; Ogata-Arakaki, D.; Feinberg, J.; Shelhamer, J.; Lane, H.C., et al. Trimetrexate-leucovorin dosage evaluation study for treatment of Pneumocystis carinii pneumonia. J Infect Dis 1990, 161, 91-96. [CrossRef]
- Zain, J.; O'Connor, O. Pralatrexate: basic understanding and clinical development. Expert Opin Pharmacother 2010, 11, 1705-1714. [CrossRef]
- Izbicka, E.; Diaz, A.; Streeper, R.; Wick, M.; Campos, D.; Steffen, R.; Saunders, M. Distinct mechanistic activity profile of pralatrexate in comparison to other antifolates in in vitro and in vivo models of human cancers. Cancer Chemother Pharmacol 2009, 64, 993-999. [CrossRef]
- Sirotnak, F.M.; DeGraw, J.I.; Colwell, W.T.; Piper, J.R. A new analogue of 10-deazaaminopterin with markedly enhanced curative effects against human tumor xenografts in mice. Cancer Chemother Pharmacol 1998, 42, 313-318. [CrossRef]
- Krug, L.M.; Ng, K.K.; Kris, M.G.; Miller, V.A.; Tong, W.; Heelan, R.T.; Leon, L.; Leung, D.; Kelly, J.; Grant, S.C., et al. Phase I and pharmacokinetic study of 10-propargyl-10-deazaaminopterin, a new antifolate. Clin Cancer Res 2000, 6, 3493-3498.
- Marchi, E.; O'Connor, O.A. Safety and efficacy of pralatrexate in the treatment of patients with relapsed or refractory peripheral T-cell lymphoma. Ther Adv Hematol 2012, 3, 227-235. [CrossRef]
- Huang, Y.S.; Yang, J.J.; Lee, N.Y.; Chen, G.J.; Ko, W.C.; Sun, H.Y.; Hung, C.C. Treatment of Pneumocystis jirovecii pneumonia in HIV-infected patients: a review. Expert Rev Anti Infect Ther 2017, 15, 873-892. [CrossRef]
- Fulton, B.; Wagstaff, A.J.; McTavish, D. Trimetrexate. A review of its pharmacodynamic and pharmacokinetic properties and therapeutic potential in the treatment of Pneumocystis carinii pneumonia. Drugs 1995, 49, 563-576. [CrossRef]
- Opdam, M.A.A.; Benoy, S.; Verhoef, L.M.; Van Bijnen, S.; Lamers-Karnebeek, F.; Traksel, R.A.M.; Vos, P.; den Broeder, A.A.; Broen, J. Identification of Risk Factors for COVID-19 Hospitalization in Patients With Anti-Rheumatic Drugs: Results From a Multicenter Nested Case Control Study. Clin Pharmacol Ther 2022, 111, 1061-1065. [CrossRef]
- Gianfrancesco, M.; Hyrich, K.L.; Al-Adely, S.; Carmona, L.; Danila, M.I.; Gossec, L.; Izadi, Z.; Jacobsohn, L.; Katz, P.; Lawson-Tovey, S., et al. Characteristics associated with hospitalisation for COVID-19 in people with rheumatic disease: data from the COVID-19 Global Rheumatology Alliance physician-reported registry. Ann Rheum Dis 2020, 79, 859-866. [CrossRef]
- Yousaf, A.; Gayam, S.; Feldman, S.; Zinn, Z.; Kolodney, M. Clinical outcomes of COVID-19 in patients taking tumor necrosis factor inhibitors or methotrexate: A multicenter research network study. J Am Acad Dermatol 2021, 84, 70-75. [CrossRef]
- Sadeghinia, A.; Daneshpazhooh, M. Immunosuppressive drugs for patients with psoriasis during the COVID-19 pandemic era. A review. Dermatol Ther 2021, 34, e14498. [CrossRef]
- Armesto, S.; Gonzalez Vela, C.; Sanchez, J.; Illaro, A.; Mayorga, J.; Lopez Sundh, A.E.; Naharro Fernandez, C.; Palmou, N.; Gomez-Fernandez, C.; Gonzalez Lopez, M.A., et al. Treating multidrug-resistant psoriasis with brodalumab, apremilast, methotrexate and prednisone combination therapy in the COVID-19 pandemic. Dermatol Ther 2020, 33, e14464. [CrossRef]
- Ghazawi, F.M.; Lim, M.; Dutz, J.P.; Kirchhof, M.G. Infection risk of dermatologic therapeutics during the COVID-19 pandemic: an evidence-based recalibration. Int J Dermatol 2020, 59, 1043-1056. [CrossRef]
- Arora, H.; Boothby-Shoemaker, W.; Braunberger, T.; Lim, H.W.; Veenstra, J. Safety of conventional immunosuppressive therapies for patients with dermatological conditions and coronavirus disease 2019: A review of current evidence. J Dermatol 2022, 49, 317-329. [CrossRef]
- Frohman, E.M.; Villemarette-Pittman, N.R.; Cruz, R.A.; Longmuir, R.; Rowe, V.; Rowe, E.S.; Varkey, T.C.; Steinman, L.; Zamvil, S.S.; Frohman, T.C. Part II. high-dose methotrexate with leucovorin rescue for severe COVID-19: An immune stabilization strategy for SARS-CoV-2 induced 'PANIC' attack. J Neurol Sci 2020, 415, 116935. [CrossRef]
- Seif, F.; Aazami, H.; Khoshmirsafa, M.; Kamali, M.; Mohsenzadegan, M.; Pornour, M.; Mansouri, D. JAK Inhibition as a New Treatment Strategy for Patients with COVID-19. Int Arch Allergy Immunol 2020, 181, 467-475. [CrossRef]
- Russell, B.; Moss, C.; George, G.; Santaolalla, A.; Cope, A.; Papa, S.; Van Hemelrijck, M. Associations between immune-suppressive and stimulating drugs and novel COVID-19-a systematic review of current evidence. Ecancermedicalscience 2020, 14, 1022. [CrossRef]
- D'Silva, K.M.; Wallace, Z.S. COVID-19 and Disease-Modifying Anti-rheumatic Drugs. Curr Rheumatol Rep 2021, 23, 28. [CrossRef]
- Ganjei, Z.; Faraji Dana, H.; Ebrahimi-Dehkordi, S.; Alidoust, F.; Bahmani, K. Methotrexate as a safe immunosuppressive agent during the COVID-19 pandemic. Int Immunopharmacol 2021, 101, 108324. [CrossRef]
- Caccuri, F.; Zani, A.; Messali, S.; Giovanetti, M.; Bugatti, A.; Campisi, G.; Filippini, F.; Scaltriti, E.; Ciccozzi, M.; Fiorentini, S., et al. A persistently replicating SARS-CoV-2 variant derived from an asymptomatic individual. J Transl Med 2020, 18, 362. [CrossRef]
- Caccuri, F.; Bugatti, A.; Zani, A.; De Palma, A.; Di Silvestre, D.; Manocha, E.; Filippini, F.; Messali, S.; Chiodelli, P.; Campisi, G., et al. SARS-CoV-2 Infection Remodels the Phenotype and Promotes Angiogenesis of Primary Human Lung Endothelial Cells. Microorganisms 2021, 9. [CrossRef]
- Caccuri, F.; Bugatti, A.; Meini, A.; Bonfanti, C.; Motta, M.; Savare, L.; Arrighini, A.; Bondioni, M.P.; Lougaris, V.; Caruso, A., et al. Temporal viral loads in respiratory and gastrointestinal tract and serum antibody responses during SARS-CoV-2 infection in an Italian pediatric cohort. Clin Immunol 2021, 225, 108695. [CrossRef]
- Kuzikov, M.; Woens, J.; Zaliani, A.; Hambach, J.; Eden, T.; Fehse, B.; Ellinger, B.; Riecken, K. High-throughput drug screening allowed identification of entry inhibitors specifically targeting different routes of SARS-CoV-2 Delta and Omicron/BA.1. Biomed Pharmacother 2022, 151, 113104. [CrossRef]
- Newman, J.A.; Douangamath, A.; Yadzani, S.; Yosaatmadja, Y.; Aimon, A.; Brandao-Neto, J.; Dunnett, L.; Gorrie-Stone, T.; Skyner, R.; Fearon, D., et al. Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase. Nat Commun 2021, 12, 4848. [CrossRef]
- Nizi, M.G.; Persoons, L.; Corona, A.; Felicetti, T.; Cernicchi, G.; Massari, S.; Manfroni, G.; Vangeel, L.; Barreca, M.L.; Esposito, F., et al. Discovery of 2-Phenylquinolines with Broad-Spectrum Anti-coronavirus Activity. ACS Med Chem Lett 2022, 13, 855-864. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).