Submitted:
04 May 2023
Posted:
04 May 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and methods
2.1. Data collection
2.2. Species-specific TE library construction
2.3. TE annotation and statistical analysis
2.4. TE propagation activity
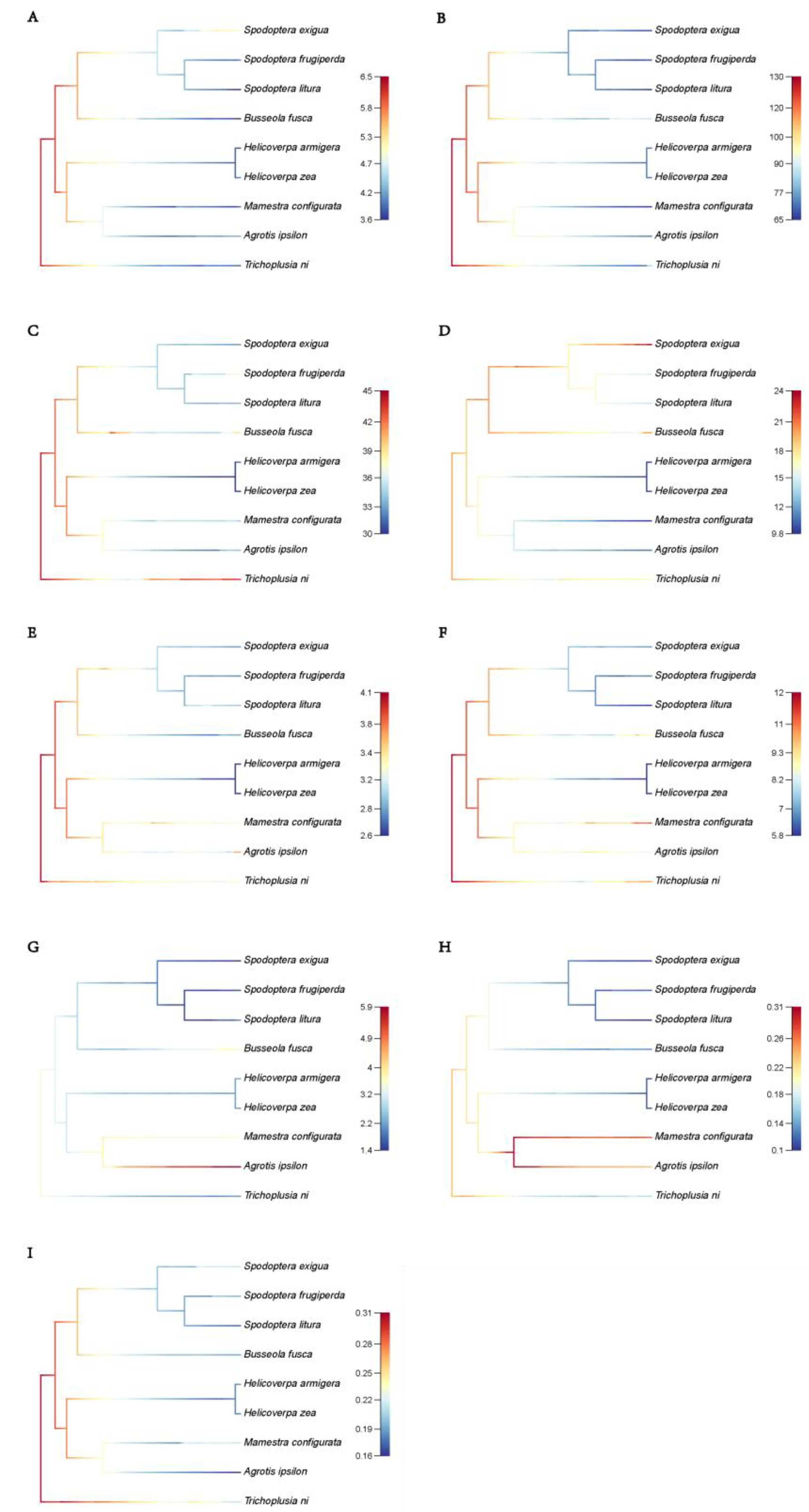
2.5. Calculation of phylogenetic signal
2.6. Ancestral node reconstruction and predicting the change rate of TEs
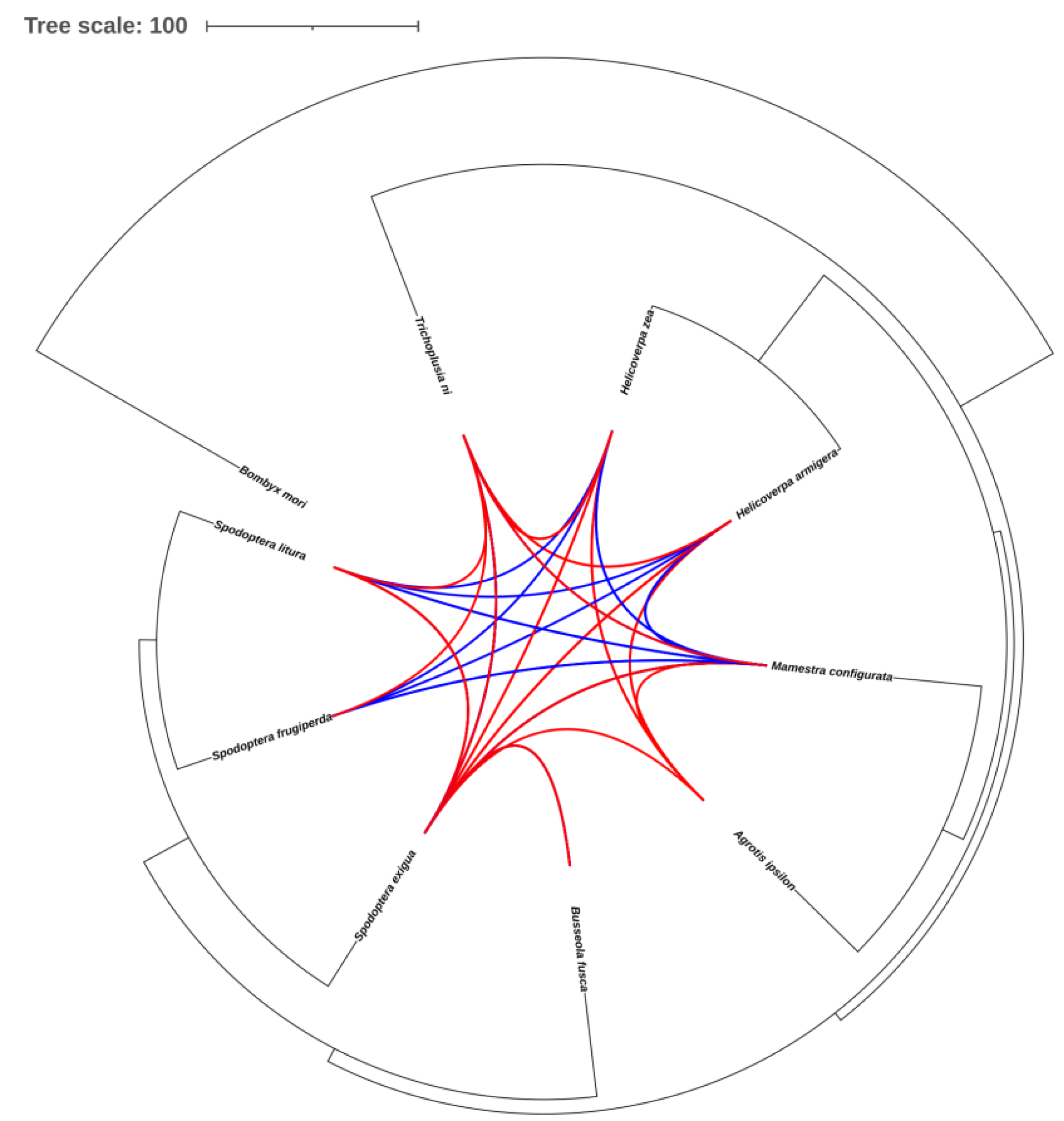
2.7. Identification of TE horizontal transfer events between Noctuidae species
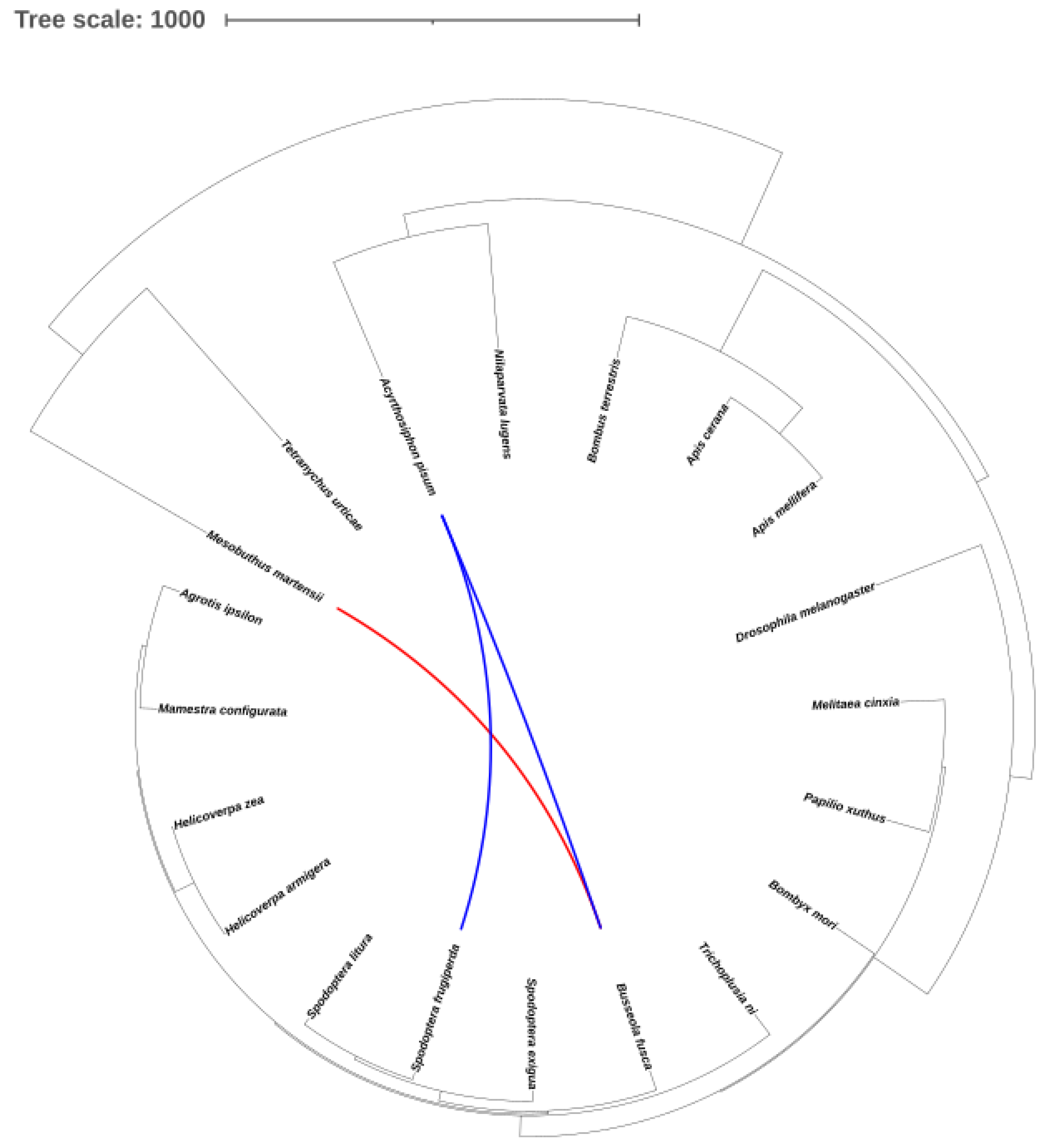
2.8. Identification of TE horizontal transfer events between Noctuidae species and other arthropods
3. Results
3.1. Construction of TE libraries in the ten Noctuidae species
3.2. TEcontents
3.3. TE subfamilies and copy numbers
3.4. Propagation activity of TE subfamilies
3.5. Relatedness of TE content with genome size of noctuid species
3.6. Phylogenetic signals and changes of TE load
3.7. Transposon horizontal transfer events among Noctuidae species
3.8. Transposon horizontal transfer events between 9 Noctuidae species and other arthropods
4. Discussion
4.1. TEs shape the genome diversity of Noctuidae species
4.2. TE expansion activity correlated with phylogeny of Noctuidae species
4.3. HTT events on the genomes of the noctuid moths
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Keegan, K.L.;Rota, J.;Zahiri, R.;Zilli, A.;Wahlberg, N.;Schmidt, B.C.;Lafontaine, J.D.;Goldstein, P.Z.;Wagner, D.L. Toward a Stable Global Noctuidae (Lepidoptera) Taxonomy. Insect Systematics and Diversity. 2021, 5, 3. [CrossRef]
- Le Goff, G.;Nauen, R. Recent Advances in the Understanding of Molecular Mechanisms of Resistance in Noctuid Pests. Insects. 2021, 12, 8. [CrossRef]
- Di Lelio, I.;Coppola, M.;Comite, E.;Molisso, D.;Lorito, M.;Woo, S.L.;Pennacchio, F.;Rao, R.;Digilio, M.C. Temperature Differentially Influences the Capacity of Trichoderma Species to Induce Plant Defense Responses in Tomato Against Insect Pests. Front Plant Sci. 2021, 12, 678830. [CrossRef]
- de Almeida, L.M.;Carareto, C.M.A. Multiple events of horizontal transfer of the Minos transposable element between Drosophila species. Molecular Phylogenetics and Evolution. 2005, 35, 3, 583-594.
- Hof, A.E.v.t.;Campagne, P.;Rigden, D.J.;Yung, C.J.;Lingley, J.;Quail, M.A.;Hall, N.;Darby, A.C.;Saccheri, I.J. The industrial melanism mutation in British peppered moths is a transposable element. Nature. 2016, 534, 7605, 102-105. [CrossRef]
- Peng, C.;Niu, L.;Deng, J.;Yu, J.;Zhang, X.;Zhou, C.;Xing, J.;Li, J.J.M.D. Can-SINE dynamics in the giant panda and three other Caniformia genomes. 2018, 9, 1, 1-14. [CrossRef]
- Talla, V.;Suh, A.;Kalsoom, F.;Dincă, V.;Vila, R.;Friberg, M.;Wiklund, C.;Backström, N.J.G.B.;Evolution Rapid increase in genome size as a consequence of transposable element hyperactivity in wood-white (Leptidea) butterflies. 2017, 9, 10, 2491-2505.
- Cordaux, R.;Batzer, M.A. The impact of retrotransposons on human genome evolution. Nat Rev Genet. 2009, 10, 10, 691-703. [CrossRef]
- Wu, C.;Lu, J. Diversification of Transposable Elements in Arthropods and Its Impact on Genome Evolution. Genes (Basel). 2019, 10, 5. [CrossRef]
- Sessegolo, C.;Burlet, N.;Haudry, A. Strong phylogenetic inertia on genome size and transposable element content among 26 species of flies. Biology Letters. 2016, 12, 8. [CrossRef]
- Petersen, M.;Armisen, D.;Gibbs, R.A.;Hering, L.;Khila, A.;Mayer, G.;Richards, S.;Niehuis, O.;Misof, B. Diversity and evolution of the transposable element repertoire in arthropods with particular reference to insects. BMC Evol Biol. 2019, 19, 1, 11.
- Schaack, S.;Gilbert, C.;Feschotte, C. Promiscuous DNA: horizontal transfer of transposable elements and why it matters for eukaryotic evolution. Trends Ecol Evol. 2010, 25, 9, 537-46. [CrossRef]
- Bartolome, C.;Bello, X.;Maside, X. Widespread evidence for horizontal transfer of transposable elements across Drosophila genomes. Genome Biol. 2009, 10, 2, R22. [CrossRef]
- Daniels, S.B.;Peterson, K.R.;Strausbaugh, L.D.;Kidwell, M.G.;Chovnick, A.J.G. Evidence for horizontal transmission of the P transposable element between Drosophila species. 1990, 124, 2, 339-355. [CrossRef]
- Dotto, B.R.;Carvalho, E.L.;Silva, A.F.;Duarte Silva, L.F.;Pinto, P.M.;Ortiz, M.F.;Wallau, G.L. HTT-DB: horizontally transferred transposable elements database. Bioinformatics. 2015, 31, 17, 2915-7. [CrossRef]
- Kofler, R.;Hill, T.;Nolte, V.;Betancourt, A.J.;Schlotterer, C. The recent invasion of natural Drosophila simulans populations by the P-element. Proc Natl Acad Sci U S A. 2015, 112, 21, 6659-63. [CrossRef]
- Reiss, D.;Mialdea, G.;Miele, V.;de Vienne, D.M.;Peccoud, J.;Gilbert, C.;Duret, L.;Charlat, S. Global survey of mobile DNA horizontal transfer in arthropods reveals Lepidoptera as a prime hotspot. PLoS genetics. 2019, 15, 2, e1007965-e1007965. [CrossRef]
- Bao, W.;Kojima, K.K.;Kohany, O. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob DNA. 2015, 6, 11. [CrossRef]
- Kumar, S.;Stecher, G.;Suleski, M.;Hedges, S.B. TimeTree: A Resource for Timelines, Timetrees, and Divergence Times. Mol Biol Evol. 2017, 34, 7, 1812-1819. [CrossRef]
- Letunic, I.;Bork, P. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W1, W293-W296. [CrossRef]
- Behere, G.T.;Tay, W.T.;Russell, D.A.;Heckel, D.G.;Appleton, B.R.;Kranthi, K.R.;Batterham, P. Mitochondrial DNA analysis of field populations of Helicoverpa armigera (Lepidoptera: Noctuidae) and of its relationship to H. zea. BMC Evol Biol. 2007, 7, 117. [CrossRef]
- Kergoat, G.J.;Prowell, D.P.;Le Ru, B.P.;Mitchell, A.;Dumas, P.;Clamens, A.-L.;Condamine, F.L.;Silvain, J.-F.J.M.P.;Evolution Disentangling dispersal, vicariance and adaptive radiation patterns: a case study using armyworms in the pest genus Spodoptera (Lepidoptera: Noctuidae). Molecular Phylogenetics and Evolution. 2012, 65, 3, 855-870. [CrossRef]
- Flynn, J.M.;Hubley, R.;Goubert, C.;Rosen, J.;Clark, A.G.;Feschotte, C.;Smit, A.F.J.P.o.t.N.A.o.S. RepeatModeler2 for automated genomic discovery of transposable element families. 2020, 117, 17, 9451-9457. [CrossRef]
- Ellinghaus, D.;Kurtz, S.;Willhoeft, U. LTRharvest, an efficient and flexible software for de novo detection of LTR retrotransposons. BMC Bioinformatics. 2008, 9, 18. [CrossRef]
- Ou, S.;Jiang, N. LTR_retriever: A Highly Accurate and Sensitive Program for Identification of Long Terminal Repeat Retrotransposons. Plant Physiol. 2018, 176, 2, 1410-1422.
- Price, A.L.;Jones, N.C.;Pevzner, P.A. De novo identification of repeat families in large genomes. Bioinformatics. 2005, 21, I351-I358. [CrossRef]
- Bao, Z.;Eddy, S.R. Automated de novo identification of repeat sequence families in sequenced genomes. Genome Res. 2002, 12, 8, 1269-76. [CrossRef]
- Hoede, C.;Arnoux, S.;Moisset, M.;Chaumier, T.;Inizan, O.;Jamilloux, V.;Quesneville, H. PASTEC: an automatic transposable element classification tool. PLoS One. 2014, 9, 5, e91929. [CrossRef]
- Abrusan, G.;Grundmann, N.;DeMester, L.;Makalowski, W. TEclass--a tool for automated classification of unknown eukaryotic transposable elements. Bioinformatics. 2009, 25, 10, 1329-30. [CrossRef]
- Chan, P.P.;Lin, B.Y.;Mak, A.J.;Lowe, T.M. tRNAscan-SE 2.0: improved detection and functional classification of transfer RNA genes. Nucleic Acids Res. 2021, 49, 16, 9077-9096. [CrossRef]
- Kalvari, I.;Nawrocki, E.P.;Ontiveros-Palacios, N.;Argasinska, J.;Lamkiewicz, K.;Marz, M.;Griffiths-Jones, S.;Toffano-Nioche, C.;Gautheret, D.;Weinberg, Z.;Rivas, E.;Eddy, S.R.;Finn, R.D.;Bateman, A.;Petrov, A.I. Rfam 14: expanded coverage of metagenomic, viral and microRNA families. Nucleic Acids Res. 2021, 49, D1, D192-D200. [CrossRef]
- Bailly-Bechet, M.;Haudry, A.;Lerat, E.J.M.D. “One code to find them all”: a perl tool to conveniently parse RepeatMasker output files. 2014, 5, 1, 1-15.
- Virtanen, P.;Gommers, R.;Oliphant, T.E.;Haberland, M.;Reddy, T.;Cournapeau, D.;Burovski, E.;Peterson, P.;Weckesser, W.;Bright, J.;van der Walt, S.J.;Brett, M.;Wilson, J.;Millman, K.J.;Mayorov, N.;Nelson, A.R.J.;Jones, E.;Kern, R.;Larson, E.;Carey, C.J.;Polat, I.;Feng, Y.;Moore, E.W.;VanderPlas, J.;Laxalde, D.;Perktold, J.;Cimrman, R.;Henriksen, I.;Quintero, E.A.;Harris, C.R.;Archibald, A.M.;Ribeiro, A.H.;Pedregosa, F.;van Mulbregt, P.;SciPy, C. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat Methods. 2020, 17, 3, 261-272.
- Keightley, P.D.;Pinharanda, A.;Ness, R.W.;Simpson, F.;Dasmahapatra, K.K.;Mallet, J.;Davey, J.W.;Jiggins, C.D. Estimation of the spontaneous mutation rate in Heliconius melpomene. Mol Biol Evol. 2015, 32, 1, 239-43. [CrossRef]
- Revell, L.J.J.M.i.e.;evolution phytools: an R package for phylogenetic comparative biology (and other things). 2012, 3, 2, 217-223.
- Pennell, M.W.;Eastman, J.M.;Slater, G.J.;Brown, J.W.;Uyeda, J.C.;FitzJohn, R.G.;Alfaro, M.E.;Harmon, L.J. geiger v2.0: an expanded suite of methods for fitting macroevolutionary models to phylogenetic trees. Bioinformatics. 2014, 30, 15, 2216-8. [CrossRef]
- Rabosky, D.L.;Donnellan, S.C.;Grundler, M.;Lovette, I.J. Analysis and visualization of complex macroevolutionary dynamics: an example from Australian scincid lizards. Syst Biol. 2014, 63, 4, 610-27. [CrossRef]
- Rabosky, D.L.;Goldberg, E.E. Model inadequacy and mistaken inferences of trait-dependent speciation. Syst Biol. 2015, 64, 2, 340-55. [CrossRef]
- Rabosky, D.L.;Santini, F.;Eastman, J.;Smith, S.A.;Sidlauskas, B.;Chang, J.;Alfaro, M.E. Rates of speciation and morphological evolution are correlated across the largest vertebrate radiation. Nat Commun. 2013, 4, 1958. [CrossRef]
- Rabosky, D.L.;Grundler, M.;Anderson, C.;Title, P.;Shi, J.J.;Brown, J.W.;Huang, H.;Larson, J.G.J.M.i.E.;Evolution BAMM tools: an R package for the analysis of evolutionary dynamics on phylogenetic trees. 2014, 5, 7, 701-707.
- Wang, D.;Zhang, Y.;Zhang, Z.;Zhu, J.;Yu, J. KaKs_Calculator 2.0: a toolkit incorporating gamma-series methods and sliding window strategies. Genomics Proteomics Bioinformatics. 2010, 8, 1, 77-80. [CrossRef]
- Manni, M.;Berkeley, M.R.;Seppey, M.;Simao, F.A.;Zdobnov, E.M. BUSCO Update: Novel and Streamlined Workflows along with Broader and Deeper Phylogenetic Coverage for Scoring of Eukaryotic, Prokaryotic, and Viral Genomes. Mol Biol Evol. 2021, 38, 10, 4647-4654. [CrossRef]
- Peccoud, J.;Loiseau, V.;Cordaux, R.;Gilbert, C. Massive horizontal transfer of transposable elements in insects. Proc Natl Acad Sci U S A. 2017, 114, 18, 4721-4726. [CrossRef]
- Zhang, H.H.;Peccoud, J.;Xu, M.R.;Zhang, X.G.;Gilbert, C. Horizontal transfer and evolution of transposable elements in vertebrates. Nat Commun. 2020, 11, 1, 1362. [CrossRef]
- Shao, F.;Han, M.;Peng, Z. Evolution and diversity of transposable elements in fish genomes. Sci Rep. 2019, 9, 1, 15399. [CrossRef]
- Pasyukova, E.G.;Nuzhdin, S.V.;Morozova, T.V.;Mackay, T.F.C. Accumulation of transposable elements in the genome of Drosophila melanogaster is associated with a decrease in fitness. Journal of Heredity. 2004, 95, 4, 284-290.
- Peccoud, J.;Cordaux, R.;Gilbert, C. Analyzing Horizontal Transfer of Transposable Elements on a Large Scale: Challenges and Prospects. Bioessays. 2018, 40, 2. [CrossRef]
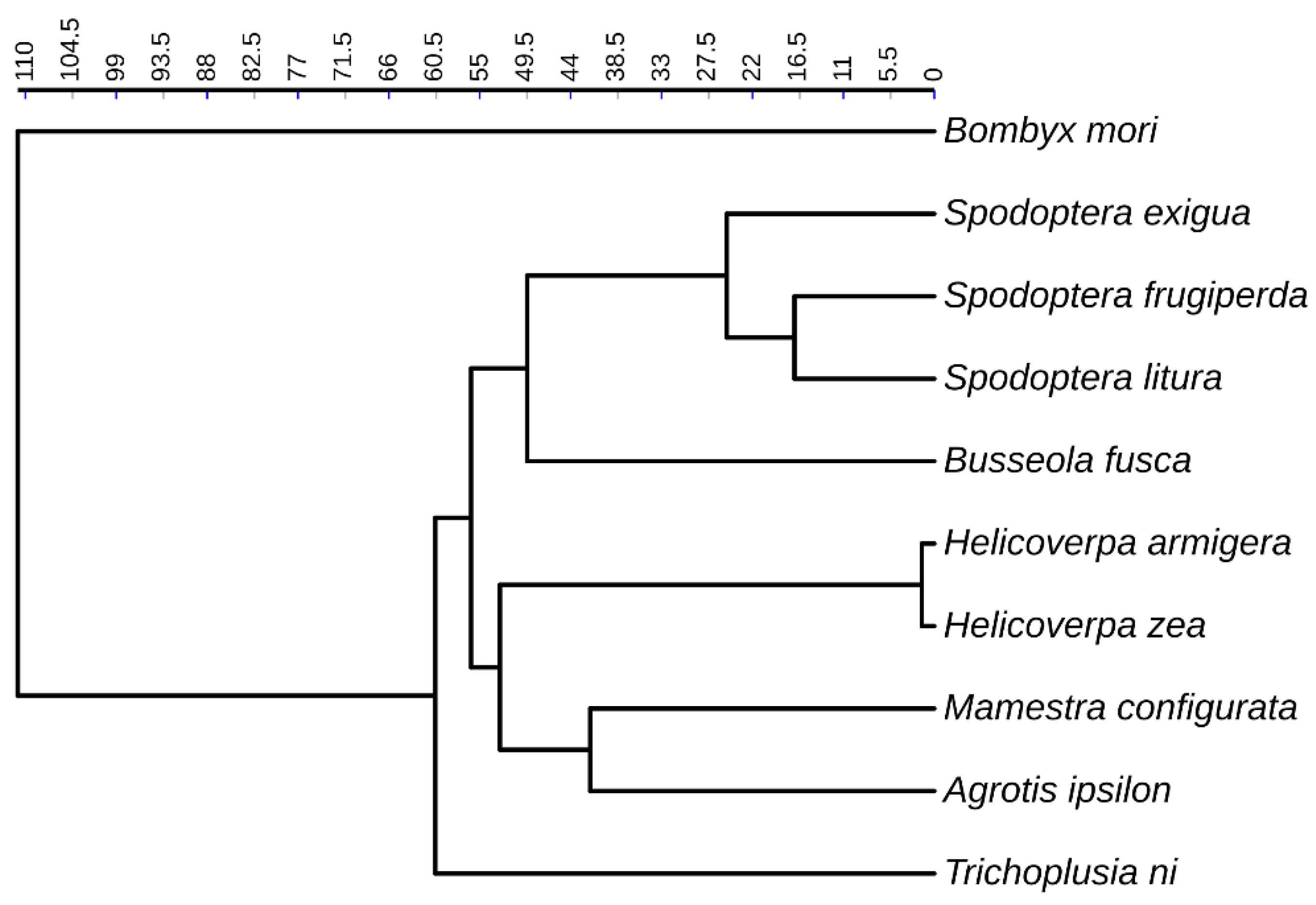
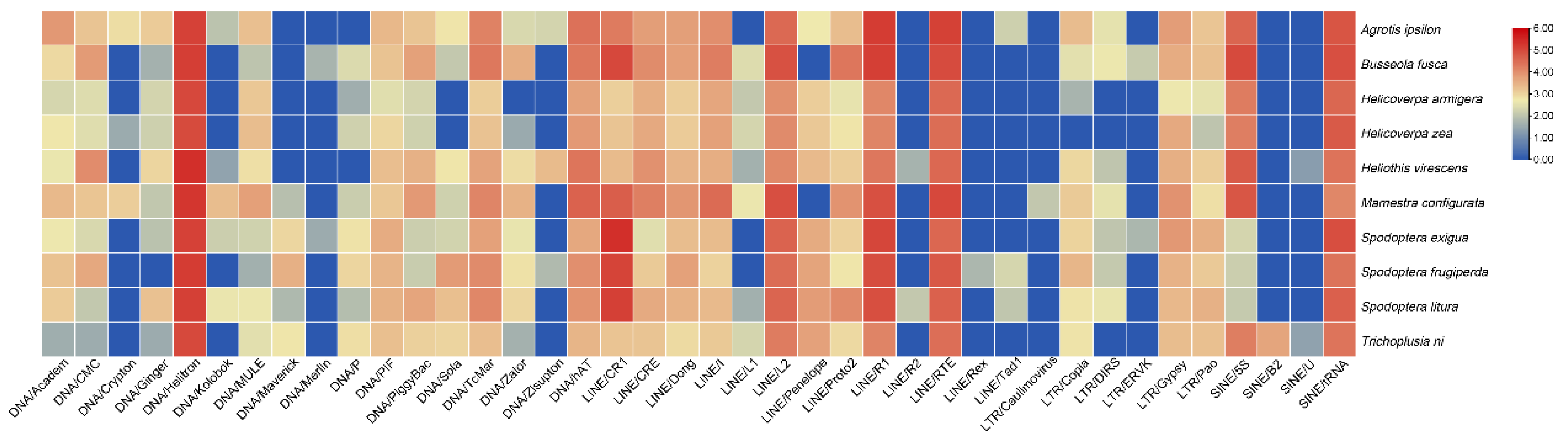
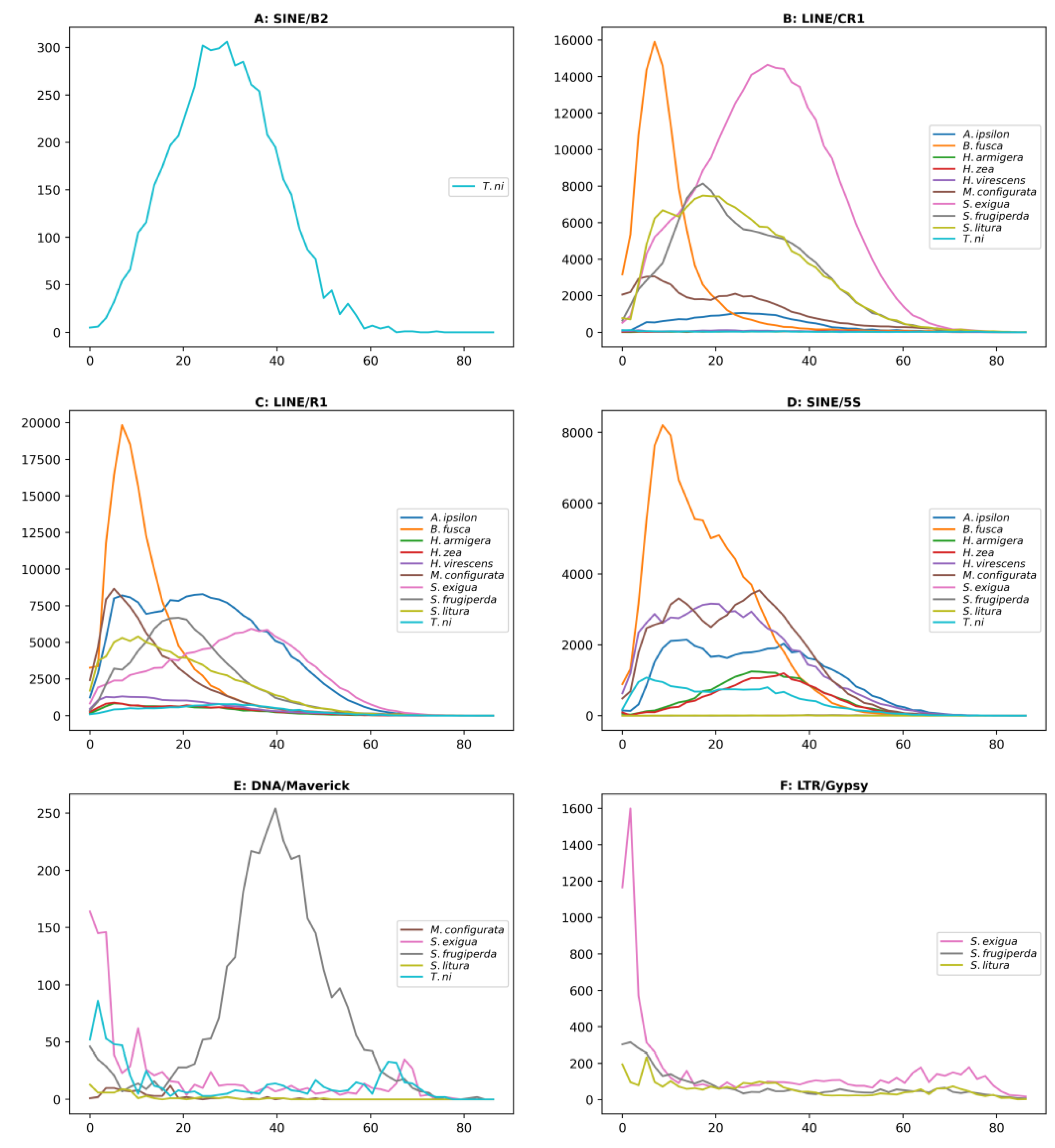
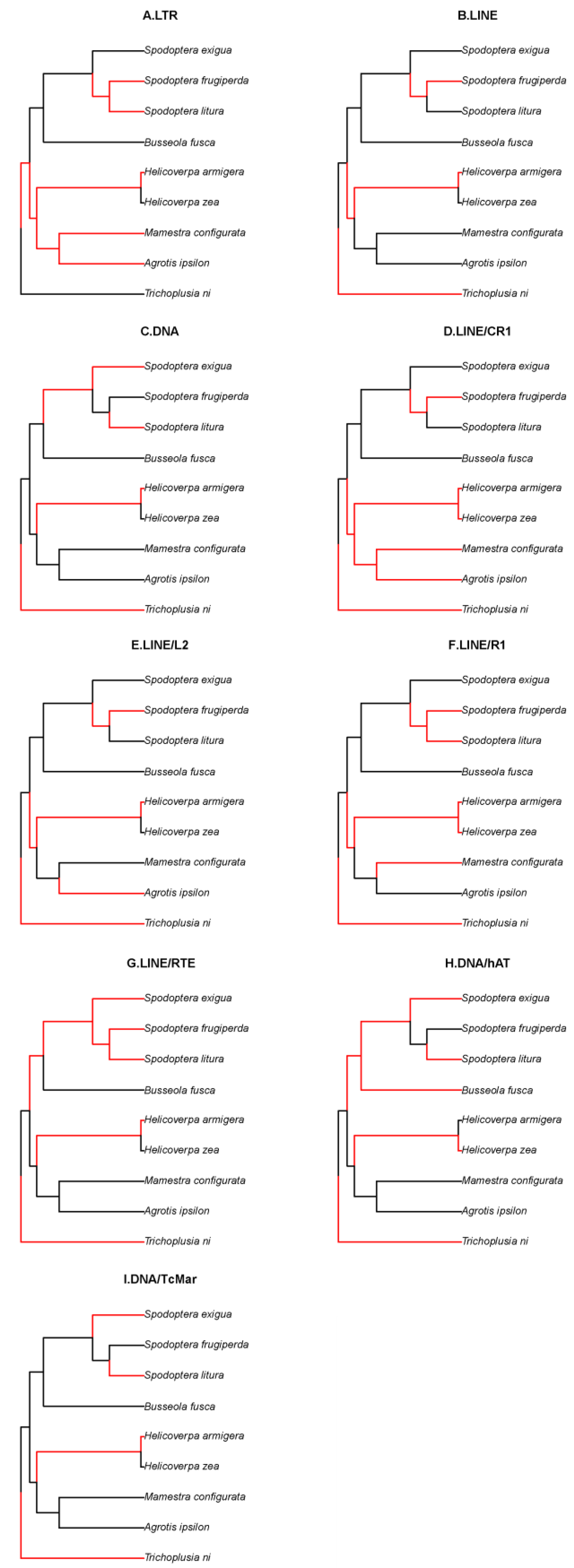
| Level | Species name | Genome size(Mb) | RTE | DTE | UnC | Total |
|---|---|---|---|---|---|---|
| Scaffold | Agrotis ipsilon | 486.92 | 1238 | 1120 | 131 | 2489 |
| Scaffold | Busseola fusca | 490.17 | 1617 | 1067 | 136 | 2820 |
| Scaffold | Helicoverpa armigera | 299.98 | 562 | 607 | 52 | 1221 |
| Scaffold | Helicoverpa zea | 306.41 | 638 | 637 | 67 | 1342 |
| Scaffold | Heliothis virescens | 403.15 | 790 | 855 | 85 | 1730 |
| Scaffold | Mamestra configurata | 559.39 | 1509 | 1186 | 131 | 2826 |
| Chromosome | Spodoptera exigua | 446.8 | 831 | 456 | 43 | 1330 |
| Chromosome | Spodoptera frugiperda | 486.23 | 743 | 644 | 63 | 1450 |
| Chromosome | Spodoptera litura | 428.03 | 1006 | 556 | 70 | 1632 |
| Chromosome | Trichoplusia ni | 367.26 | 578 | 428 | 32 | 1038 |
| Species | DTE (%) | LTR (%) | LINE (%) | SINE (%) | UnC(%) | All (%) | Genome size (Mb) |
|---|---|---|---|---|---|---|---|
| Trichoplusia ni | 4.59 | 2.63 | 4.99 | 2.25 | 1.4 | 15.86 | 367.2 |
| Helicoverpa armigera | 4.01 | 0.74 | 3.46 | 2.06 | 1.06 | 11.33 | 299.98 |
| Helicoverpa zea | 4.82 | 1.07 | 4.05 | 2.63 | 1.19 | 13.76 | 306.41 |
| Spodoptera exigua | 4.74 | 4.22 | 17.64 | 2.56 | 1.48 | 30.64 | 446.80 |
| Spodoptera litura | 5.87 | 2.4 | 14.81 | 3.39 | 2.51 | 28.98 | 428.03 |
| Spodoptera frugiperda | 9.12 | 1.94 | 12.54 | 0.98 | 2.17 | 26.75 | 486.23 |
| Agrotis ipsilon | 11.8 | 2.08 | 13.81 | 3.16 | 3.42 | 34.27 | 486.92 |
| Mamestra configurata | 11.24 | 2.12 | 15.59 | 2.37 | 3.4 | 34.72 | 559.39 |
| Busseola fusca | 12.1 | 3.16 | 20.16 | 6.11 | 3.57 | 45.10 | 490.17 |
| Heliothis virescens | 8.39 | 1.3 | 5.64 | 2.92 | 2.29 | 20.54 | 403.15 |
| Classes of TEs | Subfamily | R value | P value |
|---|---|---|---|
| DNA transposon | 0.81 | 4.21E-03 | |
| DNA/CMC | 0.31 | 0.4 | |
| DNA/TcMar | 0.97 | 2.17E-06 | |
| DNA/Zator | 0.74 | 0.01 | |
| DNA/hAT | 0.46 | 0.18 | |
| DNA/PiggyBac | 0.21 | 0.55 | |
| LINE | 0.83 | 3.2E-03 | |
| LINE/Dong | 0.91 | 2.56E-04 | |
| LINE/L2 | 0.77 | 9.6E-0.3 | |
| LINE/R1 | 0.75 | 0.01 | |
| LINE/RTE | 0.58 | 0.08 | |
| LINE/Jockey | 0.54 | 0.1 | |
| LINE/Proto2 | 0.44 | 0.21 | |
| LINE/CR1 | 0.41 | 0.24 | |
| LINE/CRE | 0.34 | 0.34 | |
| LINE/Rex | 0.25 | 0.49 | |
| LTR | 0.49 | 0.15 | |
| LTR/Copia | 0.4 | 0.26 | |
| SINE | 0.2 | 0.57 | |
| SINE/tRNA | 0.2 | 0.58 |
| Classes of TEs | Subfamily | Lambda value | P value(lambda) | K value | P value(K) |
|---|---|---|---|---|---|
| DNA | 0.93 | 0.23 | 0.75 | 0.16 | |
| DNA/hAT | 1.02 | 7.1E-03 | 1.13 | 0.01 | |
| DNA/Helitron | 0.89 | 0.72 | 0.6 | 0.22 | |
| DNA/MULE | 0.84 | 0.22 | 0.66 | 0.16 | |
| DNA/TcMar | 0.97 | 0.03 | 1.13 | 0.03 | |
| LINE* | 0.99 | 0.01 | 1.33 | 0.02 | |
| LINE/CR1 | 1.01 | 5.7E-03 | 1.62 | 4E-03 | |
| LINE/Dong | 0.99 | 0.07 | 0.99 | 0.02 | |
| LINE/Jockey | 0.73 | 0.32 | 0.52 | 0.24 | |
| LINE/L2 | 0.99 | 0.03 | 1.19 | 0.03 | |
| LINE/R1 | 1.02 | 6.6E-03 | 1.49 | 0.01 | |
| LINE/RTE | 1.01 | 0.03 | 1.18 | 0.03 | |
| LTR | 0.79 | 0.12 | 0.57 | 0.24 | |
| LTR/Pao | 0.70 | 0.17 | 0.39 | 0.33 | |
| SINE | 6.8E-05 | 1 | 0.42 | 0.43 | |
| SINE/tRNA | 1.01 | 0.21 | 0.82 | 0.1 |
| Transposon type | Range of change rates |
|---|---|
| LTR | 3.6 ~ 6.5 |
| LINE | 65 ~ 130 |
| LINE/CR1 | 9.8 ~ 24 |
| LINE/L2 | 2.6 ~ 4.1 |
| LINE/R1 | 5.8 ~ 12 |
| LINE/RTE | 1.4 ~ 5.9 |
| DNA Transposon | 30 ~ 45 |
| DNA/hAT | 0.1 ~ 0.31 |
| DNA/TcMar | 0.16 ~ 0.31 |
| Subfamily | Frequency of HTTs in every thousand TE copies | Number of HTTs | Copy number of TEs |
|---|---|---|---|
| LTR/Gypsy | 0.04 | 1 | 22547 |
| LTR/Copia | 0.25 | 1 | 3978 |
| LINE/RTE-RTE | 0.05 | 10 | 183155 |
| LINE/RTE-BovB | 0.03 | 4 | 130583 |
| LINE/R1 | 6.1E-03 | 3 | 485681 |
| LINE/Proto2 | 0.05 | 1 | 21637 |
| LINE/L2 | 0.03 | 6 | 209382 |
| LINE/Dong-R4 | 0.13 | 3 | 22186 |
| LINE/CR1-Zenon | 2.6E-03 | 1 | 380795 |
| LINE/CR1 | 0.25 | 4 | 15874 |
| DNA/Zator | 0.18 | 1 | 5418 |
| DNA/TcMar-Tc1 | 0.44 | 12 | 27452 |
| DNA/TcMar-Mariner | 0.38 | 5 | 13007 |
| DNA/RC | 1.2E-03 | 1 | 774822 |
| DNA/Maverick | 1.72 | 3 | 1741 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





