Submitted:
27 April 2023
Posted:
28 April 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Chicken Embryo Fibroblast Cells
2.3. Virus
2.4. Tissue Culture Infection Doses (TCID50)
2.5. Viral Infection
2.6. RNA Extraction and cDNA Synthesis
2.7. Primer Pairs Design
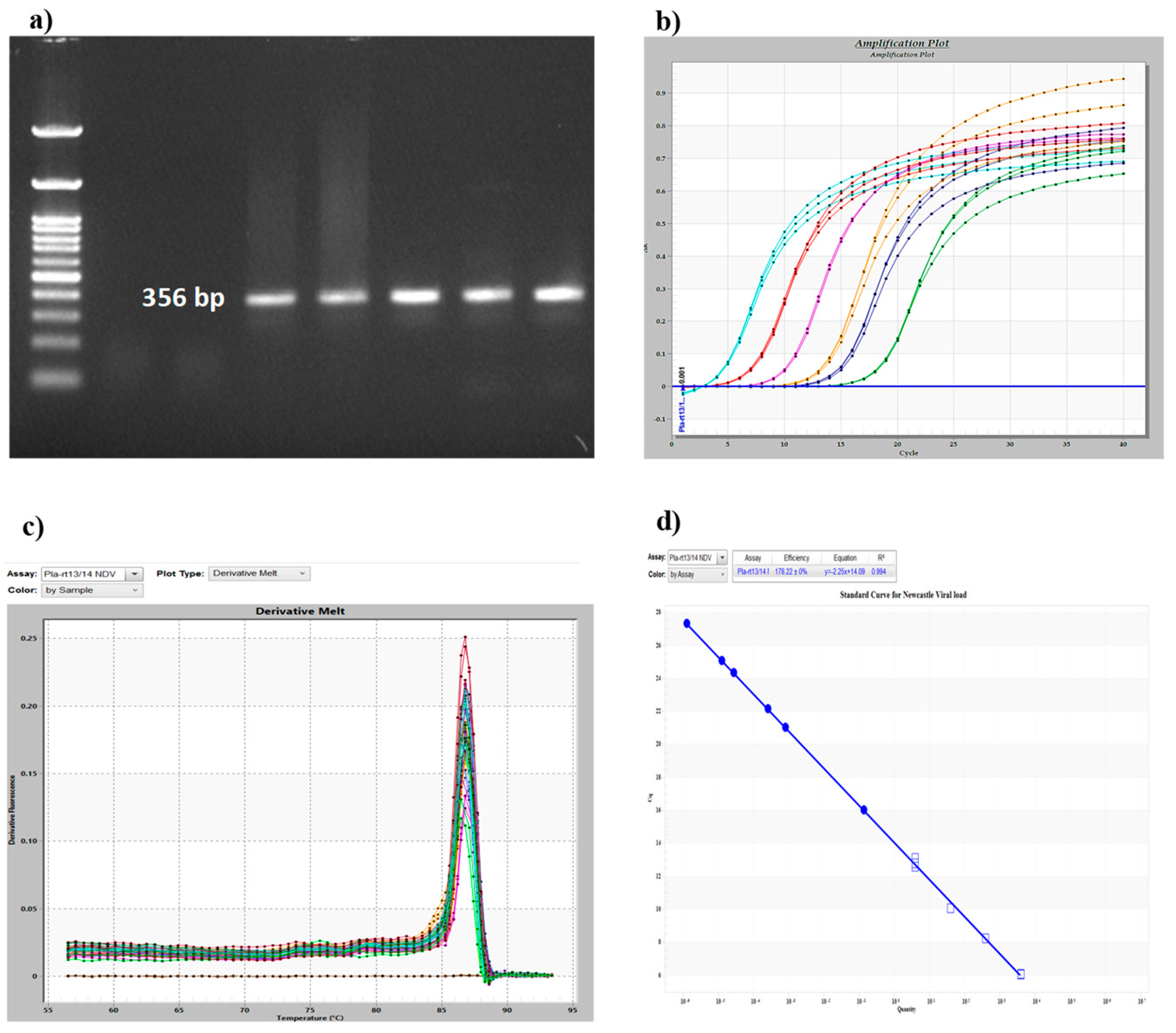
2.8. Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
2.9. Standard Curve Analysis for Detection of Viral Load
2.10. Statistical Analysis
3. Results
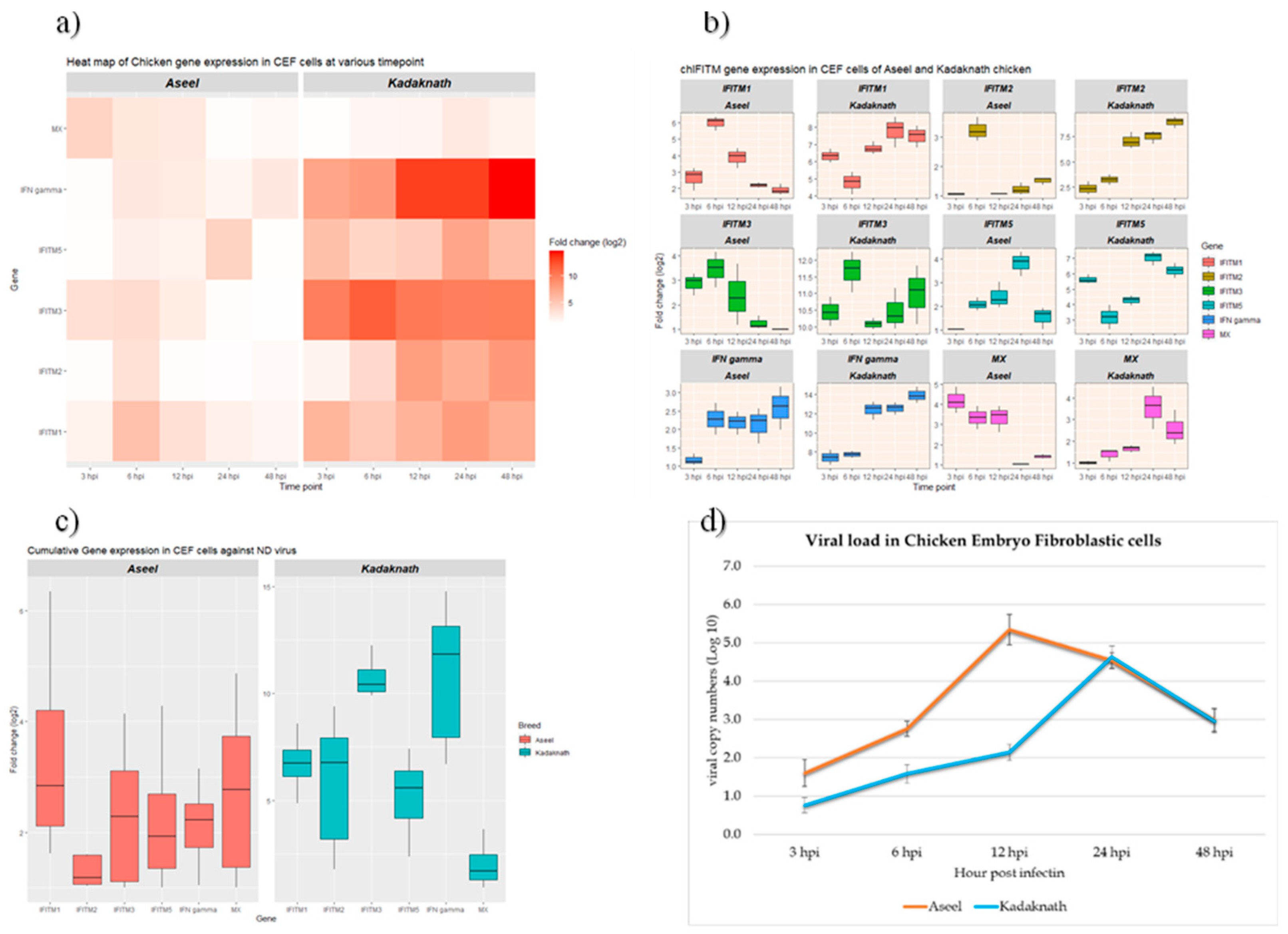
3.1. NDV Infection-Induced Cytopathic Changes and Viral Load
3.2. Expression Analysis of chIFITM Gene in Newcastle Disease Virus Infected CEF Cells
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chambers, P.; Millar, N.S.; Bingham, R.W.; Emmerson, P.T. Molecular Cloning of Complementary DNA to Newcastle Disease Virus, and Nucleotide Sequence Analysis of the Junction between the Genes Encoding the Haemagglutinin-Neuraminidase and the Large Protein. J. Gen. Virol. 1986, 67 Pt 3, 475–486. [Google Scholar] [CrossRef]
- O.I.E. Newcastle Disease. Chapter 2.3.14; In, Ed.; World Organisation for Animal Health: Paris, 2012. [Google Scholar]
- Qosimah, D.; Murwani, S.; Sudjarwo, E.; Lesmana, M.A. Effect of Newcastle Disease Virus Level of Infection on Embryonic Length, Embryonic Death, and Protein Profile Changes. Vet. World 2018, 11, 1316–1320. [Google Scholar] [CrossRef] [PubMed]
- Lamb, R.A.; Parks, G.D. Paramyxoviridae: The Viruses and Their Replication. In Fields virology; Fields, B.N., Knipe, D.N., Howley, P.M., Eds.; Lippincott, Williams, and Wilkins, 2007; pp. 1449–1496.
- Kalaria, V.A.; Prajapati, K.S.; Javia, B.B.; Bhadaniya, A.R.; Fefar, D.T.; Vagh, A.A.; Trangadiya, B.J.; Padodara, R.J.; Mokaria, K.N.; Kumbhani, T.R. An Economical Impact of Newcastle Disease Outbreaks in Various Commercial Broiler Chicken Farms During 2020- 21 in Gujarat, India. Int J Curr Microbiol App Sci 2021, 10, 411–420. [Google Scholar]
- Aristizábal, B.; González, Á. Innate Immune System; Anaya, J.M., Shoenfeld, Y., Rojas-Villarraga, A., Eds.; Autoimmunity: Bogota (Colombia, 2013. [Google Scholar]
- Dai, M.; Xie, T.; Liao, M. Systematic Identification of Chicken Type I, II and III Interferon-Stimulated Genes. Vet Res 2020, 51, 70. [Google Scholar] [CrossRef] [PubMed]
- Brass, A.L.; Huang, I.-C.; Benita, Y.; John, S.P.; Krishnan, M.N.; Feeley, E.M.; Ryan, B.J.; Weyer, J.L.; van der Weyden, L.; Fikrig, E.; et al. The IFITM Proteins Mediate Cellular Resistance to Influenza A H1N1 Virus, West Nile Virus, and Dengue Virus. Cell 2009, 139, 1243–1254. [Google Scholar] [CrossRef] [PubMed]
- Jiang, D.; Weidner, J.M.; Qing, M.; Pan, X.-B.; Guo, H.; Xu, C.; Zhang, X.; Birk, A.; Chang, J.; Shi, P.-Y.; et al. Identification of Five Interferon-Induced Cellular Proteins That Inhibit West Nile Virus and Dengue Virus Infections. J. Virol. 2010, 84, 8332–8341. [Google Scholar] [CrossRef] [PubMed]
- Huang, I.-C.; Bailey, C.C.; Weyer, J.L.; Radoshitzky, S.R.; Becker, M.M.; Chiang, J.J.; Brass, A.L.; Ahmed, A.A.; Chi, X.; Dong, L.; et al. Distinct Patterns of IFITM-Mediated Restriction of Filoviruses, SARS Coronavirus, and Influenza A Virus. PLOS Pathog. 2011, 7, e1001258. [Google Scholar] [CrossRef]
- Feeley, E.M.; Sims, J.S.; John, S.P.; Chin, C.R.; Pertel, T.; Chen, L.-M.; Gaiha, G.D.; Ryan, B.J.; Donis, R.O.; Elledge, S.J.; et al. IFITM3 Inhibits Influenza A Virus Infection by Preventing Cytosolic Entry. PLoS Pathog. 2011, 7, e1002337. [Google Scholar] [CrossRef]
- Fife, M.; Moore, J. IFITM Knockdown/Knockout Technology for Vaccine Production.
- Steyn, A.; Keep, S.; Bickerton, E.; Fife, M. The Characterization of ChIFITMs in Avian Coronavirus Infection In Vivo, Ex Vivo and In Vitro. Charact. ChIFITMs Avian Coronavirus Infect. Vivo Ex Vivo Vitro Genes 2020, 11, 918. [Google Scholar] [CrossRef]
- Smith, S.E.; Gibson, M.S.; Wash, R.S.; Ferrara, F.; Wright, E.; Temperton, N.; Kellam, P.; Fife, M. Chicken Interferon-Inducible Transmembrane Protein 3 Restricts Influenza Viruses and Lyssaviruses In Vitro. J. Virol. 2013, 87, 12957–12966. [Google Scholar] [CrossRef]
- Lanz, C.; Yángüez, E.; Andenmatten, D.; Stertz, S. Correction for Lanz et al., Swine Interferon-Inducible Transmembrane Proteins Potently Inhibit Influenza A Virus Replication. J. Virol. 2015, 89, 2988. [Google Scholar] [CrossRef] [PubMed]
- Kidani, S.; Okuzaki, Y.; Kaneoka, H.; Asai, S.; Murakami, S.; Murase, Y.; Iijima, S.; Nishijima, K. Expression of Interferon-Inducible Transmembrane Proteins in the Chicken and Possible Role in Prevention of Viral Infections. Cytotechnology 2017, 69, 477–484. [Google Scholar] [CrossRef] [PubMed]
- Hitchner, B.; Donermuch, H.; Graham Purchas, H.; William, E. Isolation and Identification of Pathogens, 2nd ed.American Association of Avian Pathologists, Inc, 1980. [Google Scholar]
- Office International des Epizooties (OIE) OIE Terrestrial Manual 2021 2021.
- Toyoda, T.; Sakaguchi, T.; Hirota, H.; Gotoh, B.; Kuma, K.; Miyataj, T.; Nagai, Y. Newcastle Disease Virus Evolution: II. Lack of Gene Recombination in Generating Virulent and Avirulent Strains. Virology 1989, 169, 273–282. [Google Scholar] [CrossRef] [PubMed]
- Ogbole, O.O.; Akinleye, T.E.; Segun, P.A.; Faleye, T.C.; Adeniji, A.J. In Vitro Antiviral Activity of Twenty-Seven Medicinal Plant Extracts from Southwest Nigeria against Three Serotypes of Echoviruses. Virol. J. 2018, 15, 110. [Google Scholar] [CrossRef] [PubMed]
- Gowthaman, V.; Ganesan, V.; Gopala Krishna Murthy, T.R.; Nair, S.; Yegavinti, N.; Saraswathy, P.V.; Suresh Kumar, G.; Udhayavel, S.; Senthilvel, K.; Subbiah, M. Molecular Phylogenetics of Newcastle Disease Viruses Isolated from Vaccinated Flocks during Outbreaks in Southern India Reveals Circulation of a Novel Sub-Genotype. Transbound. Emerg. Dis. 2019, 66, 363–372. [Google Scholar] [CrossRef] [PubMed]
- Qiu, X.; Yu, Y.; Yu, S.; Zhan, Y.; Wei, N.; Song, C.; Sun, Y.; Tan, L.; Ding, C. Development of Strand-Specific Real-Time RT-PCR to Distinguish Viral RNAs during Newcastle Disease Virus Infection. Sci. World J. 2014, 934851. [Google Scholar] [CrossRef] [PubMed]
- Pfaf, M.W. A New Mathematical Model for Relative Quantification in Real-Time RT–PCR. Nucleic Acids Res 2001, 29, 45. [Google Scholar] [CrossRef]
- Ramakrishnan, S.; Annamalai, A.; Sachan, S.; Kumar, A.; Sharma, B.K.; Govindaraj, E.; Chellappa, M.M.; Dey, S.; Krishnaswamy, N. Synergy of Lipopolysaccharide and Resiquimod on Type I Interferon, pro-Inflammatory Cytokine, Th1 and Th2 Response in Chicken Peripheral Blood Mononuclear Cells. Mol. Immunol. 2015, 64, 177–182. [Google Scholar] [CrossRef]
- Susta, L.; Cornax, I.; Diel, D.G.; Garcia, S.C.; Miller, P.J.; Liu, X.; Hu, S.; Brown, C.C.; Afonso, C.L. Expression of Interferon Gamma by a Highly Virulent Strain of Newcastle Disease Virus Decreases Its Pathogenicity in Chickens. Microb Pathog 2013, 61–62, 73–83. [Google Scholar] [CrossRef]
- Yang, X.; Arslan, M.; Liu, X.; Song, H.; Du, M.; Li, Y.; Zhang, Z. IFN-γ Establishes Interferon-Stimulated Gene-Mediated Antiviral State against Newcastle Disease Virus in Chicken Fibroblasts. Acta Biochim Biophys Sin Shanghai 2020, 52, 268–280. [Google Scholar] [CrossRef]
- Bashir, K.; Kappala, D.; Singh, Y. Combination of TLR2 and TLR3 Agonists Derepress Infectious Bursal Disease Virus Vaccine-Induced Immunosuppression in the Chicken. Sci Rep 2019, 9, 8197. [Google Scholar] [CrossRef] [PubMed]
- Li, X.Y.; Qu, L.J.; Hou, Z.C.; Yao, J.F.; Xu, G.Y.; Yang, N. Genomic Structure and Diversity of the Chicken Mx Gene. Poult. Sci. 2007, 86, 786–789. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Li, J.; Winkler, C.A.; P, A.; Guo, J.-T. IFITM Genes, Variants, and Their Roles in the Control and Pathogenesis of Viral Infections. Front Microbiol 2019, 9, 3228. [Google Scholar] [CrossRef] [PubMed]
- Q., W.; X., Y.; C., L.; H., D.; C, W. Distribution of IFITM3 in Yellow-Feathered Broilers and Inhibition of Avian Reovirus Multiplication by IFITM3. Braz. J. Poult. Sci. 2018, 20, 377–386. [CrossRef]
- Friedlová, N.; Zavadil Kokáš, F.; Hupp, T.R.; Vojtěšek, B.; Nekulová, M. IFITM Protein Regulation and Functions: Far beyond the Fight against Viruses. Front. Immunol. 2022, 13. [Google Scholar] [CrossRef] [PubMed]
- Whitehead, T.J. Characterisation of Chicken Interferon-Inducible Transmembrane Proteins: Locus Architecture, Gene Expression and Viral Restriction. Doctoral thesis (Ph. D), 2018.
- Haller, O.; Kochs, G. Mx Genes: Host Determinants Controlling Influenza Virus Infection and Trans-Species Transmission. Hum. Genet. 2020, 139, 695–705. [Google Scholar] [CrossRef] [PubMed]
- Blyth, G.A.D.; Chan, W.F.; Webster, R.G.; Magor, K.E. Duck Interferon-Inducible Transmembrane Protein 3 Mediates Restriction of Influenza Viruses. J. Virol. 2016, 90, 103–116. [Google Scholar] [CrossRef]
- Perreira, J.M.; Chin, C.R.; Feeley, E.M.; Brass, A.L. IFITMs Restrict the Replication of Multiple Pathogenic Viruses. J. Mol. Biol. 2013, 425, 4937–4955. [Google Scholar] [CrossRef]
- Desai, T.M.; Marin, M.; Chin, C.R.; Savidis, G.; Brass, A.L.; Melikyan, G.B. IFITM3 Restricts Influenza A Virus Entry by Blocking the Formation of Fusion Pores Following Virus-Endosome Hemifusion. PLoS Pathog 2014, 10, 1004048. [Google Scholar] [CrossRef]
- Amini, B.O.S.; Choi, Y.J.; Lee, J.H.; Shi, M.; Huang, I.C.; Farzan, M.; Jung, J.U. The Antiviral Effector IFITM3 Disrupts Intracellular Cholesterol Homeostasis to Block Viral Entry. 2013, 452–464.
- Ravindra, P.V.; Tiwari, A.K.; Ratta, B.; Chaturvedi, U.; Palia, S.K.; Chauhan, R.S. Newcastle Disease Virus-Induced Cytopathic Effect in Infected Cells Is Caused by Apoptosis. Virus Res 2009, 141, 13–20. [Google Scholar] [CrossRef]
- Li, X.; Su, S.; Cui, N.; Zhou, H.; Liu, X.; Cui, Z. Transcriptome Analysis of Chicken Embryo Fibroblast Cell Infected with Marek’s Disease Virus of GX0101∆LTR. Braz. J. Poult. Sci. 2017, 19, 179–184. [Google Scholar] [CrossRef]
- Siegrist, F.; Ebeling, M.; Certa, U. The Small Interferon-Induced Transmembrane Genes and Proteins. J Interferon Cytokine Res 2011, 31, 183–197. [Google Scholar] [CrossRef] [PubMed]
- Anjum, F.R.; Rahman, S.U.; Aslam, M.A.; S, Q.A. Antiviral Potential and Stability Analysis of Chicken Interferon-α Produced by Newcastle Disease Virus in Chicken Embryo Fibroblast Cells. Vet Med-Czech 2021, 66, 197–207. [Google Scholar] [CrossRef]
| Gene | Primer | Sequence (5'->3') | Reference |
|---|---|---|---|
| chIFITM1 | FP | GCAGGATGTGACCACCACTA | NM_001350059.2 |
| RP | CTTCGCTGTCCTCCCATAGC | ||
| chIFITM2 | FP | AACAGGCGGAGGTGAGCAT | NM_001350058.2 |
| RP | AAGATGAGCGAGGGGAAGCA | ||
| chIFITM3 | FP | CGTGAAGTCCAGGGATCGCA | NM_001350061.2 |
| RP | GCAACCAGGGCGATGATGAG | ||
| chIFITM5 | FP | CCAACCCCACTTCTGGACGA | NM_001199498.1 |
| RP | ATCACTCCGAAGGGCACGAC | ||
| chMx | FP | GTCCAAGAGGCTGAATAACAGAG | NM_204609 |
| RP | GTCGGATCTTTCTGTCATATTGG | ||
| chIFN-γ | FP | TGAGCCAGATTGTTTCGATG | [24] |
| RP | CTTGGCCAGGTCCATGATA | ||
| chβ-Actin | FP | TATGTGCAAGGCCGGTTTC | |
| RP | TGTCTTTCTGGCCCATACCAA | ||
| NDV-NP | Pla-rt13 | CAACAATAGGAGTGGAGTGTCTGA | [22] |
| Pla-rt14 | CAGGGTATCGGTGATGTCTTCT |
| Breed | Time point | IFITM1 | IFITM2 | IFITM3 | IFITM5 | IFN-γ | MX |
|---|---|---|---|---|---|---|---|
| 3 hpi | 2.64±0.40c | 1.06±0.01c | 2.87±0.26b | 1.01±0.00b | 1.17±0.09c | 4.18±0.38a | |
| 6 hpi | 6.00±0.25a | 3.25±0.24a | 3.46±0.42a | 2.07±0.16b | 2.28±0.24ab | 3.35±0.33b | |
| Aseel | 12 hpi | 3.88±0.35b | 1.06±0.00c | 2.38±0.71b | 2.41±0.32b | 2.18±0.18b | 3.34±0.38b |
| 24 hpi | 2.19±0.09c | 1.22±0.12bc | 1.26±0.16c | 3.82±0.30a | 2.13±0.28b | 1.03±0.01c | |
| 48 hpi | 1.91±0.20cd | 1.51±0.08b | 1.00±0.00c | 1.53±0.27c | 2.59±0.34a | 1.43±0.06c | |
| 3 hpi | 6.33±0.24b | 2.39±0.38d | 10.44±0.26b | 5.63±0.17b | 7.46±0.45c | 1.01±0.05c | |
| 6 hpi | 4.77±0.38c | 3.25±0.29c | 11.68±0.36a | 3.18±0.46c | 7.77±0.21c | 1.38±0.16c | |
| Kadaknath | 12 hpi | 6.77±0.21b | 7.02±0.47b | 10.08±0.10b | 4.28±0.19bc | 12.38±0.54b | 1.66±0.10c |
| 24 hpi | 7.79±0.52a | 7.48±0.36b | 10.47±0.36ab | 7.04±0.27a | 12.56±0.39b | 3.57±0.56a | |
| 48 hpi | 7.48±0.36a | 8.89±0.33a | 11.00±0.51a | 6.22±0.27a | 13.9±0.49a | 2.56±0.46b |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).





