Submitted:
03 May 2023
Posted:
04 May 2023
You are already at the latest version
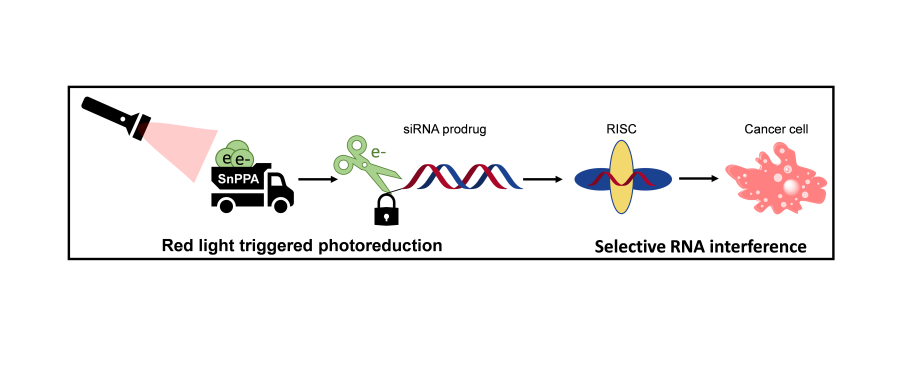
Abstract
Keywords:
1. Introduction

2. Results
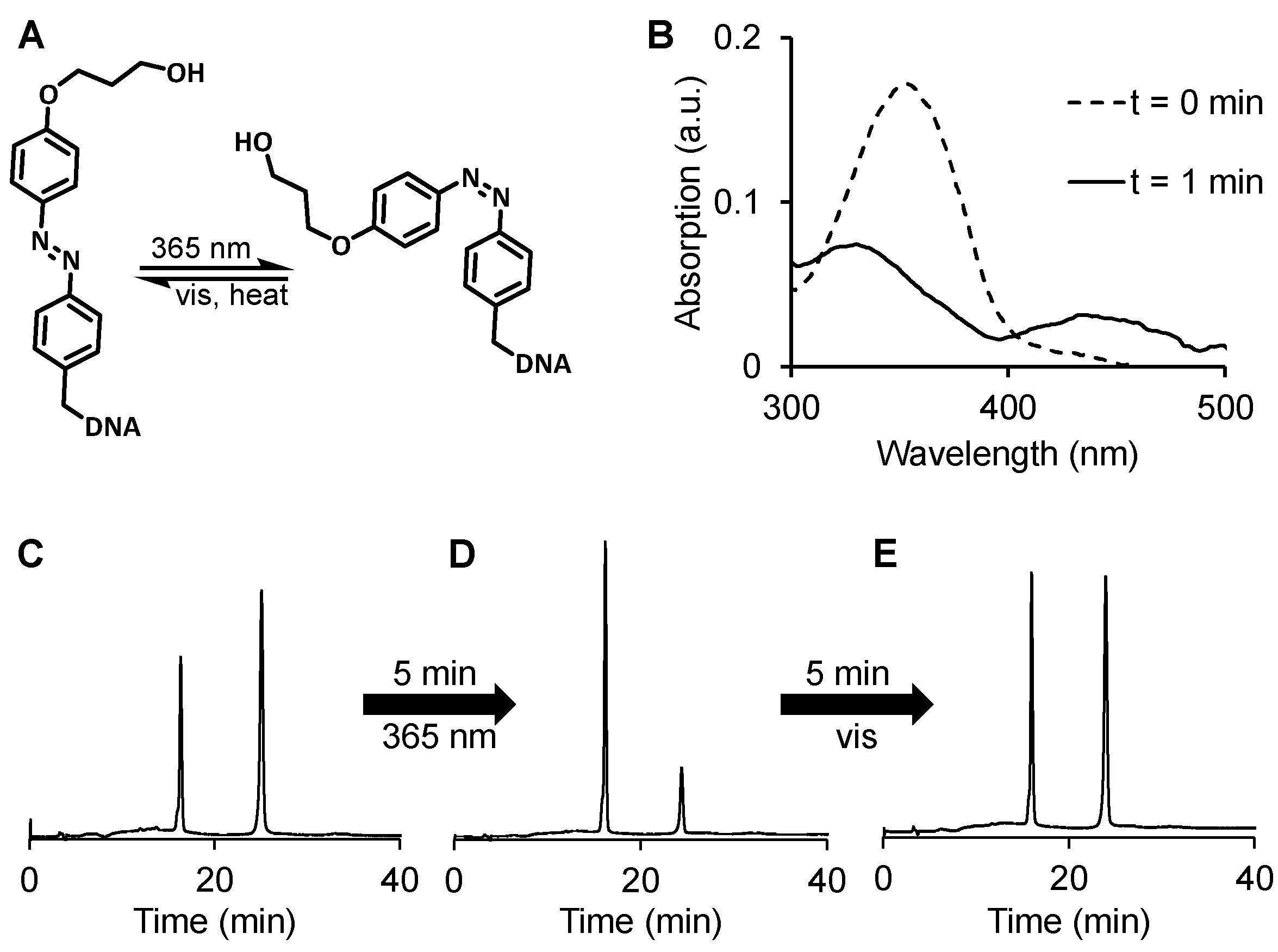
2.1. Design, Synthesis, and Characterization of DNA 1 and RNA 2
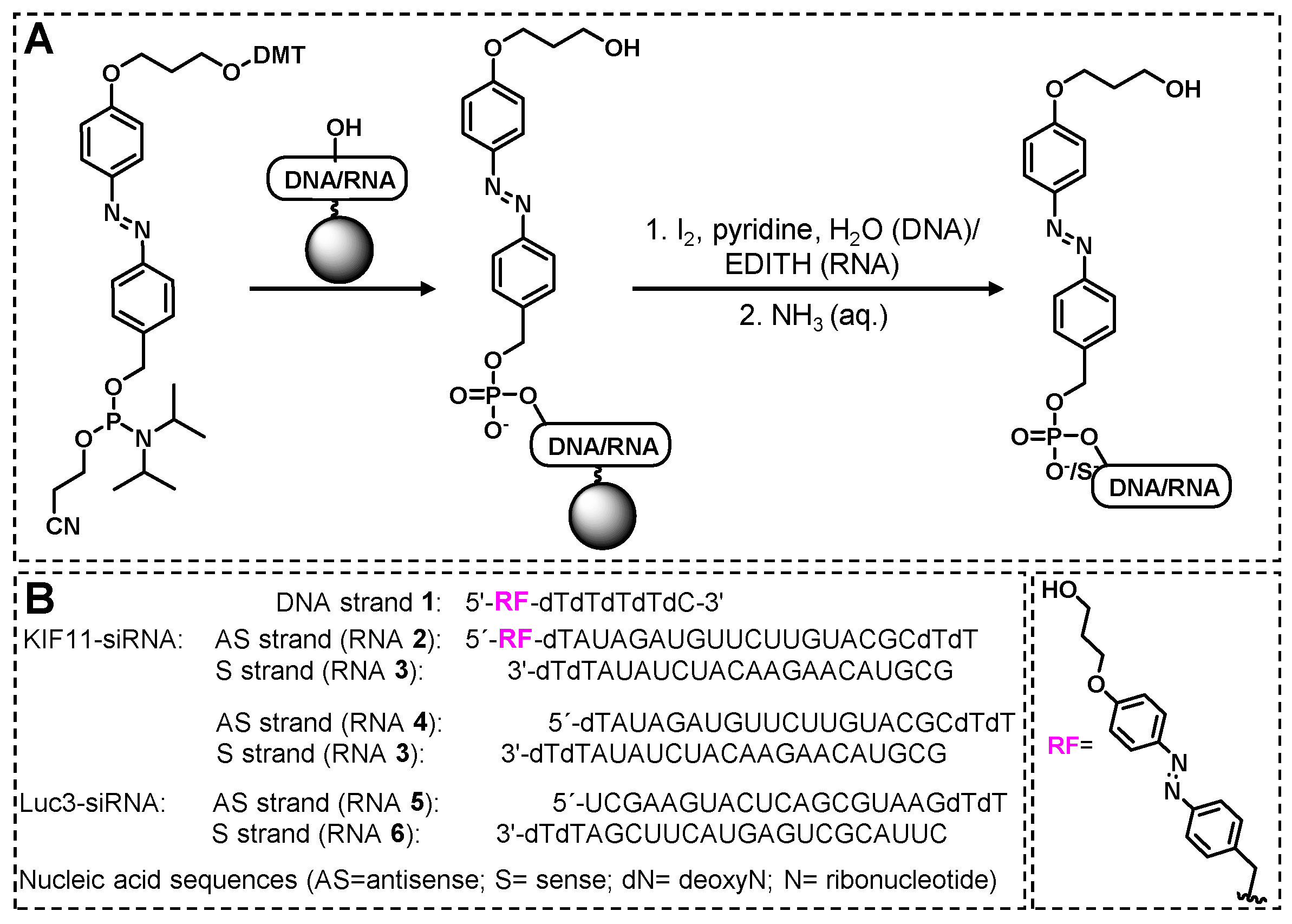
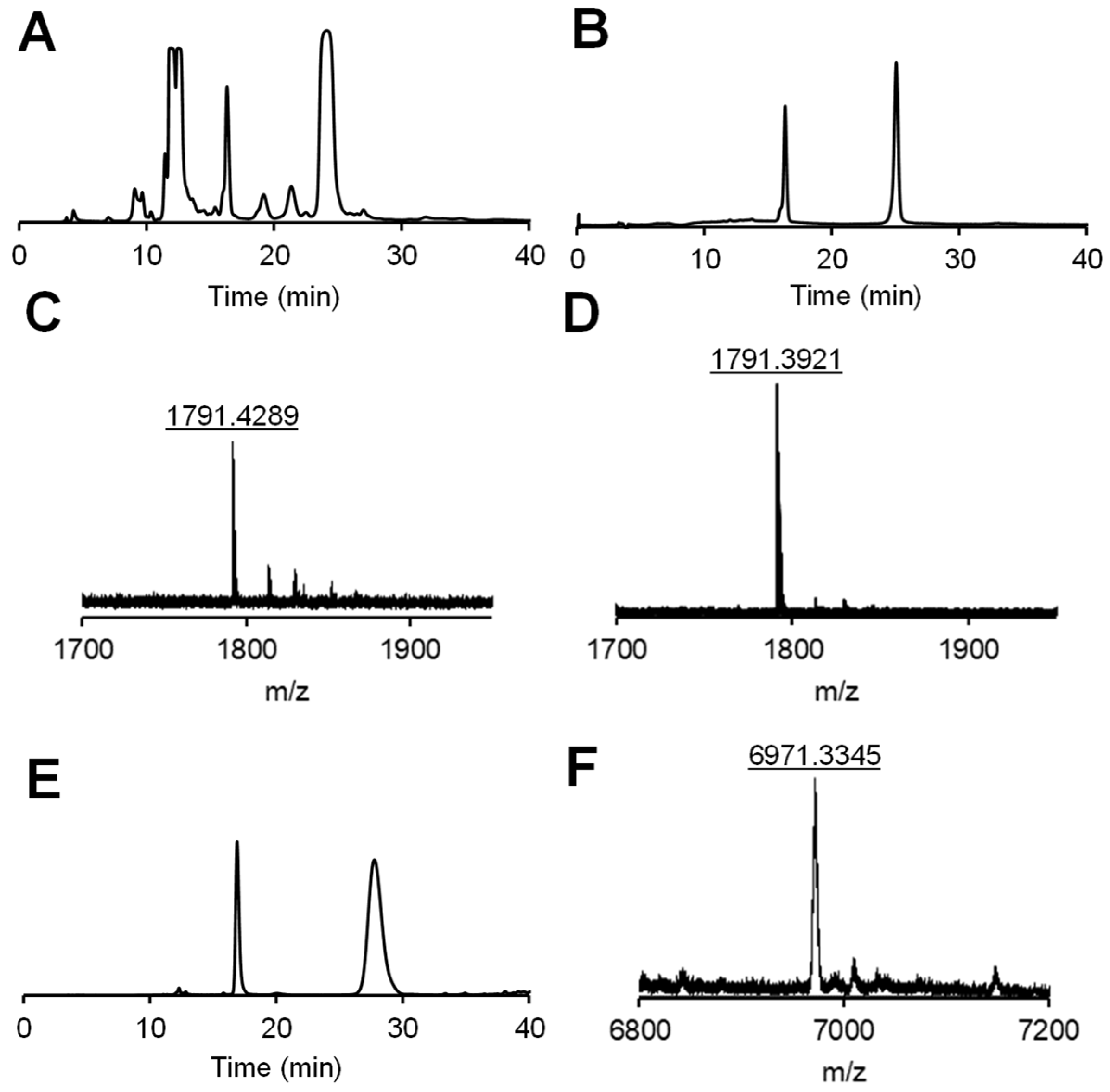
2.2. Cis-trans isomerization of DNA 1 and its photoreduction.
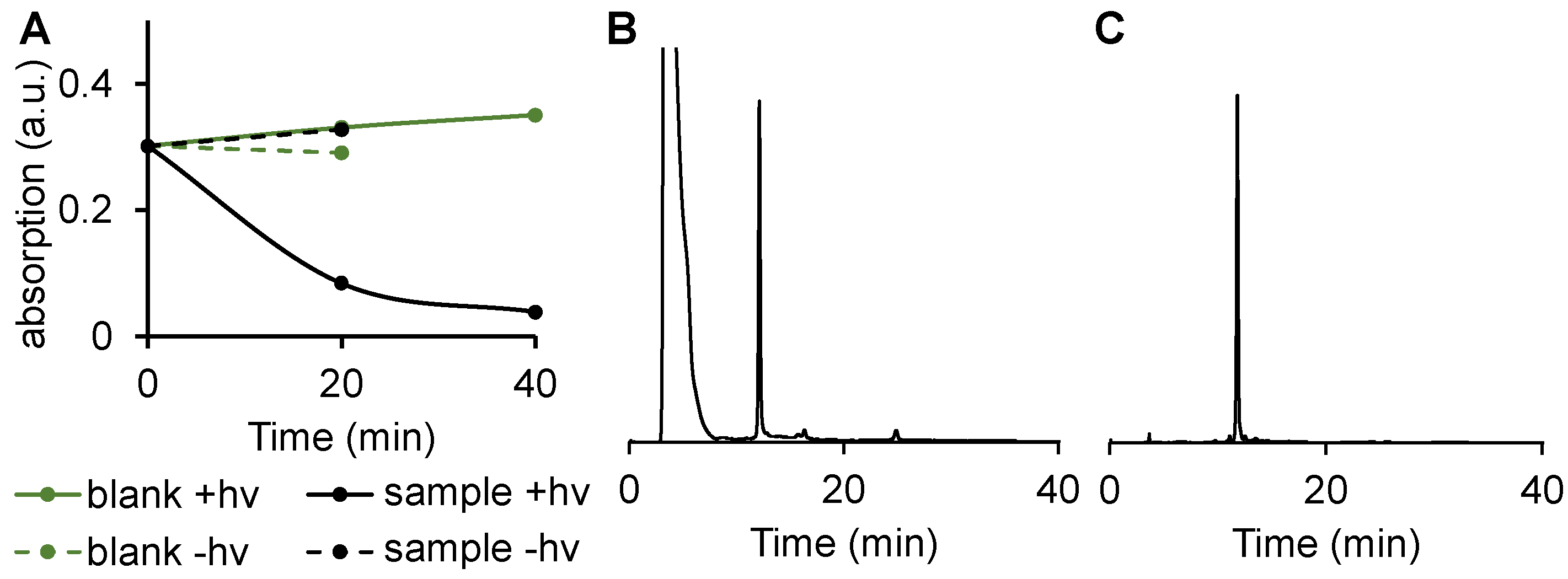
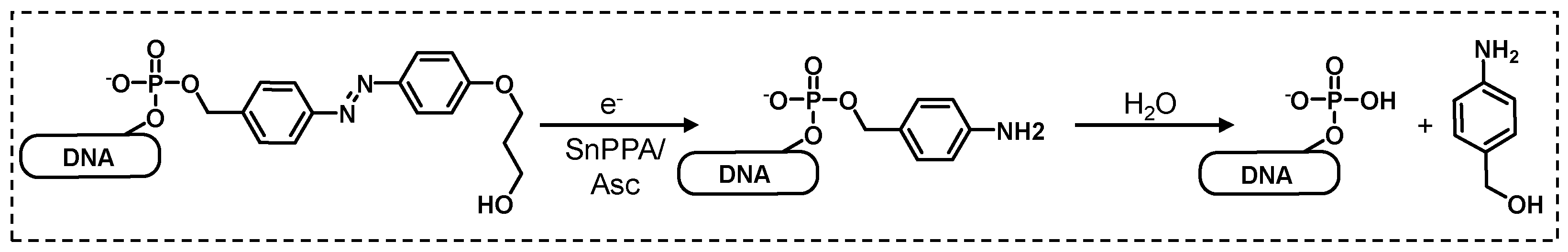
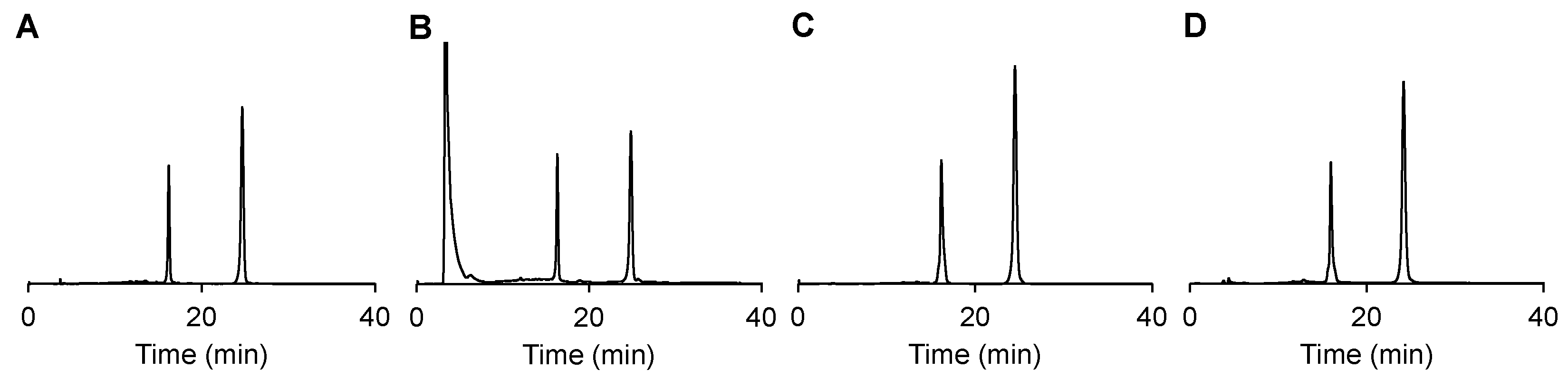
2.3. DNA 1 cleavage selectivity and RNA 2 photoreduction.
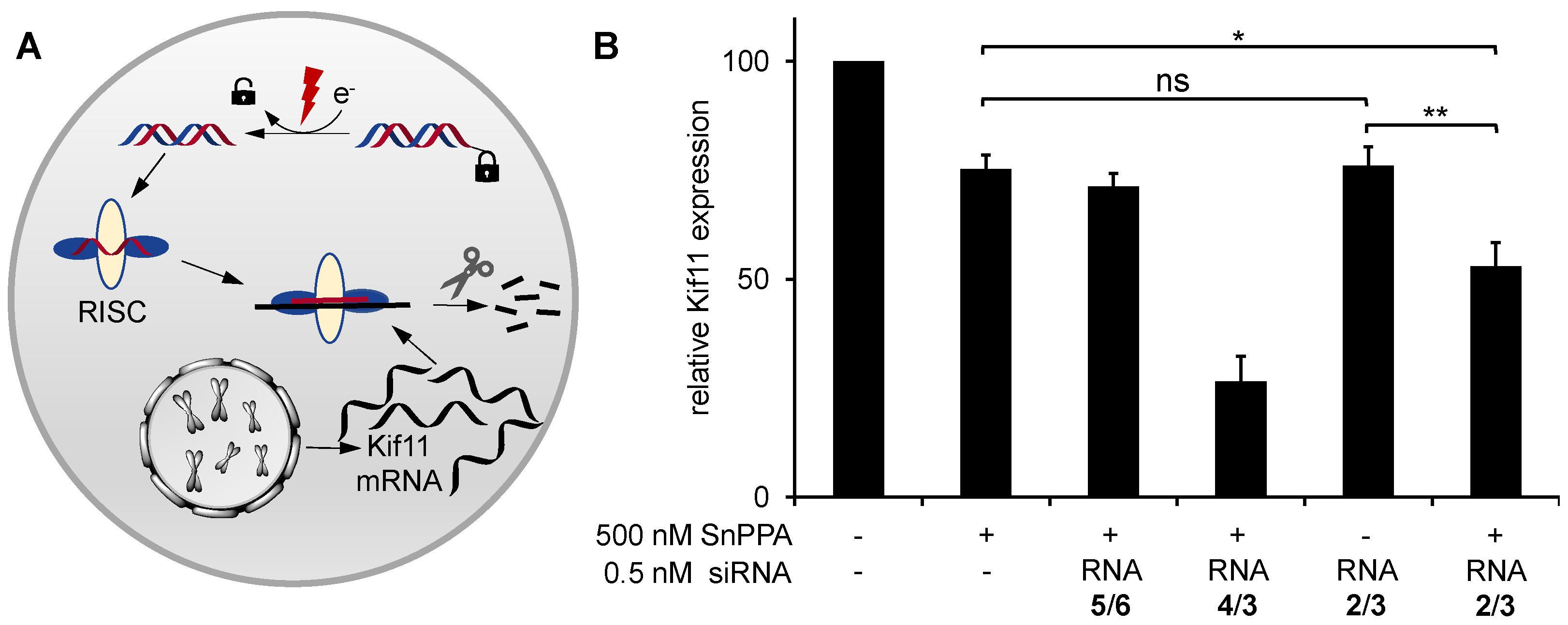
2.4. Knockdown efficacy of RNA 2/3
3. Conclusion
4. Materials and Methods
4.1. Synthesis of modified oligonucleotides
4. 2 Cis-trans isomerization of DNA 1
4.3 DNA 1 cleavage by red-light-induced photoreduction
4.4 Stability in presence of oxidizing/reducing agents
4.5 RNA 2 cleavage by red-light-induced photoreduction
4.6 Transfection
4.7 Relative gene expression quantification by RT-qPCR
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Scott, L.J., Givosiran: First Approval. Drugs, 2020. 80(3): p. 335-339. [CrossRef]
- Scott, L.J. and S.J. Keam, Lumasiran: First Approval. Drugs, 2021. 81(2): p. 277-282. [CrossRef]
- Wood, H., FDA approves patisiran to treat hereditary transthyretin amyloidosis. Nat Rev Neurol, 2018. 14(10): p. 570. [CrossRef]
- Lamb, Y.N., Inclisiran: First Approval. Drugs, 2021. 81(3): p. 389-395.
- Keam, S.J., Vutrisiran: First Approval. Drugs, 2022. 82(13): p. 1419-1425. [CrossRef]
- Han, S.P., et al., Programmable siRNA pro-drugs that activate RNAi activity in response to specific cellular RNA biomarkers. Mol Ther Nucleic Acids, 2022. 27: p. 797-809. [CrossRef]
- Hayashi, J., et al., Effective gene silencing activity of prodrug-type 2'-O-methyldithiomethyl siRNA compared with non-prodrug-type 2'-O-methyl siRNA. Bioorg Med Chem Lett, 2018. 28(12): p. 2171-2174. [CrossRef]
- Mikat, V. and A. Heckel, Light-dependent RNA interference with nucleobase-caged siRNAs. Rna, 2007. 13(12): p. 2341-7. [CrossRef]
- Rühle, J., et al., Reactive oxygen species-responsive RNA interference. Chemical Communications, 2022. 58(27): p. 4388-4391. [CrossRef]
- Antunes, F. and E. Cadenas, Estimation of H2O2 gradients across biomembranes. FEBS Letters, 2000. 475(2): p. 121-126.
- Rybak, C.J., et al., Dinickel-Catalyzed N═N Bond Rotation. Inorg Chem, 2023. 62(15): p. 5886-5891.
- Johnson, T.G., A. Sadeghi-Kelishadi, and M.J. Langton, A Photo-responsive Transmembrane Anion Transporter Relay. J Am Chem Soc, 2022. 144(23): p. 10455-10461. [CrossRef]
- Samanta, S., et al., Photoswitching azo compounds in vivo with red light. J Am Chem Soc, 2013. 135(26): p. 9777-84. [CrossRef]
- Tian, X., et al., Near-Infrared Fluorescent Probes for Hypoxia Detection via Joint Regulated Enzymes: Design, Synthesis, and Application in Living Cells and Mice. Anal Chem, 2018. 90(22): p. 13759-13766. [CrossRef]
- Guisán-Ceinos, S., et al., Turn-on Fluorescent Biosensors for Imaging Hypoxia-like Conditions in Living Cells. J Am Chem Soc, 2022. 144(18): p. 8185-8193. [CrossRef]
- Dutta, S., et al., Red light-triggered photoreduction on a nucleic acid template. Chem Commun (Camb), 2020. 56(69): p. 10026-10029. [CrossRef]
- Jiang, M., et al., KIF11 is required for proliferation and self-renewal of docetaxel resistant triple negative breast cancer cells. Oncotarget, 2017. 8(54): p. 92106-92118. [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




