Submitted:
26 April 2023
Posted:
27 April 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
2.1. Validation of the assay for the measurement of RE-DNAm
2.2. Effects of electromagnetic radiation on RE DNAm
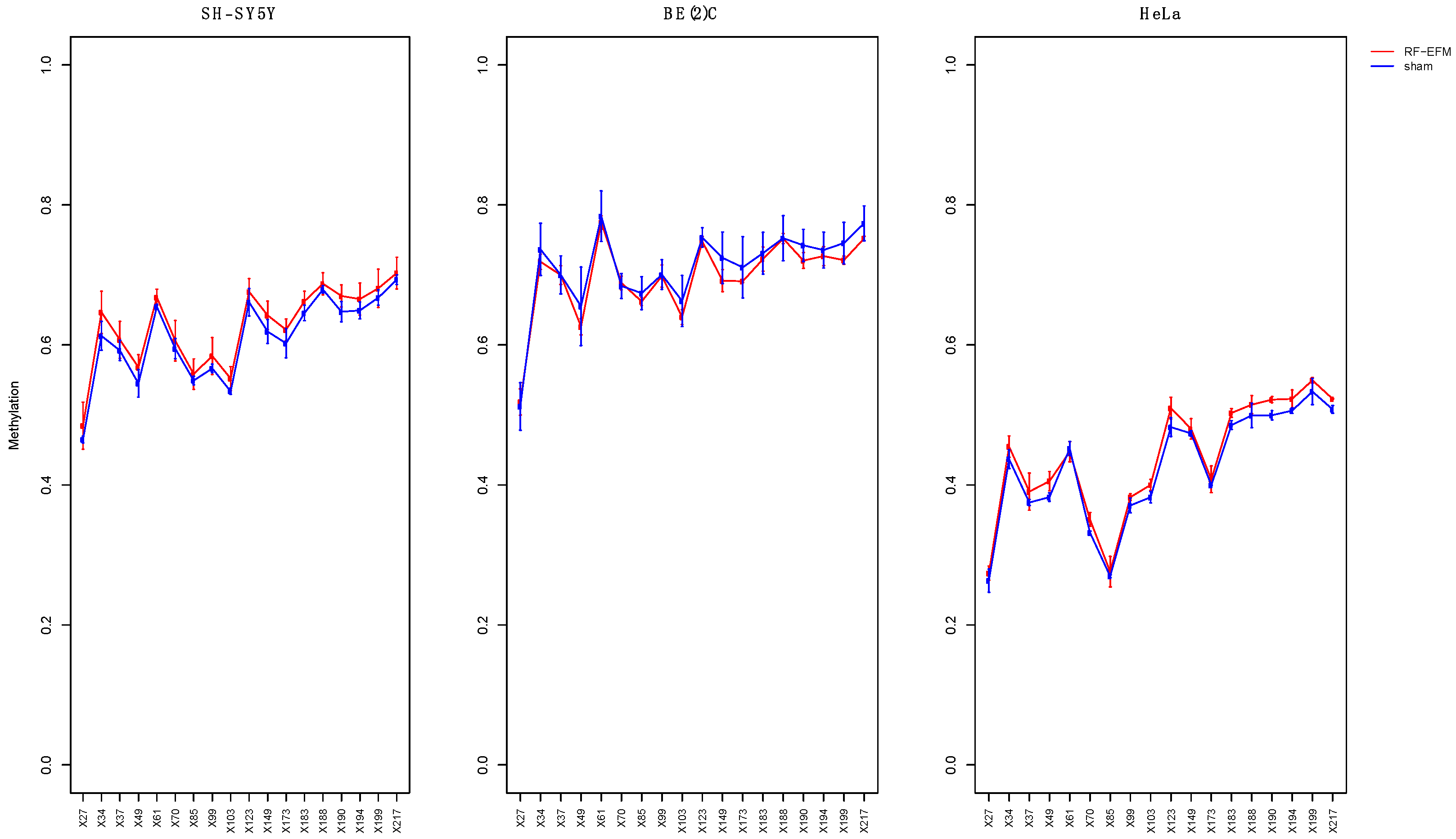
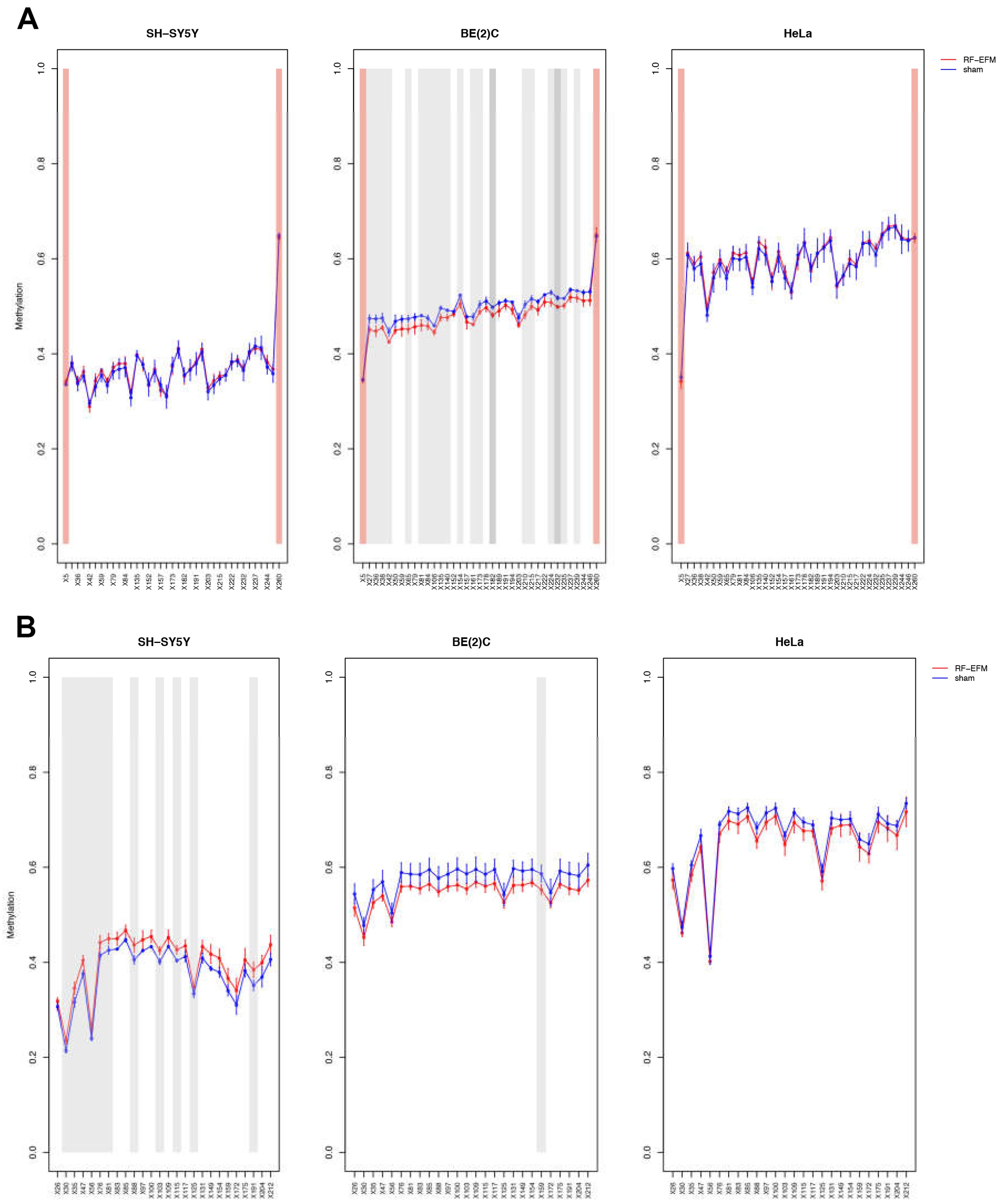
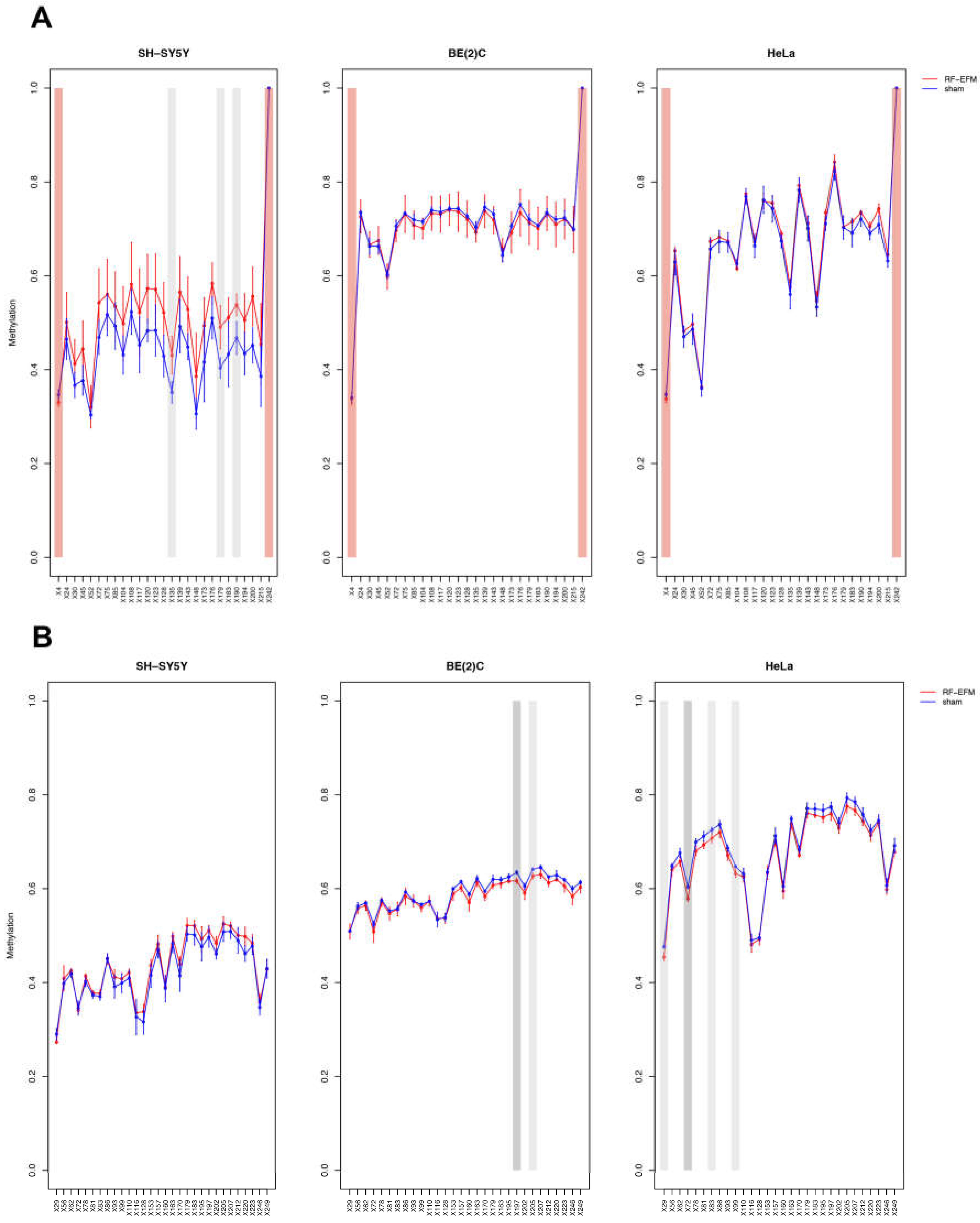
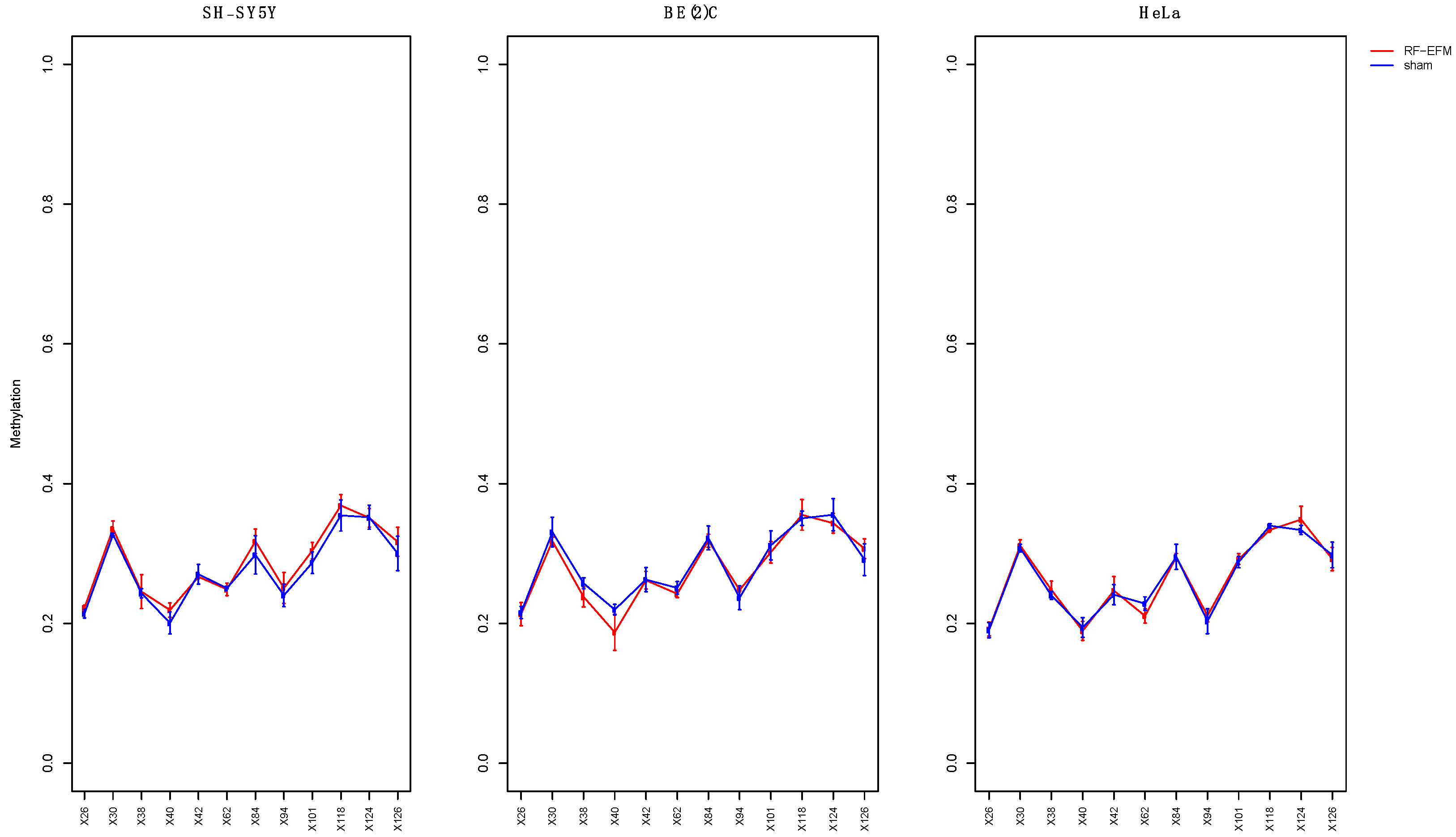
2.3. Effects of electromagnetic radiation on RE DNAm epihaplotype diversity
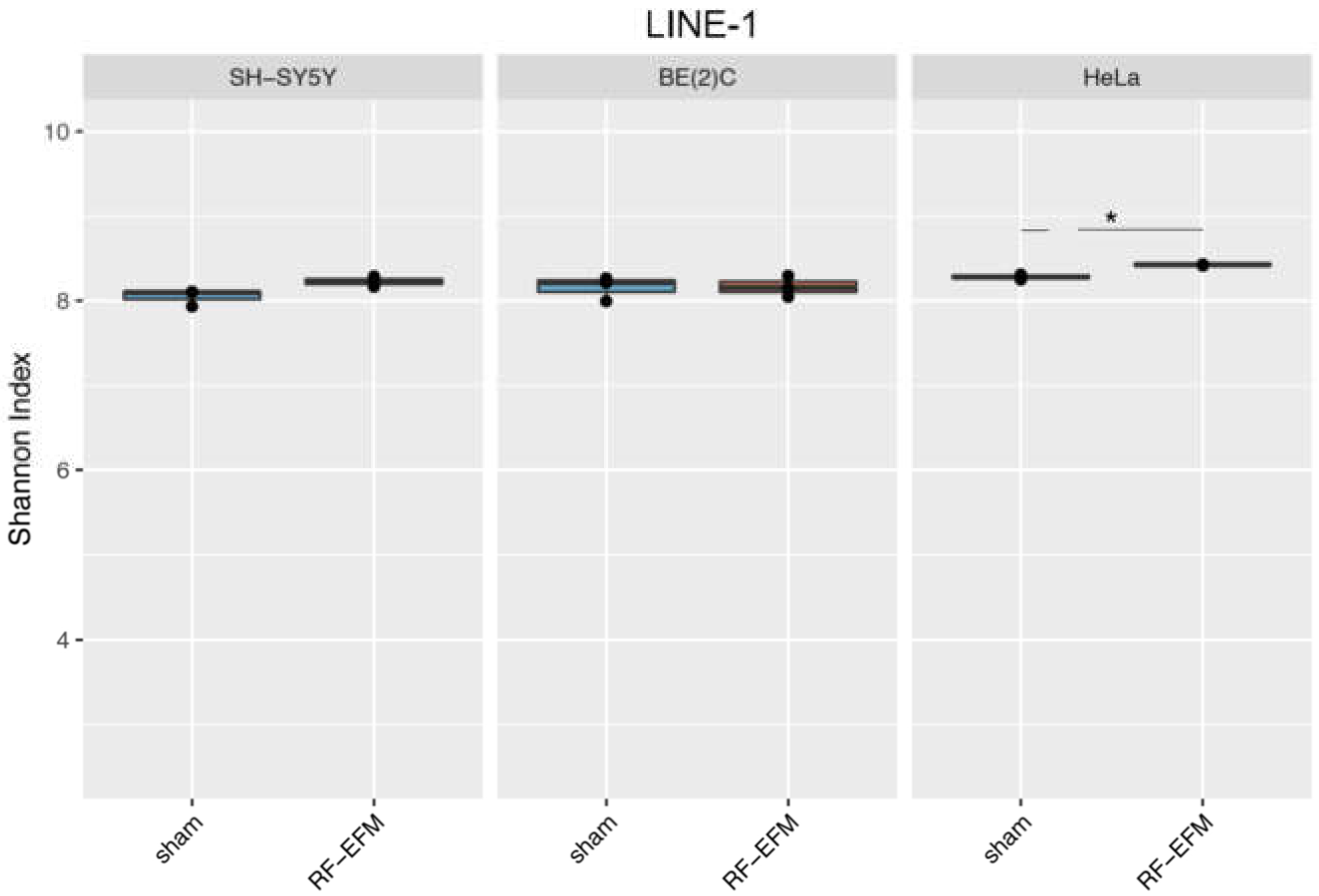
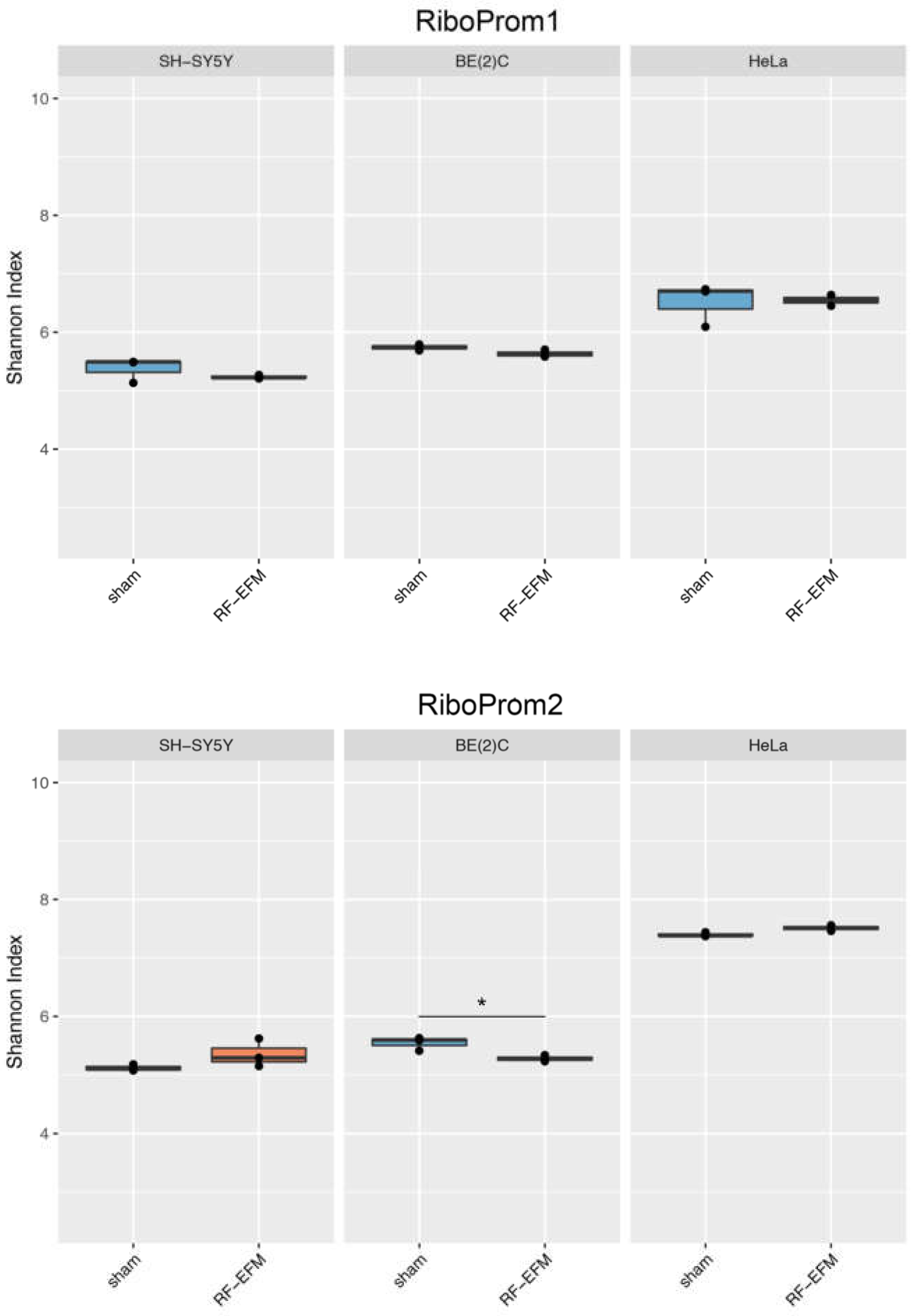
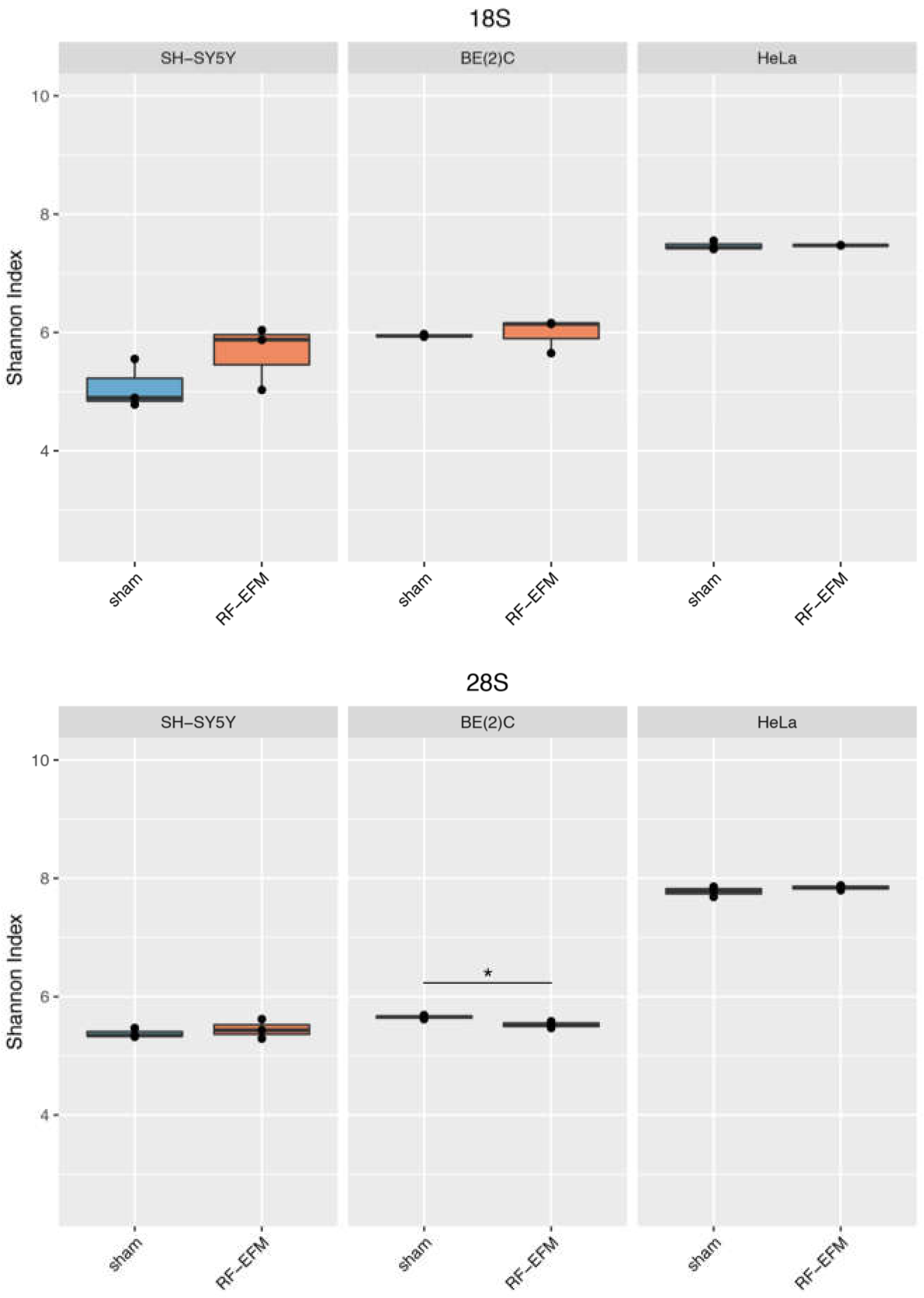
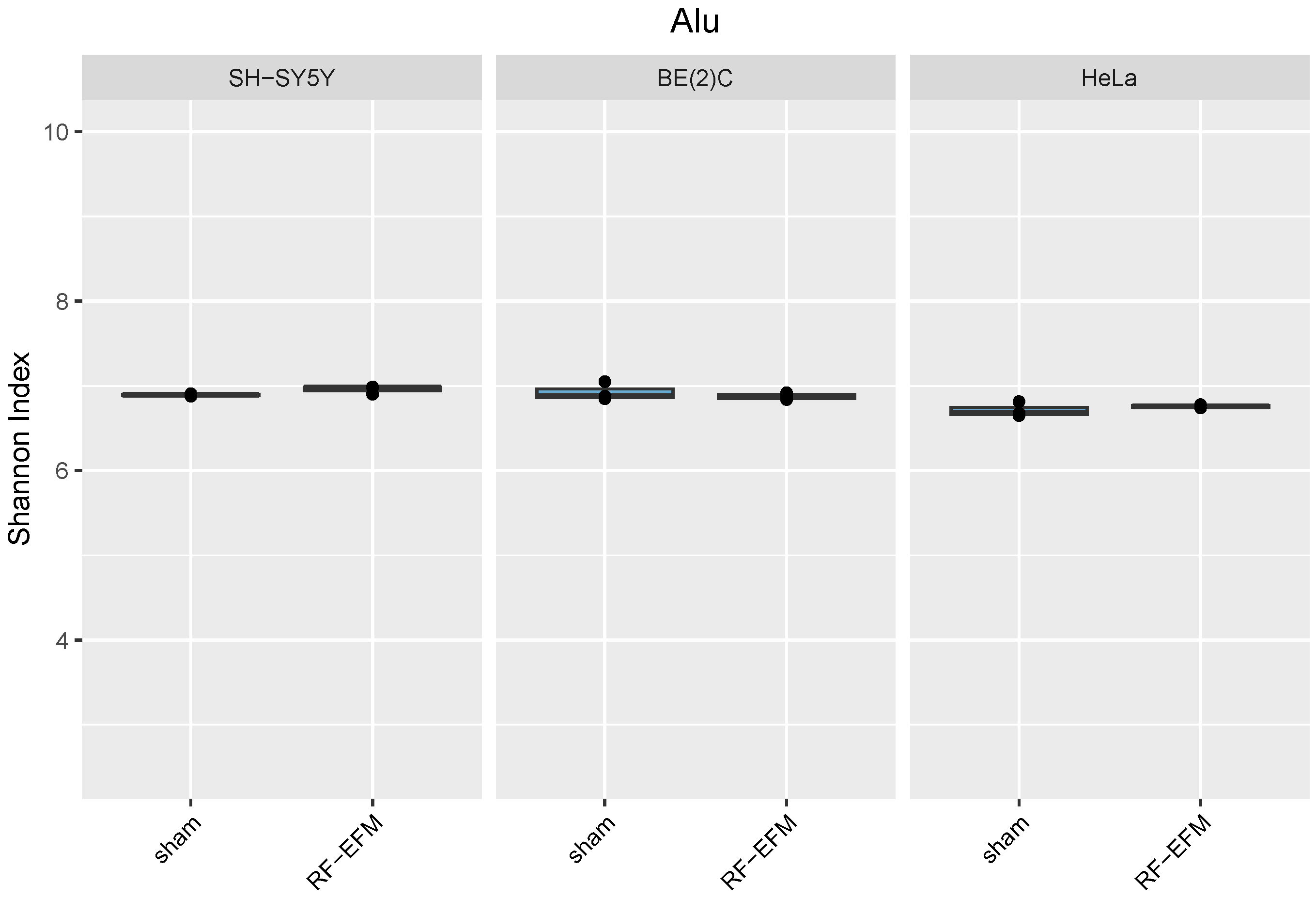
3. Discussion
4. Materials and Methods
4.1. Exposure system
4.2. Cell culture
4.3. DNA extraction and bisulfite conversion
4.4. Library Preparation and Bisulfite-targeted Sequencing
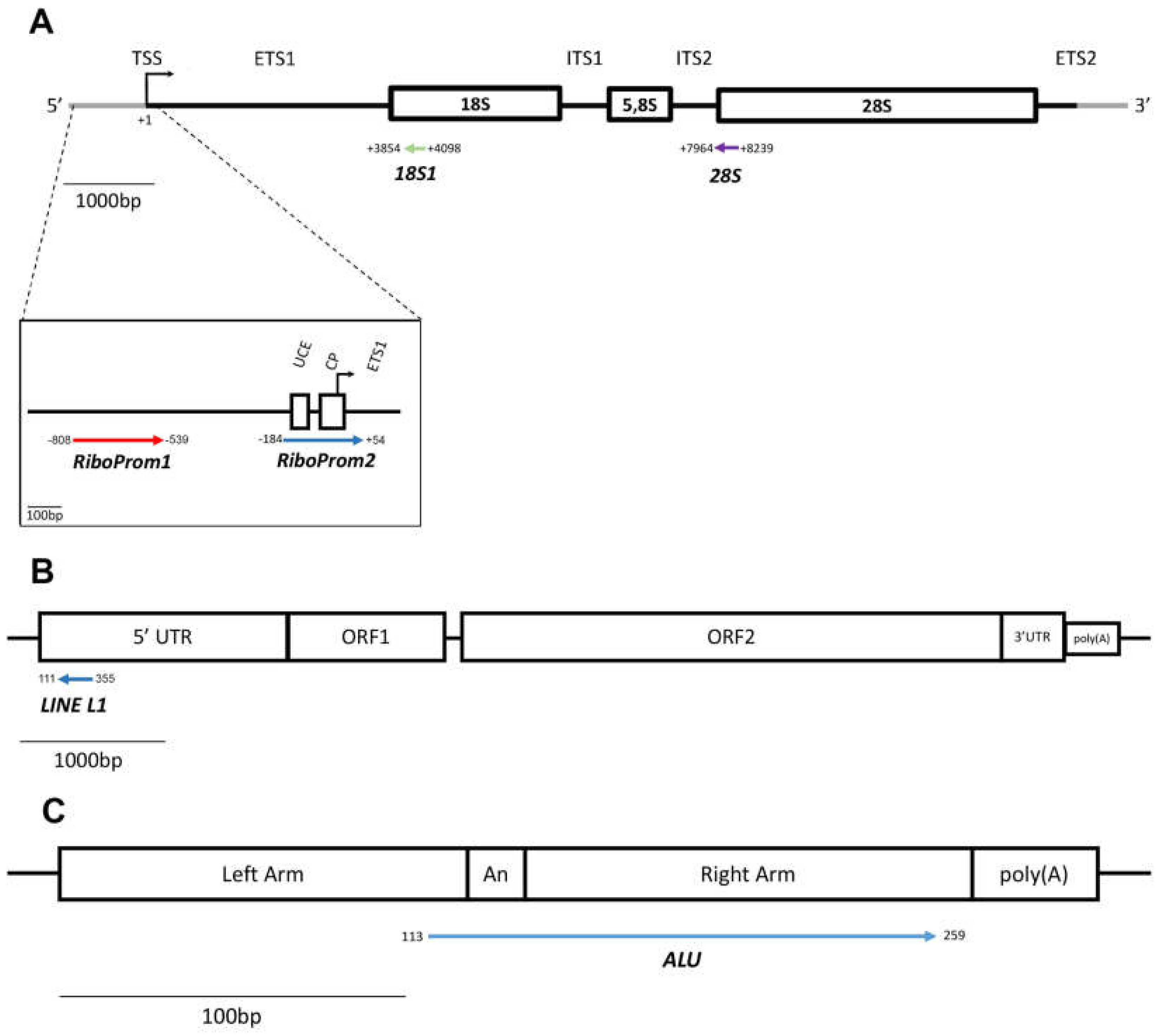
4.6. Sequencing Data Handling
4.7. DNA methylation data extraction
4.8. Epihaplotype Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Non-Ionizing Radiation; IARC Working Group on the Evaluation of Carcinogenic Risks to Humans, World Health Organization, International Agency for Research on Cancer, Eds.; IARC monographs on the evaluation of carcinogenic risks to humans; International Agency for Research on Cancer ; World Health Organization: Lyon, France : Geneva, 2013; ISBN 978-92-832-1325-3.
- Verschaeve, L. Genetic Damage in Subjects Exposed to Radiofrequency Radiation. Mutation Research/Reviews in Mutation Research 2009, 681, 259–270. [Google Scholar] [CrossRef] [PubMed]
- Vijayalaxmi; Prihoda, T.J. Genetic Damage in Human Cells Exposed to Non-Ionizing Radiofrequency Fields: A Meta-Analysis of the Data from 88 Publications (1990–2011). Mutation Research/Genetic Toxicology and Environmental Mutagenesis 2012, 749, 1–16. [CrossRef] [PubMed]
- Maffei, M.E. Magnetic Fields and Cancer: Epidemiology, Cellular Biology, and Theranostics. IJMS 2022, 23, 1339. [Google Scholar] [CrossRef] [PubMed]
- Vijayalaxmi; Prihoda, T.J. Comprehensive Review of Quality of Publications and Meta-Analysis of Genetic Damage in Mammalian Cells Exposed to Non-Ionizing Radiofrequency Fields. Radiation Research 2018, 191, 20. [CrossRef] [PubMed]
- Romeo, S.; Zeni, O.; Sannino, A.; Lagorio, S.; Biffoni, M.; Scarfì, M.R. Genotoxicity of Radiofrequency Electromagnetic Fields: Protocol for a Systematic Review of in Vitro Studies. Environment International 2021, 148, 106386. [Google Scholar] [CrossRef] [PubMed]
- Tryndyak, V.P. Role of Epigenetics in Tumor Induction by Non-Genotoxic Carcinogens. Current Opinion in Toxicology 2017, 6, 42–49. [Google Scholar] [CrossRef]
- Baek, S.; Quan, X.; Kim, S.; Lengner, C.; Park, J.-K.; Kim, J. Electromagnetic Fields Mediate Efficient Cell Reprogramming into a Pluripotent State. ACS Nano 2014, 8, 10125–10138. [Google Scholar] [CrossRef]
- Baek, S.; Choi, H.; Park, H.; Cho, B.; Kim, S.; Kim, J. Effects of a Hypomagnetic Field on DNA Methylation during the Differentiation of Embryonic Stem Cells. Sci Rep 2019, 9, 1333. [Google Scholar] [CrossRef]
- Mokarram, P.; Sheikhi, M.; Mortazavi, S.M.J.; Saeb, S.; Shokrpour, N. Effect of Exposure to 900 MHz GSM Mobile Phone Radiofrequency Radiation on Estrogen Receptor Methylation Status in Colon Cells of Male Sprague Dawley Rats. J Biomed Phys Eng 2017, 7, 79–86. [Google Scholar]
- Kumar, R.; Deshmukh, P.S.; Sharma, S.; Banerjee, B.D. Effect of Mobile Phone Signal Radiation on Epigenetic Modulation in the Hippocampus of Wistar Rat. Environmental Research 2021, 192, 110297. [Google Scholar] [CrossRef]
- Martin, E.M.; Fry, R.C. Environmental Influences on the Epigenome: Exposure- Associated DNA Methylation in Human Populations. Annu. Rev. Public Health 2018, 39, 309–333. [Google Scholar] [CrossRef] [PubMed]
- Rodic, N. LINE-1 Activity and Regulation in Cancer. Front Biosci 2018, 23, 1680–1686. [Google Scholar] [CrossRef] [PubMed]
- Ponomaryova, A.A.; Rykova, E.Y.; Gervas, P.A.; Cherdyntseva, N.V.; Mamedov, I.Z.; Azhikina, T.L. Aberrant Methylation of LINE-1 Transposable Elements: A Search for Cancer Biomarkers. Cells 2020, 9, 2017. [Google Scholar] [CrossRef] [PubMed]
- International Human Genome Sequencing Consortium; Whitehead Institute for Biomedical Research, Center for Genome Research:; Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Devon, K.; Dewar, K.; et al. Initial Sequencing and Analysis of the Human Genome. Nature 2001, 409, 860–921. [CrossRef]
- Doucet, A.J.; Wilusz, J.E.; Miyoshi, T.; Liu, Y.; Moran, J.V. A 3′ Poly(A) Tract Is Required for LINE-1 Retrotransposition. Molecular Cell 2015, 60, 728–741. [Google Scholar] [CrossRef]
- Iskow, R.C.; McCabe, M.T.; Mills, R.E.; Torene, S.; Pittard, W.S.; Neuwald, A.F.; Van Meir, E.G.; Vertino, P.M.; Devine, S.E. Natural Mutagenesis of Human Genomes by Endogenous Retrotransposons. Cell 2010, 141, 1253–1261. [Google Scholar] [CrossRef]
- Bourc’his, D.; Bestor, T.H. Meiotic Catastrophe and Retrotransposon Reactivation in Male Germ Cells Lacking Dnmt3L. Nature 2004, 431, 96–99. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Perez, J.L.; Morell, M.; Scheys, J.O.; Kulpa, D.A.; Morell, S.; Carter, C.C.; Hammer, G.D.; Collins, K.L.; O’Shea, K.S.; Menendez, P.; et al. Epigenetic Silencing of Engineered L1 Retrotransposition Events in Human Embryonic Carcinoma Cells. Nature 2010, 466, 769–773. [Google Scholar] [CrossRef]
- Sanchez-Luque, F.J.; Kempen, M.-J.H.C.; Gerdes, P.; Vargas-Landin, D.B.; Richardson, S.R.; Troskie, R.-L.; Jesuadian, J.S.; Cheetham, S.W.; Carreira, P.E.; Salvador-Palomeque, C.; et al. LINE-1 Evasion of Epigenetic Repression in Humans. Molecular Cell 2019, 75, 590–604. [Google Scholar] [CrossRef]
- Klutstein, M.; Nejman, D.; Greenfield, R.; Cedar, H. DNA Methylation in Cancer and Aging. Cancer Research 2016, 76, 3446–3450. [Google Scholar] [CrossRef]
- Cruickshanks, H.A.; Vafadar-Isfahani, N.; Dunican, D.S.; Lee, A.; Sproul, D.; Lund, J.N.; Meehan, R.R.; Tufarelli, C. Expression of a Large LINE-1-Driven Antisense RNA Is Linked to Epigenetic Silencing of the Metastasis Suppressor Gene TFPI-2 in Cancer. Nucleic Acids Research 2013, 41, 6857–6869. [Google Scholar] [CrossRef]
- Schulz, W.A. L1 Retrotransposons in Human Cancers. Journal of Biomedicine and Biotechnology 2006, 2006, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Miousse, I.R.; Chalbot, M.-C.G.; Lumen, A.; Ferguson, A.; Kavouras, I.G.; Koturbash, I. Response of Transposable Elements to Environmental Stressors. Mutation Research/Reviews in Mutation Research 2015, 765, 19–39. [Google Scholar] [CrossRef] [PubMed]
- Del Re, B.; Giorgi, G. Long INterspersed Element-1 Mobility as a Sensor of Environmental Stresses. Environ Mol Mutagen 2020, 61, 465–493. [Google Scholar] [CrossRef] [PubMed]
- Desaulniers, D.; Vasseur, P.; Jacobs, A.; Aguila, M.C.; Ertych, N.; Jacobs, M.N. Integration of Epigenetic Mechanisms into Non-Genotoxic Carcinogenicity Hazard Assessment: Focus on DNA Methylation and Histone Modifications. IJMS 2021, 22, 10969. [Google Scholar] [CrossRef] [PubMed]
- Christian, C.M.; deHaro, D.; Kines, K.J.; Sokolowski, M.; Belancio, V.P. Identification of L1 ORF2p Sequence Important to Retrotransposition Using Bipartile Alu Retrotransposition (BAR). Nucleic Acids Res 2016, 44, 4818–4834. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.-M.; Schmid, C.W. Proposed Roles for DNA Methylation in Alu Transcriptional Repression and Mutational Inactivation. Nucl Acids Res 1993, 21, 1351–1359. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Lu, X.; Xie, H. Dynamic Alu Methylation during Normal Development, Aging, and Tumorigenesis. BioMed Research International 2014, 2014, 1–12. [Google Scholar] [CrossRef]
- Henderson, A.S.; Warburton, D.; Atwood, K.C. Ribosomal DNA Connectives between Human Acrocentric Chromosomes. Nature 1973, 245, 95–97. [Google Scholar] [CrossRef]
- Gonzalez, I.L.; Sylvester, J.E. Complete Sequence of the 43-Kb Human Ribosomal DNA Repeat: Analysis of the Intergenic Spacer. Genomics 1995, 27, 320–328. [Google Scholar] [CrossRef]
- Wang, M.; Lemos, B. Ribosomal DNA Harbors an Evolutionarily Conserved Clock of Biological Aging. Genome Res. 2019, 29, 325–333. [Google Scholar] [CrossRef]
- Santoro, R.; Grummt, I. Molecular Mechanisms Mediating Methylation-Dependent Silencing of Ribosomal Gene Transcription. Molecular Cell 2001, 8, 719–725. [Google Scholar] [CrossRef]
- Huang, X.; Zhang, X.; Zong, L.; Gao, Q.; Zhang, C.; Wei, R.; Guan, Y.; Huang, L.; Zhang, L.; Lyu, G.; et al. Gene Body Methylation Safeguards Ribosomal DNA Transcription by Preventing PHF6-Mediated Enrichment of Repressive Histone Mark H4K20me3. Journal of Biological Chemistry 2021, 297, 101195. [Google Scholar] [CrossRef] [PubMed]
- D’Aquila, P.; Montesanto, A.; Mandalà, M.; Garasto, S.; Mari, V.; Corsonello, A.; Bellizzi, D.; Passarino, G. Methylation of the Ribosomal RNA Gene Promoter Is Associated with Aging and Age-Related Decline. Aging Cell 2017, 16, 966–975. [Google Scholar] [CrossRef] [PubMed]
- Shao, F.; Liu, X.; Zhang, X.; Wang, Q.; Wang, W. Methylation of 45S Ribosomal DNA (RDNA) Is Associated with Cancer and Aging in Humans. International Journal of Genomics 2021, 2021, 1–9. [Google Scholar] [CrossRef]
- Smirnov, E.; Chmúrčiaková, N.; Cmarko, D. Human RDNA and Cancer. Cells 2021, 10, 3452. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, W.; Liu, K.; Ao, L.; Zhong, J.L.; Cao, J.; Liu, J. Effect of 50 Hz Extremely Low-Frequency Electromagnetic Fields on the DNA Methylation and DNA Methyltransferases in Mouse Spermatocyte-Derived Cell Line GC-2. BioMed Research International 2015, 2015, 1–10. [Google Scholar] [CrossRef]
- Giorgi, G.; Pirazzini, C.; Bacalini, M.G.; Giuliani, C.; Garagnani, P.; Capri, M.; Bersani, F.; Del Re, B. Assessing the Combined Effect of Extremely Low-Frequency Magnetic Field Exposure and Oxidative Stress on LINE-1 Promoter Methylation in Human Neural Cells. Radiat Environ Biophys 2017, 56, 193–200. [Google Scholar] [CrossRef]
- Benassi, B.; Santangeli, S.; Merla, C.; Tarantini, L.; Bollati, V.; Butera, A.; Marino, C.; Consales, C. 50-Hz MF Does Not Affect Global DNA Methylation of SH-SY5Y Cells Treated with the Neurotoxin MPP +. Bioelectromagnetics. [CrossRef]
- Giorgi, G.; Del Re, B. Epigenetic Dysregulation in Various Types of Cells Exposed to Extremely Low-Frequency Magnetic Fields. Cell Tissue Res 2021, 386, 1–15. [Google Scholar] [CrossRef]
- Del Re, B.; Bersani, F.; Giorgi, G. Effect of Electromagnetic Field Exposure on the Transcription of Repetitive DNA Elements in Human Cells. Electromagnetic Biology and Medicine 2019, 38, 262–270. [Google Scholar] [CrossRef]
- Kresoja-Rakic, J.; Santoro, R. Nucleolus and RRNA Gene Chromatin in Early Embryo Development. Trends in Genetics 2019, 35, 868–879. [Google Scholar] [CrossRef] [PubMed]
- Faria, T.C.; Maldonado, H.L.; Santos, L.C.; DeLabio, R.; Payao, S.L.M.; Turecki, G.; Mechawar, N.; Santana, D.A.; Gigek, C.O.; Lemos, B.; et al. Characterization of Cerebellum-Specific Ribosomal DNA Epigenetic Modifications in Alzheimer’s Disease: Should the Cerebellum Serve as a Control Tissue After All? Mol Neurobiol 2020, 57, 2563–2571. [Google Scholar] [CrossRef] [PubMed]
- Gensous, N.; Bacalini, M.G.; Franceschi, C.; Meskers, C.G.M.; Maier, A.B.; Garagnani, P. Age-Related DNA Methylation Changes: Potential Impact on Skeletal Muscle Aging in Humans. Front Physiol 2019, 10, 996. [Google Scholar] [CrossRef]
- Ravaioli, F.; Zampieri, M.; Morandi, L.; Pirazzini, C.; Pellegrini, C.; De Fanti, S.; Gensous, N.; Pirazzoli, G.L.; Sambati, L.; Ghezzo, A.; et al. DNA Methylation Analysis of Ribosomal DNA in Adults With Down Syndrome. Frontiers in Genetics 2022, 13. [Google Scholar] [CrossRef] [PubMed]
- Pietrzak, M.; Rempala, G.; Nelson, P.T.; Zheng, J.-J.; Hetman, M. Epigenetic Silencing of Nucleolar RRNA Genes in Alzheimer’s Disease. PLoS ONE 2011, 6, e22585. [Google Scholar] [CrossRef]
- Horvath, S.; Raj, K. DNA Methylation-Based Biomarkers and the Epigenetic Clock Theory of Ageing. Nat Rev Genet 2018, 19, 371–384. [Google Scholar] [CrossRef]
- Yan, Q.; Paul, K.C.; Lu, A.T.; Kusters, C.; Binder, A.M.; Horvath, S.; Ritz, B. Epigenetic Mutation Load Is Weakly Correlated with Epigenetic Age Acceleration. Aging 2020, 12, 17863–17894. [Google Scholar] [CrossRef]
- Nielsen, C.; Hui, R.; Lui, W.-Y.; Solov’yov, I.A. Towards Predicting Intracellular Radiofrequency Radiation Effects. PLoS ONE 2019, 14, e0213286. [Google Scholar] [CrossRef]
- Zhao, X.; Department of Experimental Pathology, Beijing Institute of Radiation Medicine, Beijing, P. R. China; Dong, G.; Department of Experimental Pathology, Beijing Institute of Radiation Medicine, Beijing, P. R. China; Wang, C.; Department of Experimental Pathology, Beijing Institute of Radiation Medicine, Beijing, P. R. China The Non-Thermal Biological Effects and Mechanisms of Microwave Exposure. Int J Radiat Res 2021, 19, 483–494. [CrossRef]
- Mumtaz, S.; Rana, J.N.; Choi, E.H.; Han, I. Microwave Radiation and the Brain: Mechanisms, Current Status, and Future Prospects. IJMS 2022, 23, 9288. [Google Scholar] [CrossRef] [PubMed]
- Barnes, F.S.; Greenebaum, B. The Effects of Weak Magnetic Fields on Radical Pairs: Weak Magnetic Field Effects on Radicals. Bioelectromagnetics 2015, 36, 45–54. [Google Scholar] [CrossRef] [PubMed]
- Sherrard, R.M.; Morellini, N.; Jourdan, N.; El-Esawi, M.; Arthaut, L.-D.; Niessner, C.; Rouyer, F.; Klarsfeld, A.; Doulazmi, M.; Witczak, J.; et al. Low-Intensity Electromagnetic Fields Induce Human Cryptochrome to Modulate Intracellular Reactive Oxygen Species. PLoS Biol 2018, 16, e2006229. [Google Scholar] [CrossRef] [PubMed]
- Yurekli, A.I.; Ozkan, M.; Kalkan, T.; Saybasili, H.; Tuncel, H.; Atukeren, P.; Gumustas, K.; Seker, S. GSM Base Station Electromagnetic Radiation and Oxidative Stress in Rats. Electromagnetic Biology and Medicine 2006, 25, 177–188. [Google Scholar] [CrossRef] [PubMed]
- Meral, I.; Mert, H.; Mert, N.; Deger, Y.; Yoruk, I.; Yetkin, A.; Keskin, S. Effects of 900-MHz Electromagnetic Field Emitted from Cellular Phone on Brain Oxidative Stress and Some Vitamin Levels of Guinea Pigs. Brain Research 2007, 1169, 120–124. [Google Scholar] [CrossRef] [PubMed]
- De Iuliis, G.N.; Newey, R.J.; King, B.V.; Aitken, R.J. Mobile Phone Radiation Induces Reactive Oxygen Species Production and DNA Damage in Human Spermatozoa In Vitro. PLoS ONE 2009, 4, e6446. [Google Scholar] [CrossRef] [PubMed]
- Avci, B.; Akar, A.; Bilgici, B.; Tunçel, Ö.K. Oxidative Stress Induced by 1.8 GHz Radio Frequency Electromagnetic Radiation and Effects of Garlic Extract in Rats. International Journal of Radiation Biology 2012, 88, 799–805. [Google Scholar] [CrossRef] [PubMed]
- Bilgici, B.; Akar, A.; Avci, B.; Tuncel, O.K. Effect of 900 MHz Radiofrequency Radiation on Oxidative Stress in Rat Brain and Serum. Electromagnetic Biology and Medicine 2013, 32, 20–29. [Google Scholar] [CrossRef]
- Houston, B.J.; Nixon, B.; McEwan, K.E.; Martin, J.H.; King, B.V.; Aitken, R.J.; De Iuliis, G.N. Whole-Body Exposures to Radiofrequency-Electromagnetic Energy Can Cause DNA Damage in Mouse Spermatozoa via an Oxidative Mechanism. Sci Rep 2019, 9, 17478. [Google Scholar] [CrossRef]
- Sun, A.; Zhao, X.; Li, Z.; Gao, Y.; Liu, Q.; Zhou, H.; Dong, G.; Wang, C. Effects of Long-Term and Multigeneration Exposure of Caenorhabditis Elegans to 9.4 GHz Microwaves. Bioelectromagnetics 2022, 43, 336–346. [Google Scholar] [CrossRef]
- Consales, C.; Butera, A.; Merla, C.; Pasquali, E.; Lopresto, V.; Pinto, R.; Pierdomenico, M.; Mancuso, M.; Marino, C.; Benassi, B. Exposure of the SH-SY5Y Human Neuroblastoma Cells to 50-Hz Magnetic Field: Comparison Between Two-Dimensional (2D) and Three-Dimensional (3D) In Vitro Cultures. Mol Neurobiol 2021, 58, 1634–1649. [Google Scholar] [CrossRef]
- Reale, M.; Kamal, M.A.; Patruno, A.; Costantini, E.; D’Angelo, C.; Pesce, M.; Greig, N.H. Neuronal Cellular Responses to Extremely Low Frequency Electromagnetic Field Exposure: Implications Regarding Oxidative Stress and Neurodegeneration. PLoS ONE 2014, 9, e104973. [Google Scholar] [CrossRef]
- Naarala, J.; Kesari, K.K.; McClure, I.; Chavarriaga, C.; Juutilainen, J.; Martino, C.F. Direction-Dependent Effects of Combined Static and ELF Magnetic Fields on Cell Proliferation and Superoxide Radical Production. BioMed Research International 2017, 2017, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Benassi, B.; Filomeni, G.; Montagna, C.; Merla, C.; Lopresto, V.; Pinto, R.; Marino, C.; Consales, C. Extremely Low Frequency Magnetic Field (ELF-MF) Exposure Sensitizes SH-SY5Y Cells to the Pro-Parkinson’s Disease Toxin MPP+. Mol Neurobiol 2016, 53, 4247–4260. [Google Scholar] [CrossRef] [PubMed]
- Ziech, D.; Franco, R.; Pappa, A.; Panayiotidis, M.I. Reactive Oxygen Species (ROS)––Induced Genetic and Epigenetic Alterations in Human Carcinogenesis. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis 2011, 711, 167–173. [Google Scholar] [CrossRef] [PubMed]
- Bhat, A.V.; Hora, S.; Pal, A.; Jha, S.; Taneja, R. Stressing the (Epi)Genome: Dealing with Reactive Oxygen Species in Cancer. Antioxidants & Redox Signaling 2018, 29, 1273–1292. [Google Scholar] [CrossRef]
- Seddon, A.R.; Liau, Y.; Pace, P.E.; Miller, A.L.; Das, A.B.; Kennedy, M.A.; Hampton, M.B.; Stevens, A.J. Genome-Wide Impact of Hydrogen Peroxide on Maintenance DNA Methylation in Replicating Cells. Epigenetics & Chromatin 2021, 14, 17. [Google Scholar] [CrossRef]
- Hitchler, M.J.; Domann, F.E. Redox Regulation of the Epigenetic Landscape in Cancer: A Role for Metabolic Reprogramming in Remodeling the Epigenome. Free Radical Biology and Medicine 2012, 53, 2178–2187. [Google Scholar] [CrossRef]
- Domann, F.E.; Hitchler, M.J. Aberrant Redox Biology and Epigenetic Reprogramming: Co-Conspirators across Multiple Human Diseases. Free Radical Biology and Medicine 2021, 170, 2–5. [Google Scholar] [CrossRef]
- Marjanovic Cermak, A.M.; Pavicic, I.; Trosic, I. Oxidative Stress Response in SH-SY5Y Cells Exposed to Short-Term 1800 MHz Radiofrequency Radiation. Journal of Environmental Science and Health, Part A 2018, 53, 132–138. [Google Scholar] [CrossRef]
- Stefi, A.L.; Margaritis, L.H.; Skouroliakou, A.S.; Vassilacopoulou, D. Mobile Phone Electromagnetic Radiation Affects Amyloid Precursor Protein and α-Synuclein Metabolism in SH-SY5Y Cells. Pathophysiology 2019, 26, 203–212. [Google Scholar] [CrossRef]
- Martin, C.; Percevault, F.; Ryder, K.; Sani, E.; Le Cun, J.; Zhadobov, M.; Sauleau, R.; Le Dréan, Y.; Habauzit, D. Effects of Radiofrequency Radiation on Gene Expression: A Study of Gene Expressions of Human Keratinocytes From Different Origins. Bioelectromagnetics 2020, 41, 552–557. [Google Scholar] [CrossRef] [PubMed]
- Del Vecchio, G.; Giuliani, A.; Fernandez, M.; Mesirca, P.; Bersani, F.; Pinto, R.; Ardoino, L.; Lovisolo, G.A.; Giardino, L.; Calzà, L. Continuous Exposure to 900MHz GSM-Modulated EMF Alters Morphological Maturation of Neural Cells. Neuroscience Letters 2009, 455, 173–177. [Google Scholar] [CrossRef] [PubMed]
- Del Vecchio, G.; Giuliani, A.; Fernandez, M.; Mesirca, P.; Bersani, F.; Pinto, R.; Ardoino, L.; Lovisolo, G.A.; Giardino, L.; Calzà, L. Effect of Radiofrequency Electromagnetic Field Exposure on in Vitro Models of Neurodegenerative Disease. Bioelectromagnetics 2009, 30, 564–572. [Google Scholar] [CrossRef] [PubMed]
- Flunkert, J.; Maierhofer, A.; Dittrich, M.; Müller, T.; Horvath, S.; Nanda, I.; Haaf, T. Genetic and Epigenetic Changes in Clonal Descendants of Irradiated Human Fibroblasts. Experimental Cell Research 2018, 370, 322–332. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt Removes Adapter Sequences from High-Throughput Sequencing Reads. EMBnet j. 2011, 17, 10. [Google Scholar] [CrossRef]
- Zhang, J.; Kobert, K.; Flouri, T.; Stamatakis, A. PEAR: A Fast and Accurate Illumina Paired-End ReAd MergeR. Bioinformatics 2014, 30, 614–620. [Google Scholar] [CrossRef]
- Scala, G.; Affinito, O.; Palumbo, D.; Florio, E.; Monticelli, A.; Miele, G.; Chiariotti, L.; Cocozza, S. AmpliMethProfiler: A Pipeline for the Analysis of CpG Methylation Profiles of Targeted Deep Bisulfite Sequenced Amplicons. BMC Bioinformatics 2016, 17, 484. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




