Submitted:
04 May 2023
Posted:
05 May 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
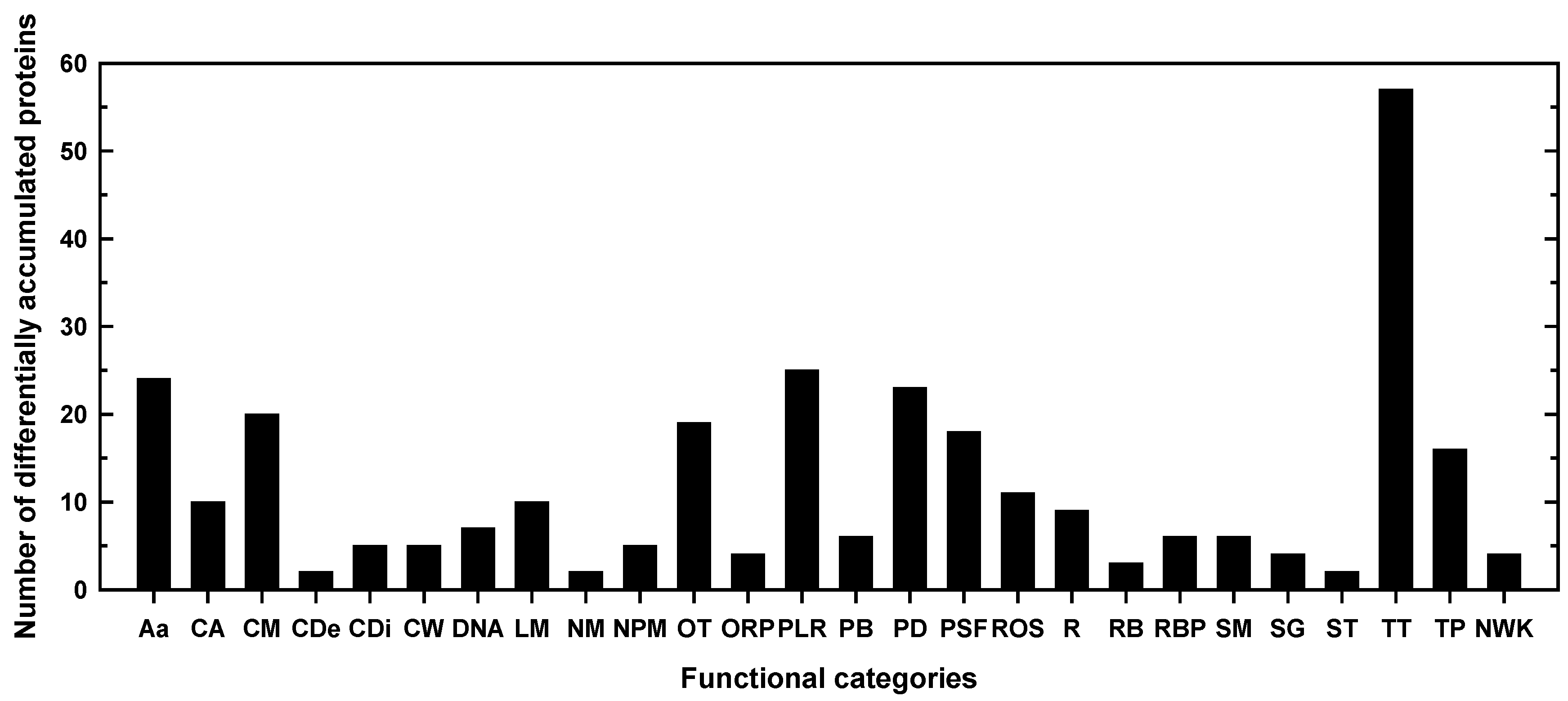
2.1. Classification into several functional categories of differentially accumulated proteins in both maize landraces and B treatments
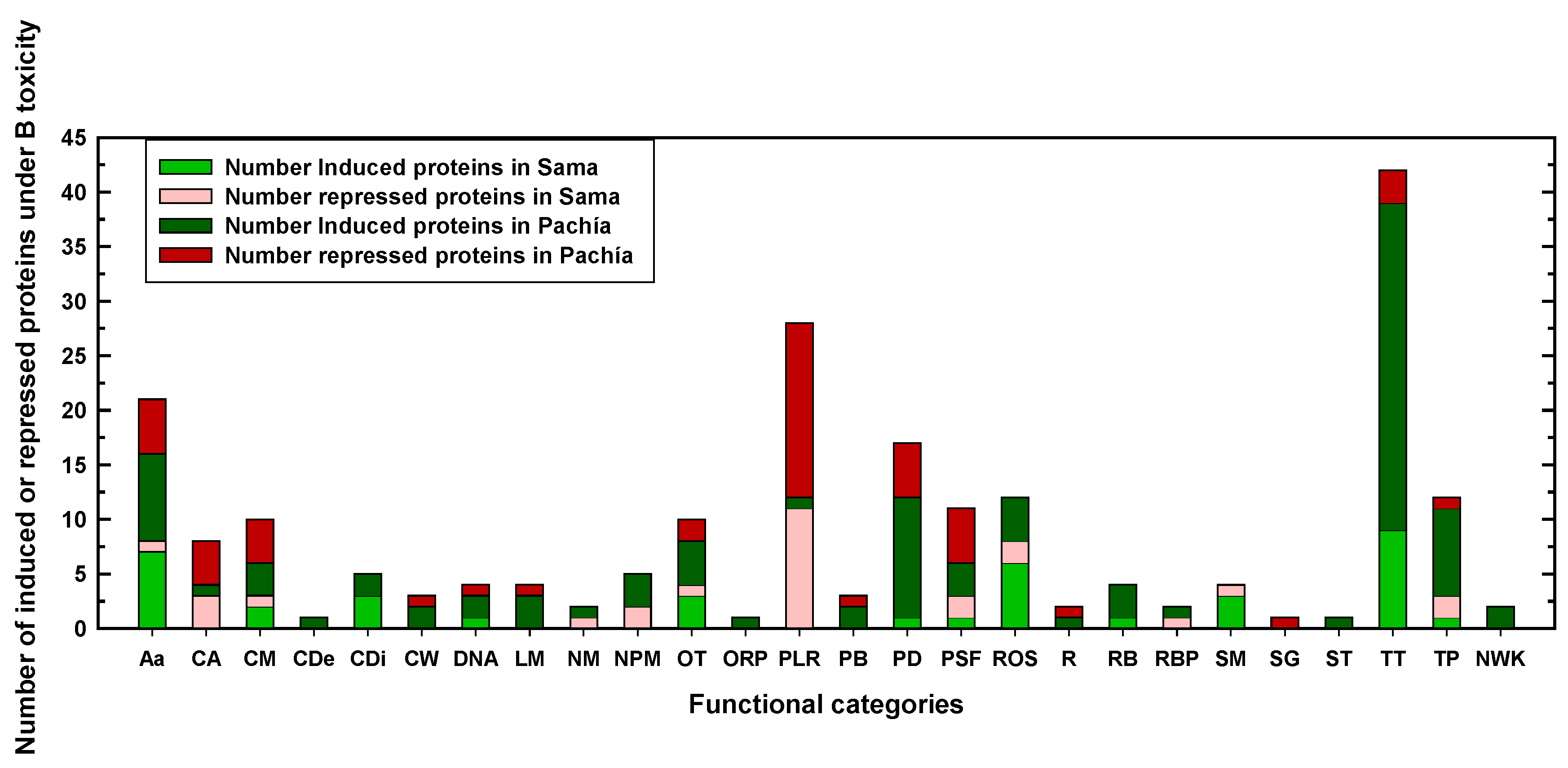
2.2. Differentially expressed proteins in Sama and Pachía in response to B toxicity
3. Discussion
3.1. Several proteases and translation-related proteins would allow Pachía to survive in media with B excess
3.2. Proteins that would confer Sama more B toxicity tolerance
3.2.1. Lower repression of photosynthesis-related proteins would enhance the B-toxicity tolerance of Sama
4. Materials and Methods
4.1. Plant materials and growth conditions
4.2. Protein extraction and digestion
4.3. Shotgun-DDA-LC-MS/MS analysis
4.4. Protein quantification
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Warington, K. The effect of boric acid and borax on the broad bean and certain other plants. Ann. Bot. 1923, 37, 629–672. [Google Scholar] [CrossRef]
- Princi, M.P.; Lupini, A.; Araniti, F.; Longo, C.; Mauceri, A.; Sunseri, F.; Abenavoli, M.R. Boron toxicity and tolerance in plants: recent advances and future perspectives. In Plant Metal Interaction: Emerging Remediation Techniques; Ahmad, P., Ed.; Elsevier Inc: Amsterdam, Netherlands, 2016; pp. 115–147. [Google Scholar]
- González-Fontes, A.; Fujiwara, T. Advances in plant boron. Int. J. Mol. Sci. 2020, 21, 4107. [Google Scholar] [CrossRef]
- González-Fontes, A. Why boron is an essential element for vascular plants. New Phytol. 2020, 226, 1228–1230. [Google Scholar] [CrossRef]
- Wimmer, M.A.; Abreu, I.; Bell, R.W.; Bienert, M.D.; Brown, P.H.; Dell, B.; Fujiwara, T.; Goldbach, H.E.; Lehto, T.; Mock, H.-P.; et al. Boron: an essential element for vascular plants. New Phytol. 2020, 226, 1232–1237. [Google Scholar] [CrossRef]
- Ishii, T.; Matsunaga, T. Isolation and characterization of a boron-rhamnogalacturonan-II complex from cell walls of sugar beet pulp. Carbohydr. Res. 1996, 284, 1–9. [Google Scholar] [CrossRef]
- Kobayashi, M.; Matoh, T.; Azuma, J. Two chains of rhamnogalacturonan II are cross-linked by borate-diol ester bonds in higher plant cell walls. Plant Physiol. 1996, 110, 1017–1020. [Google Scholar] [CrossRef]
- O’Neill, M.A.; Warrenfeltz, D.; Kates, K.; Pellerin, P.; Doco, T.; Darvill, A.G.; Albersheim, P. Rhamnogalacturonan-II, a pectic polysaccharide in the walls of growing plant cell, forms a dimer that is covalently cross-linked by a borate ester. J. Biol. Chem. 1996, 271, 22923–22930. [Google Scholar] [CrossRef]
- Cakmak, I.; Römheld, V. Boron deficiency-induced impairments of cellular functions in plants. Plant Soil 1997, 193, 71–83. [Google Scholar] [CrossRef]
- Goldbach, H.E.; Yu, Q.; Wingender, R.; Schulz, M.; Wimmer, M.; Findeklee, P.; Baluška, F. Rapid response reactions of roots to boron deprivation. J. Plant Nutr. Soil Sci. 2001, 164, 173–181. [Google Scholar] [CrossRef]
- Brown, P.H.; Bellaloui, N.; Wimmer, M.A.; Bassil, E.S.; Ruiz, J.; Hu, H.; Pfeffer, H.; Dannel, F.; Römheld, V. Boron in plant biology. Plant Biol. 2002, 4, 205–223. [Google Scholar] [CrossRef]
- Voxeur, A.; Fry, S.C. Glycosylinositol phosphorylceramides from Rosa cell cultures are boron-bridged in the plasma membrane and form complexes with rhamnogalacturonan II. Plant J. 2014, 79, 139–149. [Google Scholar] [CrossRef]
- Wang, N.; Yang, C.; Pan, Z.; Liu, Y.; Peng, S. Boron deficiency in woody plants: various responses and tolerance mechanisms. Front. Plant Sci. 2015, 6, 916. [Google Scholar] [CrossRef] [PubMed]
- Blevins, D.G.; Lukaszewski, K.M. Boron in plant structure and function. Annu. Rev. Plant Physiol. Plant Molec. Biol. 1998, 49, 481–500. [Google Scholar] [CrossRef] [PubMed]
- Camacho-Cristóbal, J.J.; Navarro-Gochicoa, M.T.; Rexach, J.; González-Fontes, A.; Herrera-Rodríguez, M.B. Plant response to boron deficiency and boron use efficiency in crop plants. In Plant Micronutrient Use Efficiency. Molecular and Genomic Perspectives in Crop Plants; Hossain, M.A., Kamiya, T., Burritt, D.J., Phan Tran, L.-S., Fujiwara., T., Eds.; Elsevier Inc., 2018; pp. 109–121. [Google Scholar]
- Shireen, F.; Nawaz, M.A.; Chen, C.; Zhang, Q.; Zheng, Z.; Sohail, H.; Sun, J.; Cao, H.; Huang, Y.; Bie, Z. Boron: functions and approaches to enhance its availability in plants for sustainable agriculture. Int. J. Mol. Sci. 2018, 19, 1856. [Google Scholar] [CrossRef] [PubMed]
- Reid, R.J.; Hayes, J.E.; Post, A.; Stangoulis, J.C.R.; Graham, R.D. A critical analysis of the causes of boron toxicity in plants. Plant Cell Environ. 2004, 27, 1405–1414. [Google Scholar] [CrossRef]
- Camacho-Cristóbal, J.J.; Rexach, J.; González-Fontes, A. Boron in plants: deficiency and toxicity. J. Integr. Plant Biol. 2008, 50, 1247–1255. [Google Scholar] [CrossRef]
- Beato, V.M.; Rexach, J.; Navarro-Gochicoa, M.T.; Camacho-Cristóbal, J.J.; Herrera-Rodríguez, M.B.; Maldonado, J.M.; González-Fontes, A. A tobacco asparagine synthetase gene responds to carbon and nitrogen status and its root expression is affected under boron stress. Plant Sci. 2010, 178, 289–298. [Google Scholar] [CrossRef]
- Beato, V.M.; Navarro-Gochicoa, M.T.; Rexach, J.; Herrera-Rodríguez, M.B.; Camacho-Cristóbal, J.J.; Kempa, S.; Weckwerth, W.; González-Fontes, A. Expression of root glutamate dehydrogenase genes in tobacco plants subjected to boron deprivation. Plant Physiol. Biochem. 2011, 49, 1350–1354. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, J.-W.; Guo, L.-X.; Liu, Y.-Z.; Jin, L.-F.; Hussain, S.B.; Du, W.; Deng, Z.; Peng, S.-A. Transcriptome changes associated with boron deficiency in leaves of two citrus scion-rootstock combinations. Front. Plant Sci. 2017, 8, 317. [Google Scholar] [CrossRef]
- Landi, M.; Margaritopoulou, T.; Papadakis, I.E.; Araniti, F. Boron toxicity in higher plants: an update. Planta 2019, 250, 1011–1032. [Google Scholar] [CrossRef]
- Brdar-Jokanovi´c, M. Boron toxicity and deficiency in agricultural plants. Int. J. Mol. Sci. 2020, 21, 1424. [Google Scholar] [CrossRef]
- Kabay, N.; Güler, E.; Bryjak, M. Boron in seawater and methods for its separation—a review. Desalination 2010, 261, 212–217. [Google Scholar] [CrossRef]
- Chatzistathis, T.; Fanourakis, D.; Aliniaeifard, S.; Kotsiras, A.; Delis, C.; Tsaniklidis, G. Leaf age-dependent effects of boron toxicity in two Cucumis melo varieties. Agronomy. 2021, 11, 759. [Google Scholar] [CrossRef]
- Uluisik, I.; Karakaya, H.C.; Koc, A. The importance of boron in biological systems. J. Trace Elem. Med. Biol. 2018, 45, 156–162. [Google Scholar] [CrossRef]
- Mamani-Huarcaya, B.M.; González-Fontes, A.; Navarro-Gochicoa, M.T.; Camacho-Cristóbal, J.J.; Ceacero, C.J.; Herrera-Rodríguez, M.B.; Fernández Cutire, Ó.; Rexach, J. Characterization of two Peruvian maize landraces differing in boron toxicity tolerance. Plant Physiol. Biochem. 2022, 185, 167–177. [Google Scholar] [CrossRef] [PubMed]
- Behera, B.; Kancheti, M.; Raza, M.B.; Shiv, A.; Mangal, V.; Rathod, G.; Altaf, M.A.; Kumar, A.; Aftab, T.; Kumar, R.; et al. Mechanistic insight on boron-mediated toxicity in plant vis-a-vis its mitigation strategies: a review. Int. J. Phytoremediat. 2023, 25, 9–26. [Google Scholar] [CrossRef]
- Ralston, N.V.C.; Hunt, C.D. Diadenosine phosphates and S-adenosylmethionine: novel boron binding biomolecules detected by capillary electrophoresis. Biochim. Biophys. Acta 2001, 1527, 20–30. [Google Scholar] [CrossRef] [PubMed]
- Liang, Z.; Pandey, P.; Stoerger, V.; Xu, Y.; Qiu, Y.; Ge, Y.; Schnable, J.C. Conventional and hyperspectral timeseries imaging of maize lines widely used in field trials. GigaScience 2018, 72, 1–11. [Google Scholar]
- Andorf, C.; Beavis, W.D.; Hufford, M.; Smith, S.; Suza, W.P.; Wang, K.; Woodhouse, M.; Yu, J.; Lübberstedt, T. Technological advances in maize breeding: past, present, and future. Theor. Appl. Genet. 2019, 132, 817–849. [Google Scholar] [CrossRef]
- Kausch, A.P.; Wang, K.; Kaeppler, H.F.; Gordon-Kamm, W. Maize transformation: history, progress, and perspectives. Mol. Breed. 2021, 41, 38. [Google Scholar] [CrossRef]
- Xi, Y.; Hu, W.; Zhou, Y.; Liu, X.; Qian, Y. Genome-wide identification and functional analysis of polyamine oxidase genes in maize reveal essential roles in abiotic stress tolerance. Front. Plant Sci. 2022, 13, 950064. [Google Scholar] [CrossRef]
- Ogunwole, A.A.; Otusanya, O.O.; Oloyede, F.A.; Olabamiji, T.M. Comparative effects of boron toxicity and deficiency on the growth, chlorophyll, protein and some cations accumulation in Zea mays seedlings. Int. J. Sci. Res. Innov. 2015, 17, 316–335. [Google Scholar]
- Gotz, L.F.; Silvestrin, F.; Motta, A.C.V.; Pauletti, V. Response to application and tissue diagnosis of boron deficiency and toxicity in maize. Commun. Soil Sci. Plant Anal. 2021, 52, 2898–2911. [Google Scholar] [CrossRef]
- Sang, W.; Huang, Z.-R.; Yang, L.-T.; Guo, P.; Ye, X.; Chen, L.-S. Effects of high toxic boron concentration on protein profiles in roots of two citrus species differing in boron-tolerance revealed by a 2-DE based MS approach. Front. Plant Sci. 2017, 8, 180. [Google Scholar] [CrossRef]
- Fan, T.; Bykova, N.V.; Rampitsch, C.; Xing, T. Identification and characterization of a serine protease from wheat leaves. Eur. J. Plant Pathol. 2016, 146, 293–304. [Google Scholar] [CrossRef]
- Malefo, M.B.; Mathibela, E.O.; Crampton, B.G.; Makgopa, M.E. Investigating the role of Bowman-Birk serine protease inhibitor in Arabidopsis plants under drought stress. Plant Physiol. Biochem. 2020, 149, 286–293. [Google Scholar] [CrossRef]
- D’Ippolito, S.; Rey-Burusco, M.F.; Feingold, S.E.; Guevara, M.G. Role of proteases in the response of plants to drought. Plant Physiol. Biochem. 2021, 168, 1–9. [Google Scholar] [CrossRef]
- Sharma, P.; Gayen, D. Plant protease as regulator and signaling molecule for enhancing environmental stress-tolerance. Plant Cell Reports. 2021, 40, 2081–2095. [Google Scholar] [CrossRef]
- Moin, M.; Bakshi, A.; Saha, A.; Dutta, M.; Madhav, S.M.; Kirti, P.B. Rice ribosomal protein large subunit genes and their spatio-temporal and stress regulation. Front. Plant Sci. 2016, 7, 1284. [Google Scholar] [CrossRef]
- Nozawa, A.; Miwa, K.; Kobayashi, M.; Fujiwara, T. Isolation of Arabidopsis thaliana cDNAs that confer yeast boric acid tolerance. Biosci. Biotechnol. Biochem. 2006, 70, 1724–1730. [Google Scholar] [CrossRef]
- Tanaka, M.; Sotta, N.; Yamazumi, Y.; Yamashita, Y.; Miwa, K.; Murota, K.; Chiba, Y.; Hirai, M.Y.; Akiyama, T.; Onouchi, H.; et al. The minimum open reading frame, AUG-stop, induces boron-dependent ribosome stalling and mRNA degradation. Plant Cell 2016, 28, 2830–2849. [Google Scholar] [CrossRef] [PubMed]
- Sotta, N.; Chiba, Y.; Miwa, K.; Takamatsu, S.; Tanaka, M.; Yamashita, Y.; Naito, S.; Fujiwara, T. Global analysis of boron-induced ribosome stalling reveals its effects on translation termination and unique regulation by AUG-stops in Arabidopsis shoots. Plant J. 2021, 106, 1455–1467. [Google Scholar] [CrossRef]
- Saidi, A.; Hajibarat, Z. In-silico analysis of eukaryotic translation initiation factors (eIFs) in response to environmental stresses in rice (Oryza sativa). Biologia 2020, 75, 1731–1738. [Google Scholar] [CrossRef]
- Pakdel, H.; Hassani, S.B.; Ghotbi-Ravandi, A.A.; Bernard, F. Contrasting the expression pattern change of polyamine oxidase genes and photosynthetic efficiency of maize (Zea mays L.) genotypes under drought stress. J. Biosci. 2020, 45, 73. [Google Scholar] [CrossRef]
- Shu, S.; Guo, S.-R.; Sun, J.; Yuan, L.-Y. Effects of salt stress on the structure and function of the photosynthetic apparatus in Cucumis sativus and its protection by exogenous putrescine. Physiol. Plant. 2012, 146, 285–296. [Google Scholar] [CrossRef]
- Hamdani, S.; Yaakoubi, H.; Carpentier, R. Polyamines interaction with thylakoid proteins during stress. J. Photochem. Photobiol. B-Biol. 2011, 104, 314–331. [Google Scholar] [CrossRef]
- Chen, M.; Blankenship, R.E. Expanding the solar spectrum used by photosynthesis. Trends Plant Sci. 2011, 16, 427–431. [Google Scholar] [CrossRef]
- Zhang, D.; Chang, E.; Yu, X.; Chen, Y.; Yang, Q.; Cao, Y.; Li, X.; Wang, Y.; Fu, A.; Xu, M. Molecular characterization of magnesium chelatase in soybean [Glycine max (L.) Merr.]. Front. Plant Sci. 2018, 9, 720. [Google Scholar] [CrossRef]
- Sasi, S.; Venkatesh, J.; Daneshi, R.F.; Gururani, M.A. Photosystem II extrinsic proteins and their putative role in abiotic stress tolerance in higher plants. Plants, 2018, 7, 100. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Portis, A.R. A novel nucleus-encoded chloroplast protein, PIFI, is involved in NAD(P)H dehydrogenase complex-mediated chlororespiratory electron transport in Arabidopsis. Plant Physiol. 2007, 144, 1742–1752. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, N.; Takabayashi, A.; Noguchi, K.; Tazoe, Y.; Yamamoto, H.; von Caemmerer, S.; Sato, F.; Endo, T. NDH-mediated cyclic electron flow around photosystem I is crucial for C4 photosynthesis. Plant Cell Physiol. 2016, 57, 2020–2028. [Google Scholar] [CrossRef] [PubMed]
- Ma, M.; Liu, Y.; Bai, C.; Yong, J.W.H. The significance of chloroplast NAD(P)H dehydrogenase complex and its dependent cyclic electron transport in photosynthesis. Front. Plant Sci. 2021, 12, 661863. [Google Scholar] [CrossRef]
- Yamori, W.; Shikanai, T. Physiological functions of cyclic electron transport around photosystem I in sustaining photosynthesis and plant growth. Annu. Rev. Plant Biol. 2016, 67, 81–106. [Google Scholar] [CrossRef]
- Zhu, D.; Luo, F.; Zou, R.; Liu, J.; Yan, Y. Integrated physiological and chloroplast proteome analysis of wheat seedling leaves under salt and osmotic stresses. J. Proteomics 2021, 234, 104097. [Google Scholar] [CrossRef] [PubMed]
- Murata, N.; Nishiyama, Y. ATP is a driving force in the repair of photosystem II during photoinhibition. Plant Cell Environ. 2018, 41, 285–299. [Google Scholar] [CrossRef]
- Araniti, F.; Miras-Moreno, B.; Lucini, L.; Landi, M.; Abenavoli, M.R. Metabolomic, proteomic and physiological insights into the potential mode of action of thymol, a phytotoxic natural monoterpenoid phenol. Plant Physiol. Biochem. 2020, 153, 141–153. [Google Scholar] [CrossRef]
- Wang, W.; Vignani, R.; Scali, M.; Cresti, M. A universal and rapid protocol for protein extraction from recalcitrant plant tissues for proteomic analysis. Electrophoresis 2006, 27, 2782–2786. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of proteins utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef]
- Cox, J.; Hein, M.Y.; Luber, C.A.; Paron, I.; Nagaraj, N.; Mann, M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics 2014, 13, 2513–2526. [Google Scholar] [CrossRef] [PubMed]
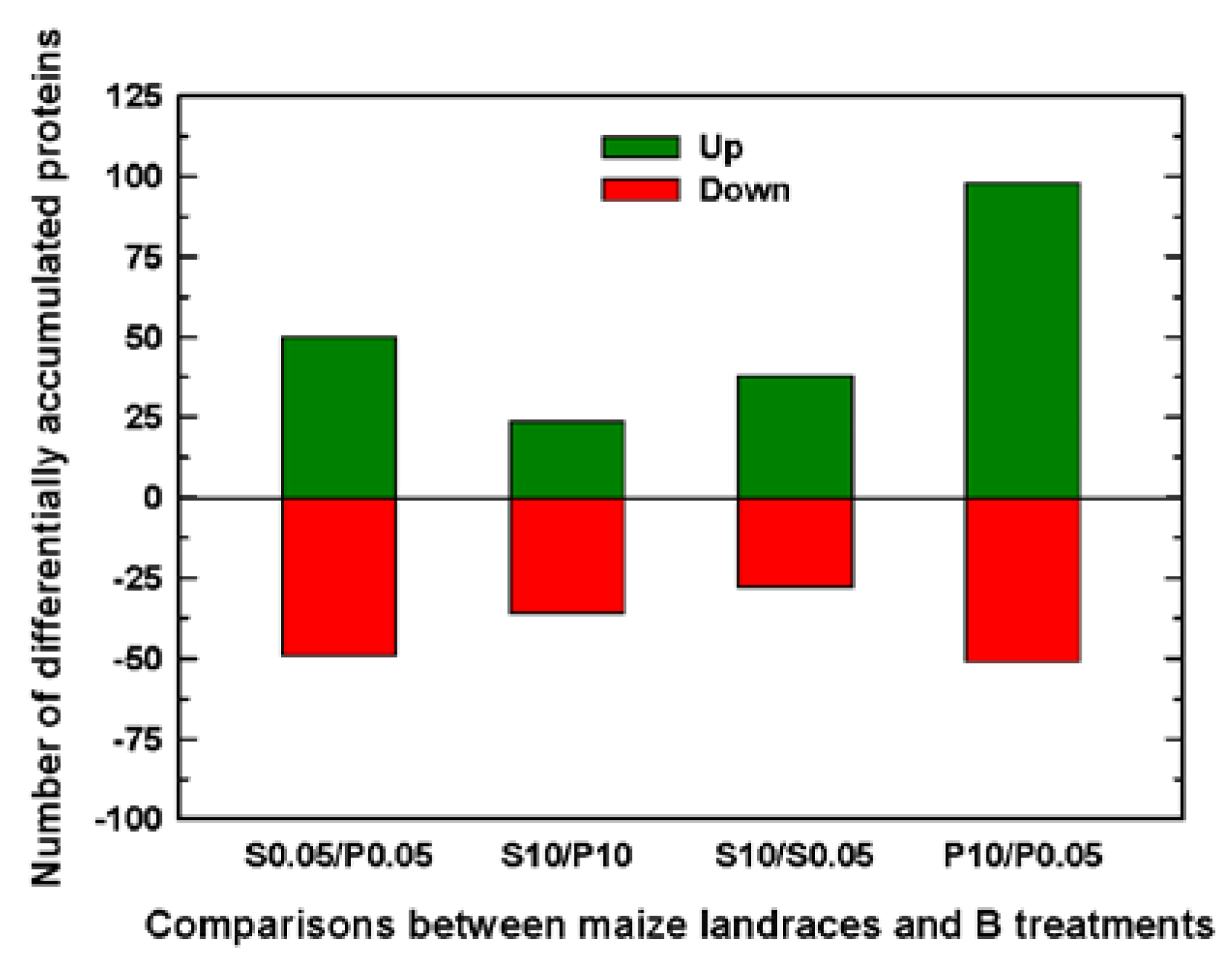
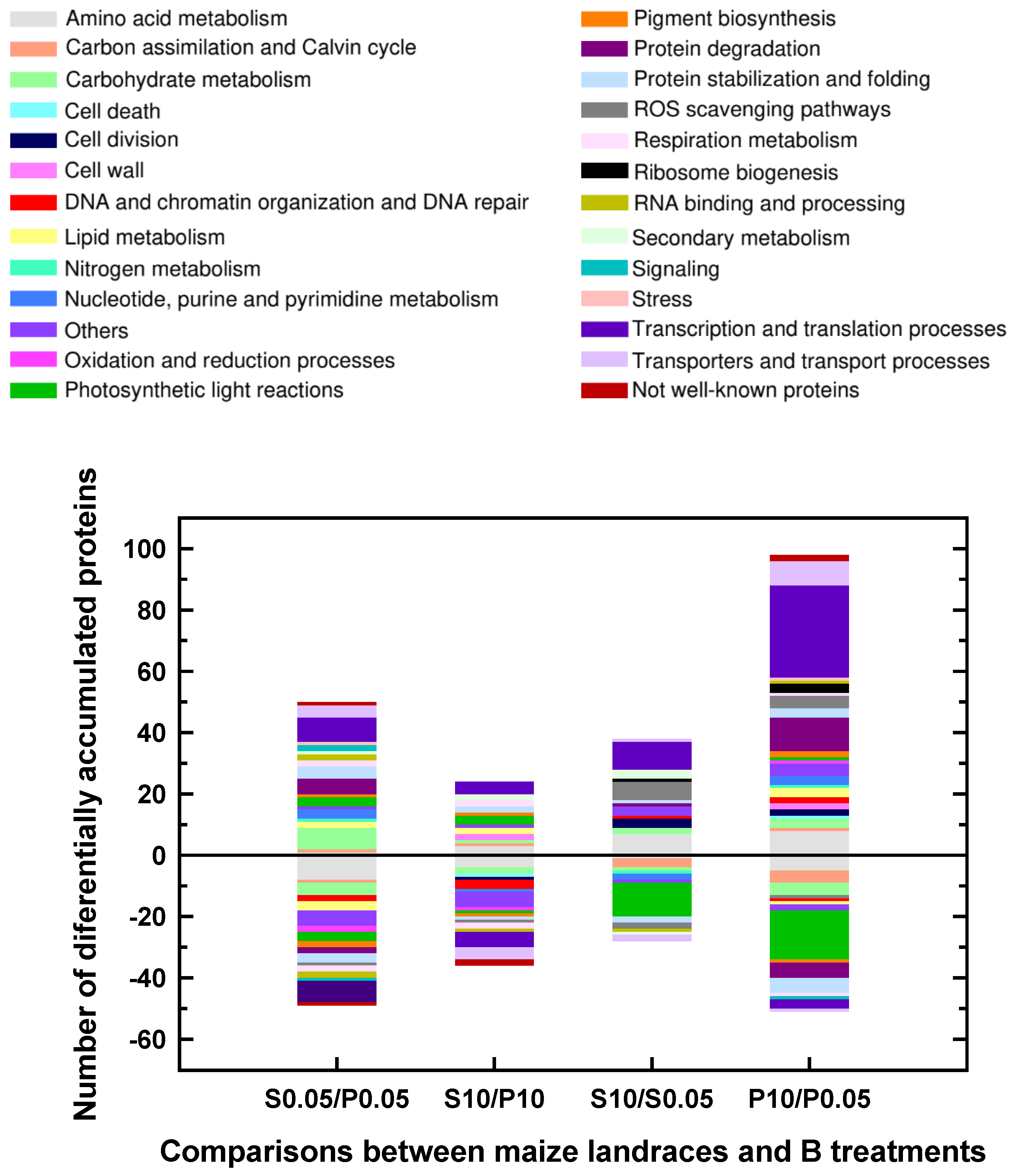
| P0.05 mM (Control) | P10 mM B (B toxicity) | S0.05 mM (control) | S10 mM (B toxicity) | |||
|---|---|---|---|---|---|---|
| Number of detected proteins1 | 1100 | 1040 | 1111 | 1145 | ||
|
S0.05 versus P0.05 (Control conditions) |
S10 versus P10 (B toxicity conditions) |
|||||
| Number of significant DAPs between Sama and Pachía | 99 | 60 | ||||
|
Sama S10 versus S0.05 |
Pachía P10 versus P0.05 |
|||||
| Number of significant DAPs by B toxicity | 66 | 149 | ||||
| Pachía | Sama | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Protein ID1 | Gene Name/ID2 | Protein name / Annotation |
FC3 | P-value4 | FC3 |
P- value4 |
FCSA/ FCPA5 |
Function/Biological process6 | |||||||
| Amino acid metabolism | |||||||||||||||
| B6SKB7 | Zm00001d031013 | Methylcrotonoyl-CoA carboxylase subunit α | 4.44 | 0.0022 | 3.56 | 0.0049 | 0.80 | Leucine degradation | |||||||
| A0A1D6K836 | Zm00001d029848 | Branched-chain-amino-acid aminotransferase | 2.35 | 0.0272 | 1.65 | 0.0241 | 0.70 | Branched-chain amino acid biosynthesis | |||||||
| B4G011 | Zm00001d046923 | D-3-phosphoglycerate dehydrogenase chloroplastic | 2.31 | 0.0154 | 1.52 | 0.0202 | 0.66 | Serine biosynthesis | |||||||
| A0A1D6DW07 | Zm00001d002051 | D-3-phosphoglycerate dehydrogenase | 1.78 | 0.0494 | 1.69 | 0.0175 | 0.95 | Serine biosynthesis | |||||||
| Carbon assimilation / Calvin cycle | |||||||||||||||
| O24574 | Zm00001d004894 | Ribulose bisphosphate carboxylase small chain | 0.38 | 0.0113 | 0.33 | 0.0466 | 0.87 | Carbon dioxide fixation | |||||||
| Carbohydrate metabolism | |||||||||||||||
| Q9FQ11 | Zm00001d010523 | Sucrose-phosphatase 1 | 1.50 | 0.0154 | 1.58 | 0.0420 | 1.05 | Sucrose biosynthesis | |||||||
| A0A1D6IJ76 | Zm00001d022107 | Glyceraldehyde-3-phosphate dehydrogenase A | 0.34 | 0.0319 | 0.51 | 0.0019 | 1.52 | Carbon metabolism | |||||||
| Cell division | |||||||||||||||
| A0A1D6FRI4 | Zm00001d010500 | ERBB-3 binding protein 1 | 1.89 | 0.0387 | 1.58 | 0.0266 | 0.84 | Cell division and cell growth regulation | |||||||
| Photosynthetic light reactions | |||||||||||||||
| A0A1D6HS38 | Zm00001d018779 | Oxygen-evolving enhancer protein 2-1 chloroplastic (OEE2-1) | 0.27 | 0.0110 | 0.48 | 0.0354 | 1.78 | Photosynthesis. Photosystem II oxygen evolving complex | |||||||
| B4FWG2 | Zm00001d048422 | Photosynthetic NDH subunit of subcomplex B 2 chloroplastic | 0.25 | 0.0047 | 0.41 | 0.0200 | 1.62 | Photosynthetic electron transport flow around photosystem I to produce ATP | |||||||
| A0A1X7YHG9 | AtpA | ATP synthase subunit α chloroplastic (ATPα) | 0.20 | 0.0166 | 0.61 | 0.0163 | 2.99 | Chloroplast ATP synthesis coupled proton transport | |||||||
| P46617 | PetA | Cytochrome f | 0.18 | 0.0193 | 0.29 | 0.0161 | 1.59 | Photosynthetic electron transport activity | |||||||
| P00827 | Zm00001d006403 | ATP synthase subunit β chloroplastic (ATPβ) | 0.15 | 0.0076 | 0.52 | 0.0274 | 3.45 | Chloroplast ATP synthesis coupled proton transport | |||||||
| Reactive Oxygen Species (ROS) Scavenging Pathways / Response to oxidative stress | |||||||||||||||
| A0A1D6MSE3 | Zm00001d040721 | Dihydrolipoyl dehydrogenase | 2.30 | 0.0273 | 1.80 | 0.0205 | 0.78 | Cell redox homeostasis | |||||||
| A0A1D6JPH3 | Zm00001d027769 | Glutathione reductase | 2.21 | 0.0053 | 1.71 | 0.0436 | 0.77 | Cell redox homeostasis. Glutathione metabolic process. Cellular oxidant detoxification | |||||||
| Ribosome biogenesis | |||||||||||||||
| K7UTH7 | Zm00001d009596 | GTPase ERA1 chloroplastic | 2.61 | 0.0108 | 1.81 | 0.0126 | 0.69 | Ribosome biogenesis. Ribosomal small subunit assembly. rRNA processing | |||||||
| Transcription and translation processes | |||||||||||||||
| A0A1D6LIV5 | Zm00001d035802 | Phenylalanine--tRNA ligase beta subunit cytoplasmic | 2.56 | 0.0314 | 2.23 | 0.0093 | 0.87 | Translation. Phenylalanyl-tRNA aminoacylation | |||||||
| Transporters and transport processes | |||||||||||||||
| B6SP43 | Zm00001d007597 | ABC family1 | 4.54 | 0.0103 | 2.69 | 0.0125 | 0.59 | ATPase-coupled transmembrane transporter activity | |||||||
| Protein ID1 | Gene Name/ID2 | Protein name/Annotation | FC3 | P-Value4 | Function/Biological process5 |
|---|---|---|---|---|---|
| AMINO ACID AND PEPTIDE METABOLISMS | |||||
| Strongly induced proteins by B toxicity in Pachía | |||||
| B6SKB7 | Zm00001d031013 | Methylcrotonoyl-CoA carboxylase subunit α | 4.44 | 0.0022 | Leucine degradation |
| B6SWZ4 | Zm00001d050336 | Methylcrotonoyl-CoA carboxylase β chain mitochondrial | 2.85 | 0.0154 | Leucine degradation |
| A0A1D6K836 | Zm00001d029848 | Branched-chain-amino-acid amino-transferase | 2.35 | 0.0272 | Branched-chain amino acid biosynthesis |
| B4G011 | Zm00001d046923 | D-3-phosphoglycerate dehydroge-nase chloroplastic | 2.31 | 0.0154 | Serine biosynthesis |
| C4J411 | Zm00001d028464 | Imidazole glycerol phosphate synthase hisHF | 2.17 | 0.0017 | Histidine biosynthesis |
| C4JBG7 | Zm00001d015088 | 3-isopropylmalate dehydratase large subunit | 2.14 | 0.0320 | Leucine biosynthesis |
| Strongly repressed proteins by B toxicity in Pachía | |||||
| B4FUH2 | Zm00001d043382 | Aspartate aminotransferase | 0.48 | 0.0195 | Amino acid metabolic process |
| B4FU01 | Zm00001d045153 | Cystathionine β-lyase chloroplas-tic | 0.44 | 0.0235 | Methionine biosynthetic. Cysteine biosynthetic process via cystathionine |
| A0A1D6ICL3 | Zm00001d021596 | Adenosine 5-phosphosulfate reductase-like1 | 0.29 | 0.0140 | Cysteine biosynthetic process. Sulfate reduction |
| B6TZD1 | Zm00001eb168430 | Methylthioribose-1-phosphate isomerase | 0.24 | 0.0461 | Methionine biosynthesis |
| CARBON ASSIMILATION AND CALVIN CYCLE | |||||
| Strongly induced proteins by B toxicity in Pachía | |||||
| A0A1D6FQE4 | Zm00001d010321 | Pyruvate phosphate dikinase | 2.31 | 0.0449 | C4 photosynthetic carbon assimilation cycle |
| Strongly repressed proteins by B toxicity in Pachía | |||||
| O24574 | Zm00001d004894 | Ribulose bisphosphate carboxylase small chain | 0.38 | 0.0113 | Carbon dioxide fixation |
| B4FQ59 | Zm00001d017711 | Phosphoribulokinase | 0.33 | 0.0004 | Calvin- Benson cycle |
| Q9ZT00 | Zm00001eb164390 | Ribulose bisphosphate carboxylase/oxygenase activase chloroplastic | 0.26 | 0.0090 | Carbon dioxide fixation. Rubisco activator activity |
| CARBOHYDRATE METABOLISM | |||||
| Strongly induced proteins by B toxicity in Pachía | |||||
| A0A1D6NE29 | Zm00001d043662 | α-amylase 3 chloroplastic | 2.05 | 0.0460 | Starch degradation |
| Strongly repressed proteins by B toxicity in Pachía | |||||
| A0A1D6M7C2 | Zm00001d038579 | Phosphoglycerate kinase cytosolic | 0.49 | 0.0136 | Glycolysis and gluconeogenesis |
| B4FRC9 | Zm00001d011965 | Transaldolase | 0.41 | 0.0407 | Pentose-phosphate shunt |
| A0A1D6IJ76 | Zm00001d022107 | Glyceraldehyde-3-phosphate dehydrogenase A | 0.34 | 0.0319 | Carbon metabolism |
| CELL DEATH | |||||
| Strongly induced proteins by B toxicity in Pachía | |||||
| B4F8B9 | Zm00001d018468 | S-(hydroxymethyl)glutathione dehydrogenase | 2.81 | 0.0027 | Cell death. Formaldehyde oxidation (glutathione-dependent) |
| CELL WALL | |||||
| Strongly induced proteins by B toxicity in Pachía | |||||
| B4F9J1 | Zm00001d046357 | β-galactosidase | 3.17 | 0.0092 | Xyloglucan degradation |
| DNA AND CHROMATIN ORGANIZATION AND DNA REPAIR | |||||
| Strongly induced proteins by B toxicity in Pachía | |||||
| B6TGH8 | Zm00001d034479 | Histone H1 | 3.60 | 0.0349 | Chromosome condensation. Nucleosome assembly. Nucleosome positioning |
| C0P6Q6 | Zm00001d040416 | DNA gyrase subunit B | 3.48 | 0.0007 | DNA topological change |
| Strongly repressed proteins by B toxicity in Pachía | |||||
| B6SK03 | Zm00001d053295 | Ubiquitin-conjugating enzyme E2 variant 1C | 0.39 | 0.0409 | DNA postreplication repair. Protein polyubiquitination |
| LIPID METABOLISM | |||||
| Strongly induced proteins by B toxicity in Pachía | |||||
| K7VQG5 | Zm00001d008727 | Phospholipase D | 2.30 | 0.0244 | Phospholipid degradation |
| A0A1D6NE81 | Zm00001d043680 | Phospholipase A1-IIδ | 2.02 | 0.0390 | Lipid degradation |
| Strongly repressed proteins by B toxicity | |||||
| B4FLS8 | Zm00001d003584 | 12-oxo-phytodienoic acid reductase 5 | 0.33 | 0.0436 | Fatty acid and oxylipin biosynthesis |
| NITROGEN METABOLISM | |||||
| Strongly induced proteins by B toxicity in Pachía | |||||
| A0A1D6PZA5 | Zm00001d049995 | Nitrate reductase | 2.19 | 0.0077 | Nitrate reductase (NADH) activity. Nitrate assimilation |
| OTHERS | |||||
| Strongly induced proteins by B toxicity in Pachía | |||||
| A0A1D6JGY3 | Zm00001d026515 | Molybdopterin molybdenum-transferase | 2.92 | 0.0023 | Molybdenum cofactor biosynthesis |
| A0A1D6HUN3 | Zm00001d019040 | D-2-hydroxyglutarate dehydrogenase mitochondrial | 2.09 | 0.0380 | Lysine degradation |
| Strongly repressed proteins by B toxicity in Pachía | |||||
| C0PDB6 | Zm00001d039535 | HXXXD-type acyl-transferase family protein | 0.40 | 0.0112 | N-acyltransferase activity |
| C0PE12 | Zm00001d009877 | Protein plastid transcriptionally active 16 chloroplastic | 0.24 | 0.0121 | Circadian rhythm |
| OXIDATION AND REDUCTION PROCESSES | |||||
| Strongly induced proteins by B toxicity in Pachía | |||||
| A0A1D6M498 | Zm00001d038189 | FAD/NAD(P)-binding oxidoreductase family protein | 2.04 | 0.0101 | Oxidoreductase activity |
| PHOTOSYNTHETIC LIGHT REACTIONS | |||||
| Strongly repressed proteins by B toxicity in Pachía | |||||
| B6SSB9 | Zm00001d035859 | Plastocyanin | 0.50 | 0.0300 | Photosynthetic electron transport |
| A0A1D6GU53 | Zm00001d014564 | Oxygen-evolving enhancer protein 1-1 chloroplastic | 0.47 | 0.0268 | Photosynthesis. Oxygen evolving activity. Photosystem II assembly and stabilization |
| B6SUC4 | Zm00001d046786 | Chlorophyll a-b binding protein, chloroplastic | 0.41 | 0.0086 | Photosynthesis. Light harvesting in photosystem I |
| B6T927 | Zm00001d014349 | NAD(P)H-quinone oxidoreductase subunit S chloroplastic (NDHS) | 0.39 | 0.0095 | Photosynthetic electron transport chain |
| P25709 | NdhH | NAD(P)H-quinone oxidoreductase subunit H, chloroplastic | 0.37 | 0.0022 | Photosynthesis, light reaction. Photosynthetic electron transport chain. Couples the photosynthetic redox reaction to proton translocation |
| B6SP99 | Zm00001d024148 | Photosynthetic NDH subunit of subcomplex B 1 chloroplastic | 0.33 | 0.0137 | Photosynthetic electron transport in photosystem I |
| B4FJP7 | Zm00001d027729 | Photosynthetic NDH subunit of subcomplex B 2 chloroplastic | 0.32 | 0.0169 | Photosynthetic electron transport in photosystem I |
| B4FR80 | Zm00001d033098 | Post-illumination chlorophyll fluorescence increase (ZmPIFI) | 0.28 | 0.0270 | Chlororespiration |
| A0A1D6HS38 | Zm00001d018779 | Oxygen-evolving enhancer protein 2-1 chloroplastic (OEE2-1) | 0.27 | 0.0110 | Photosynthesis. Photosystem II oxygen evolving complex |
| B4FWG2 | Zm00001d048422 | Photosynthetic NDH subunit of subcomplex B 2 chloroplastic | 0.25 | 0.0047 | Photosynthetic electron transport flow around photosystem I to produce ATP |
| P19124 | NdhJ | NAD(P)H-quinone oxidoreductase subunit J, chloroplastic | 0.22 | 0.0147 | Photosynthesis, light reaction, photosynthetic electron transport chain. Couples the photosynthetic redox reaction to proton translocation |
| A0A1X7YHG9 | AtpA | ATP synthase subunit α (ATPα) | 0.20 | 0.0166 | Chloroplast ATP synthesis coupled proton transport |
| P46617 | PetA | Cytochrome f | 0.18 | 0.0193 | Photosynthetic electron transport chain |
| P00827 | Zm00001d009488 | ATP synthase subunit β, chloroplastic (ATPβ) | 0.15 | 0.0076 | Chloroplast ATP synthesis coupled proton transport |
| A0A1D6JYG6 | Zm00001d028670 | Photosynthetic NDH subunit of lumenal location 1 chloroplastic | 0.13 | 0.0134 | Part of photosystem II oxygen evolving complex |
| PIGMENT BIOSYNTHESIS | |||||
| Strongly repressed proteins by B toxicity in Pachía | |||||
| A0A1D6FAV8 | Zm00001d008203 | Protoporphyrinogen oxidase | 0.38 | 0.0173 | 3,8-divinyl-chlorophyllide a and protoporphyrinogen IX biosynthesis |
| PROTEIN DEGRADATION | |||||
| Strongly induced proteins by B toxicity in Pachía | |||||
| B4FS65 | Zm00001d005391 | Cysteine protease 14 | 4.38 | 0.0146 | Proteolysis. Proteolysis involved in protein catabolic process |
| A0A1D6HM49 | Zm00001d018282 | Subtilisin-like protease SBT1.4 | 3.70 | 0.0399 | Serine protease. Serine-type endopeptidase activity. Proteolysis |
| A0A1D6H4R4 | Zm00001d015962 | Prolyl oligopeptidase family protein | 3.58 | 0.0080 | Proteolysis. Serine protease. Serine-type peptidase activity |
| Q84TL7 | Zm00001d011036 | Legumin-like protein | 2.86 | 0.0453 | Protein ubiquitination. Nutrient reservoir activity. Storage protein |
| A0A1D6KWW2 | Zm00001d033194 | Subtilisin-like protease | 2.85 | 0.0403 | Proteolysis. Serine protease. Serine-type endopeptidase activity |
| A0A1D6KV27 | Zm00001d032956 | Acylamino-acid-releasing enzyme | 2.54 | 0.0086 | Proteolysis. Serine protease. Serine-type endopeptidase activity |
| C0HI51 | Zm00001d044102 | Zn-dependent exopeptidase superfamily protein | 2.53 | 0.0131 | Proteolysis. Aminopeptidase. Metalloaminopeptidase activity |
| Q84TL6 | Zm00001d035597 | Legumin-like protein | 2.36 | 0.0386 | Protein ubiquitination. Storage protein. Nutrient reservoir activity |
| A0A1D6HL34 | Zm00001d018145 | Presequence protease 2 chloroplastic/mitochondrial | 2.22 | 0.0180 | Proteolysis. Metalloendopeptidase activity. Protein processing |
| K7VGG8 | Zm00001d010522 | ATP-dependent zinc metalloprotease FTSH 10 mitochondrial | 2.07 | 0.0359 | Proteolysis. Metalloprotease mitochondrial |
| C4JC43 | Zm00001d049100 | Target of Myb protein 1 | 2.04 | 0.0450 | Proteolysis. Protein transport to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway |
| Strongly repressed proteins by B toxicity in Pachía | |||||
| A0A1D6H558 | Zm00001d016036 | Chloroplast processing peptidase | 0.47 | 0.0438 | Protease. Serine-type endopeptidase activity |
| B4FQJ6 | Zm00001d018309 | 26S protease regulatory subunit 7 homolog A | 0.46 | 0.0249 | Proteolysis. Protein catabolic process. Peptidase activity |
| A0A1D6FKP2 | Zm00001d009613 | Protease Do-like 1 chloroplastic | 0.45 | 0.0496 | Proteolysis. Serine-type endopeptidase activity |
| K7TTX0 | Zm00001d025628 | Plant UBX domain-containing protein 4 | 0.44 | 0.0107 | Proteasome-mediated ubiquitin-dependent protein catabolic process |
| PROTEIN STABILIZATION AND FOLDING | |||||
| Strongly induced proteins by B toxicity in Pachía | |||||
| A0A1D6FN98 | Zm00001d009948 | Heat shock 70 kDa protein 14 | 2.28 | 0.0487 | Protein folding. Stress response |
| B6SZ69 | Zm00001d028630 | Heat shock cognate 70 kDa protein 2 | 2.02 | 0.0398 | Protein refolding. Stress response |
| Strongly repressed proteins by B toxicity in Pachía | |||||
| A0A1D6KC46 | Zm00001d030346 | Hsp20/alpha crystallin family protein | 0.49 | 0.0499 | Chaperone. Response to heat |
| C0PKD9 | Zm00001d052101 | Chaperonin10 | 0.42 | 0.0428 | Chaperone cofactor-dependent protein refolding Protein folding. Chaperone |
| G2XK63 | Zm00001d040257 | T-complex protein 1 subunit beta | 0.27 | 0.0065 | Protein folding. Chaperone |
| B4FR04 | Zm00001d019052 | Peptidylprolyl isomerase | 0.23 | 0.0205 | Protein folding. Rotamase |
| REACTIVE OXYGEN SPECIES (ROS) SCAVENGING PATHWAYS / RESPONSE TO OXIDATIVE STRESS | |||||
| Strongly induced proteins by B toxicity in Pachía | |||||
| A0A1D6K5D2 | Zm00001d029457 | Nucleoredoxin1 | 2.91 | 0.0117 | Protection against oxidative stress. Cellular oxidant detoxification |
| A0A1D6MSE3 | Zm00001d040721 | Dihydrolipoyl dehydrogenase | 2.30 | 0.0273 | Cell redox homeostasis |
| A0A1D6JPH3 | Zm00001d027769 | Glutathione reductase | 2.21 | 0.0053 | Cell redox homeostasis. Cellular oxidant detoxification. Glutathione metabolic process. |
| K7US39 | Zm00001d009163 | Dihydrolipoyl dehydrogenase | 2.19 | 0.0088 | Cell redox homeostasis |
| RIBOSOME BIOGENESIS | |||||
| Strongly induced proteins by B toxicity in Pachía | |||||
| B4FPB7 | Zm00001d006100 | 60S ribosomal protein L7a | 2.63 | 0.0051 | Ribosome biogenesis. Maturation of LSU-rRNA |
| K7UTH7 | Zm00001d009596 | GTPase ERA1 chloroplastic | 2.61 | 0.0108 | Ribosome biogenesis. Ribosomal small subunit assembly. rRNA processing |
| B4F7Y1 | Zm00001d031640 | 60S ribosomal protein L7a-1 | 2.39 | 0.0448 | Ribosomal protein. Maturation of LSU-rRNA |
| RNA BINDING AND PROCESSING | |||||
| Strongly induced proteins by B toxicity in Pachía | |||||
| A0A1D6HT50 | Zm00001d018891 | Chloroplast RNA processing 4 | 2.60 | 0.0142 | mRNA catabolic process |
| SIGNALING | |||||
| Strongly repressed proteins by B toxicity in Pachía | |||||
| P49235 | Zm00001eb411380 | 4-hydroxy-7-methoxy-3-oxo-3,4-dihydro-2H-1,4-benzoxazin-2-yl glucoside beta-D-glucosidase 1, chloroplastic | 0.19 | 0.0090 | Cytokinin signaling pathway |
| STRESS | |||||
| Strongly induced proteins by B toxicity in Pachía | |||||
| B4F9K2 | Zm00001d005315 | Calcium-dependent lipid-binding (CaLB domain) family protein | 2.11 | 0.0402 | Defense response. Response to stress |
| TRANSCRIPTION AND TRANSLATION PROCESSES | |||||
| Strongly induced proteins by B toxicity in Pachía | |||||
| A0A1D6LEN8 | Zm00001d035139 | MA3 domain-containing protein | 4.95 | 0.0073 | Negative regulation of transcription, DNA-templated. Regulation of translation |
| Q6R9D1 | GRMZM5G806488 | Ribosomal protein S7 | 3.89 | 0.0202 | Translation. Ribosomal small subunit assembly. Structural constituent of ribosome |
| A0A1D6IAN8 | Zm00001d021400 | Octicosapeptide/Phox/Bem1p (PB1) domain-containing protein / tetratricopeptide repeat (TPR)-containing protein | 3.47 | 0.0323 | RNA processing |
| C0P456 | Zm00001d002789 | Pentatricopeptide repeat-containing protein | 3.26 | 0.0259 | Likely involved in posttranscriptional control of gene expression in organelles |
| A0A1D6NR59 | Zm00001d044745 | Probable alanine--tRNA ligase, chloroplastic | 2.74 | 0.0097 | Translation. Alanyl-tRNA aminoacylation |
| A0A1D6LIV5 | Zm00001d035802 | Phenylalanine--tRNA ligase beta subunit cytoplasmic | 2.56 | 0.0314 | Translation. Phenylalanyl-tRNA aminoacylation |
| B6T5F2 | Zm00001d011992 | 60S ribosomal protein L13 | 2.48 | 0.0387 | Translation. Structural constituent of ribosome |
| A0A1D6HM03 | Zm00001d018274 | Isoleucine--tRNA ligase chloroplastic/mitochondrial | 2.29 | 0.0087 | Translation. Isoleucyl-tRNA aminoacylation |
| A0A1D6QAN9 | Zm00001d051885 | ATG8-interacting protein 1 | 2.28 | 0.0047 | Box C/D RNA 3'-end processing. rRNA processing |
| A0A1D6FRP3 | Zm00001d010530 | Cysteine--tRNA ligase 1 cytoplasmic | 2.26 | 0.0470 | Translation. Cysteinyl-tRNA aminoacylation |
| B4FMD3 | Zm00001d012978 | 40S ribosomal protein S23-2 | 2.13 | 0.0451 | Translation. Structural constituent of ribosome |
| K7UTZ2 | Zm00001d009761 | Spliceosome RNA helicase BAT1 isoform 1 | 2.10 | 0.0455 | RNA splicing. RNA helicase activity |
| K7TY03 | Zm00001d023741 | Alanine--tRNA ligase | 2.07 | 0.0144 | Translation. Alanyl-tRNA aminoacylation |
| B4FYR2 | Zm00001d038865 | 60S ribosomal protein L28 | 2.05 | 0.0275 | Translation. Structural constituent of ribosome |
| B6U151 | Zm00001d002104 | Glutamyl-tRNA(Gln) amidotransferase subunit A, chloroplastic/mitochondrial | 2.02 | 0.0464 | Mitochondrial translation |
| Strongly repressed proteins by B toxicity in Pachía | |||||
| C0P7X7 | Zm00001d034808 | 30S ribosomal protein S6 alpha chloroplastic | 0.50 | 0.0059 | Translation. Structural constituent of ribosome |
| B4FUZ5 | Zm00001d047581 | 30S ribosomal protein S1 | 0.46 | 0.0055 | Translation. Ribosomal protein |
| O50018 | Zm00001d046449 | Elongation factor 1-α | 0.29 | 0.0269 | Translation. Translation elongation factor activity |
| TRANSPORTERS AND TRANSPORT PROCESSES | |||||
| Strongly induced proteins by B toxicity in Pachía | |||||
| B6SP43 | Zm00001d007597 | ABC family1 | 4.54 | 0.0103 | ATPase-coupled transmembrane transporter activity |
| A0A1D6H2R4 | Zm00001d015569 | H+-exporting diphosphatase | 4.34 | 0.0050 | Ion transport. Pyrophosphate hydrolysis-driven proton transmembrane transporter activity |
| A0A1D6MS70 | Zm00001d040686 | Protein translocase subunit SECA1 chloroplastic | 4.12 | 0.0173 | Protein transport |
| A0A1D6DSW6 | Zm00001d001788 | K+ efflux antiporter 2 chloroplastic | 3.79 | 0.0414 | Chloroplast potassium ion trans-port |
| B6T5R1 | Zm00001d010504 | Ran-binding protein 1 | 3.16 | 0.0492 | Intracellular transport. Protein and mRNA transport. Nucleocytoplasmic transport |
| A0A1D6KSB0 | Zm00001d032615 | Protein TIC110 chloroplastic | 2.35 | 0.0118 | Protein import into chloroplast stroma |
| NOT WELL-KNOWN PROTEINS | |||||
| Strongly induced proteins by B toxicity in Pachía | |||||
| A0A1D6KKK1 | Zm00001d031677 | MtN19-like protein | 2.62 | 0.0464 | Not well determined |
| A0A1D6JI62 | Zm00001d026632 | Stem-specific protein TSJT1 | 2.43 | 0.0283 | Not well determined |
| Protein ID1 | Gene Name/ID2 | Protein name/Annotation | FC3 | P-value4 | Function/Biological process5 |
|---|---|---|---|---|---|
| AMINO ACID AND PEPTIDE METABOLISMS | |||||
| Strongly induced proteins by B toxicity in Sama | |||||
| B6SKB7 | Zm00001d031013 | Methylcrotonoyl-CoA carboxylase subunit alpha | 3.56 | 0.0049 | Leucine degradation |
| C4J3S6 | Zm00001d004960 | 2-isopropylmalate synthase 1 chloroplastic | 2.17 | 0.0025 | Leucine biosynthesis |
| CARBON ASSIMILATION AND CALVIN CYCLE | |||||
| Strongly repressed proteins by B toxicity in Sama | |||||
| O24574 | Zm00001d004894 | Ribulose bisphosphate carboxylase small chain | 0.33 | 0.0466 | Carbon dioxide fixation |
| P05348 | Rbcs | Ribulose bisphosphate carboxylase small chain, chloroplastic | 0.13 | 0.0096 | Carbon dioxide fixation |
| CELL DIVISION | |||||
| Strongly induced proteins by B toxicity in Sama | |||||
| C0P4T2 | Zm00001d042664 | Patellin-1 | 3.05 | 0.0149 | Cell division and cell cycle |
| NUCLEOTIDE, PURINE AND PYRIMIDINE METABOLISM | |||||
| Strongly repressed proteins by B toxicity in Sama | |||||
| A0A1D6P7V2 | Zm00001d047217 | 5-hydroxyisourate hydrolase | 0.50 | 0.0136 | Purine metabolism |
| PHOTOSYNTHETIC LIGHT REACTIONS | |||||
| Strongly repressed proteins by B toxicity in Sama | |||||
| A0A1D6HS38 | Zm00001d018779 | Oxygen-evolving enhancer protein 2-1 chloroplastic (OEE2-1) | 0.48 | 0.0354 | Photosynthesis. Photosystem II oxygen evolving complex |
| B6SSN3 | Zm00001d015385 | Chlorophyll a-b binding protein, chloroplastic | 0.43 | 0.0435 | Light harvesting in photosystem I |
| B4FWG2 | Zm00001d048422 | Photosynthetic NDH subunit of subcomplex B 2 chloroplastic | 0.41 | 0.0200 | Photosynthetic electron transport flow around photosystem I to produce ATP |
| P06670 | NdhK | NAD(P)H-quinone oxidoreductase subunit K, chloroplastic | 0.38 | 0.0149 | Photosynthetic electron transport coupled photosynthetic proton transport |
| A0A1X7YHF7 | PsbD | Photosystem II D2 protein | 0.35 | 0.0347 | Photosynthetic electron transport in photosystem II |
| P46617 | PetA | Cytochrome f | 0.29 | 0.0161 | Photosynthetic electron transport chain |
| B6SQV5 | Zm00001d049387 | Photosystem II 10 kDa polypeptide | 0.14 | 0.0438 | Photosynthesis. Photosystem II oxygen evolving complex |
| PROTEIN STABILIZATION AND FOLDING | |||||
| Strongly repressed proteins by B toxicity in Sama | |||||
| A0A1D6GJM6 | Zm00001d013455 | Peptidylprolyl isomerase | 0.40 | 0.0275 | Protein folding. Rotamase |
| C4J6Y2 | Zm00001d018077 | Peptidylprolyl isomerase | 0.18 | 0.0422 | Protein folding. Rotamase |
| REACTIVE OXYGEN SPECIES (ROS) SCAVENGING PATHWAYS / RESPONSE TO OXIDATIVE STRESS | |||||
| Strongly induced proteins by B toxicity in Sama | |||||
| B4FSM5 | Zm00001d040341 | Peroxiredoxin | 2.68 | 0.0112 | Cellular response to oxidative stress. Hydrogen peroxide catabolic process. Cell redox homeostasis |
| Strongly repressed proteins by B toxicity in Sama | |||||
| B6U038 | Zm00001d005482 | Thiol-disulfide isomerase and thioredoxins | 0.44 | 0.0223 | Antioxidant activity. Cellular oxidant detoxification. Thioredoxin-dependent peroxiredoxin activity |
| B4FZ35 | Zm00001d002240 | CHL-Zea mays chloroplastic li-pocalin | 0.31 | 0.0272 | Response to oxidative stress. Violaxanthin, antheraxanthin and zeaxanthin interconversion |
| SECONDARY METABOLISM | |||||
| Strongly induced proteins by B toxicity in Sama | |||||
| O64411 | Zm00001d024281 | Polyamine oxidase 1 (PAO1) | 3.34 | 0.0108 | Spermine degradation. Amine and polyamine degradation |
| Strongly repressed proteins by B toxicity in Sama | |||||
| B6TAE7 | Zm00001d028575 | Tropinone reductase | 0.44 | 0.0313 | Tropane alkaloid biosynthesis |
| STRESS | |||||
| Strongly induced proteins by B toxicity in Sama | |||||
| A0A1D6NJS4 | Zm00001d044222 | Tetratricopeptide repeat (TPR)-containing protein | 2.12 | 0.0428 | N-terminal peptidyl-methionine acetylation. Protein maturation |
| TRANSCRIPTION AND TRANSLATION PROCESSES | |||||
| Strongly induced proteins by B toxicity in Sama | |||||
| A0A1D6IBP5 | Zm00001d021507 | Asparagine--tRNA ligase chloroplastic/mitochondrial | 2.66 | 0.0491 | Translation. Asparaginyl-tRNA aminoacylation |
| B4FSE0 | Zm00001d033913 | Alba DNA/RNA-binding protein | 2.48 | 0.0244 | Translational initiation. RNA binding |
| B6T872 | Zm00001d021020 | 60S ribosomal protein L32 | 2.28 | 0.0415 | Translation. Structural constituent of ribosome |
| A0A1D6LIV5 | Zm00001d035802 | Phenylalanine--tRNA ligase beta subunit cytoplasmic | 2.23 | 0.0093 | Translation. Phenylalanyl-tRNA aminoacylation |
| B4FJ27 | Zm00001d011741 | 40S ribosomal protein S24 | 2.13 | 0.0363 | Translation. Structural constituent of ribosome |
| TRANSPORTERS AND TRANSPORT PROCESSES | |||||
| Strongly induced proteins by B toxicity in Sama | |||||
| B6SP43 | Zm00001d007597 | ABC family1 | 2.69 | 0.0125 | ATPase-coupled transmembrane transporter activity |
| Protein ID1 | Gene Name/ID2 | Protein name/Annotation | FC3 | P-value4 | Function/Biological process5 | ||
|---|---|---|---|---|---|---|---|
| AMINO ACID AND PEPTIDE METABOLISMS | |||||||
| Strongly up-accumulated proteins in Sama in media with 10 mM B | |||||||
| A0A1D6ICL3 | Zm00001d021596 | Adenosine 5-phosphosulfate reductase-like1 | 2.33 | 0.0417 | Cysteine biosynthetic process. Sulfate reduction | ||
| CARBON ASSIMILATION AND CALVIN CYCLE | |||||||
| Strongly up-accumulated proteins in Sama in media with 10 mM B | |||||||
| A0A1D6EXF1 | Zm00001d006520 | PDK regulatory protein1 | 2.16 | 0.0167 | Regulation of C4 photosynthetic carbon assimilation cycle | ||
| CARBOHYDRATE METABOLISM | |||||||
| Strongly up-accumulated proteins in Sama in media with 10 mM B | |||||||
| Q9SYS1 | Zm00001d021702 | β-amylase | 2.63 | 0.0499 | β-amylase activity. Starch degradation | ||
| Strongly down-accumulated proteins in Sama in media with 10 mM B | |||||||
| A0A1D6K5L6 | Zm00001d029502 | Glucose-6-phosphate 1-dehydrogenase | 0.36 | 0.0411 | Pentose phosphate pathway | ||
| A0A1D6LY56 | Zm00001d037480 | Alkaline α galactosidase 2 | 0.33 | 0.0438 | Carbohydrate metabolic process | ||
| CELL DEATH | |||||||
| Strongly down-accumulated proteins in Sama in media with 10 mM B | |||||||
| A0A1D6JNJ8 | Zm00001d027656 | Lethal leaf-spot 1 | 0.32 | 0.0016 | Cell death. Chlorophyll catabolic process | ||
| CELL DIVISION | |||||||
| Strongly down-accumulated proteins in Sama in media with 10 mM B | |||||||
| A0A1D6JH24 | Zm00001d026532 | Protein RCC2 | 0.42 | 0.0214 | Cell division | ||
| CELL WALL | |||||||
| Strongly up-accumulated proteins in Sama in media with 10 mM B | |||||||
| A0A1D6MWZ7 | Zm00001d041578 | Glossy6 | 3.27 | 0.0403 | Epicuticular wax accumulation. Intracellular trafficking of cuticular waxes | ||
| DNA AND CHROMATIN ORGANIZATION AND DNA REPAIR | |||||||
| Strongly down-accumulated proteins in Sama in media with 10 mM B | |||||||
| B4FQA5 | Zm00001d018981 | Histone1a | 0.35 | 0.0318 | Chromosome condensation. Nucleosome assembly | ||
| B6TGH8 | Zm00001d034479 | Histone H1 | 0.31 | 0.0138 | Chromosome condensation. Nucleosome assembly. Nucleosome positioning | ||
| LIPID METABOLISM | |||||||
| Strongly up-accumulated proteins in Sama in media with 10 mM B | |||||||
| Q8W0V2 | Zm00001d033623 | Lipoxygenase 3 | 5.06 | 0.0455 | Fatty acid and oxylipin biosynthesis | ||
| Q06XS3 | Zm00001d053675 | Lipoxygenase 10 | 3.44 | 0.0247 | Fatty acid and oxylipin biosynthesis | ||
| OTHERS | |||||||
| Strongly down-accumulated proteins in Sama in media with 10 mM B | |||||||
| B6TY16 | Zm00001d040331 | SUN domain protein2 | 0.41 | 0.0262 | Nuclear envelope organization | ||
| B4F7V3 | Zm00001d021582 | Protein phosphatase 2C isoform ε | 0.39 | 0.0214 | Protein dephosphorylation | ||
| A0A1D6HUN3 | Zm00001d019040 | D-2-hydroxyglutarate dehydrogenase mitochondrial | 0.33 | 0.0024 | Photorespiration | ||
| OXIDATION AND REDUCTION PROCESSES | |||||||
| Strongly down-accumulated proteins in Sama in media with 10 mM B | |||||||
| B4F987 | Zm00001d020984 | Putative sarcosine oxidase | 0.23 | 0.0321 | Sarcosine oxidase activity | ||
| PHOTOSYNTHETIC LIGHT REACTIONS | |||||||
| Strongly up-accumulated proteins in Sama in media with 10 mM B | |||||||
| B4FR80 | Zm00001d033098 | Post-illumination chlorophyll fluorescence increase (ZmPIFI) | 2.52 | 0.0097 | Chlororespiration | ||
| A0A1D6HS38 | Zm00001d018779 | Oxygen-evolving enhancer protein 2-1 chloroplastic (OEE2-1) | 2.31 | 0.0325 | Photosynthesis. Photosystem II oxygen evolving complex | ||
| PIGMENT BIOSYNTHESIS | |||||||
| Strongly up-accumulated proteins in Sama in media with 10 mM B | |||||||
| A0A1D6JHX0 | Zm00001d026603 | Magnesium-chelatase subunit ChlH1 chloroplastic (ChlH1) | 2.90 | 0.0484 | Chlorophyll biosynthetic process | ||
| PROTEIN STABILIZATION AND FOLDING | |||||||
| Strongly down-accumulated proteins in Sama in media with 10 mM B | |||||||
| A0A1D6KE29 | Zm00001d030725 | Heat shock protein 70 | 0.43 | 0.0406 | Protein refolding. Protein folding chaperone. Cellular response to unfolded protein | ||
| RESPIRATION (GLYCOLISIS, TCA CYCLE AND MITOCHONDRIAL ELECTRON TRANSFER) | |||||||
| Strongly up-accumulated proteins in Sama in media with 10 mM B | |||||||
| B4G1C9 | Zm00001d023606 | Dihydrolipoamide acetyltransferase component of pyruvate dehydrogenase complex | 2.04 | 0.0332 | Acetyl-CoA biosynthetic process from pyruvate | ||
| Strongly down-accumulated proteins in Sama in media with 10 mM B | |||||||
| A0A1D6MAK9 | Zm00001d038792 | Phosphotransferase | 0.49 | 0.0331 | Glycolysis | ||
| SECONDARY METABOLISM | |||||||
| Strongly up-accumulated proteins in Sama in media with 10 mM B | |||||||
| O64411 | Zm00001d024281 | Polyamine oxidase 1 (PAO1) | 5.15 | 0.0007 | Spermine degradation. Amine and polyamine degradation | ||
| TRANSCRIPTION AND TRANSLATION | |||||||
| Strongly up-accumulated proteins in Sama in media with 10 mM B | |||||||
| B4FP25 | Zm00001d047296 | 40S ribosomal protein S19 | 6.38 | 0.0289 | Translation. Structural constituent of ribosome. Ribosomal small subunit assembly | ||
| B6TDF7 | Zm00001d019898 | Plastid-specific 30S ribosomal protein 2 | 2.31 | 0.0243 | Ribosomal protein. Ribonucleoprotein complex. RNA-binding | ||
| C0PEC4 | Zm00001d032420 | 30S ribosomal protein S5 chloroplastic | 2.12 | 0.0487 | Translation. Structural constituent of ribosome | ||
| Strongly down-accumulated proteins in Sama in media with 10 mM B | |||||||
| B6SX73 | Zm00001d016549 | 60S ribosomal protein L35 | 0.42 | 0.0284 | Translation. Structural constituent of ribosome | ||
| Q6R9D1 | GRMZM5G806488 | Ribosomal protein S7 | 0.35 | 0.0426 | Translation. Structural constituent of ribosome. Ribosomal small subunit assembly | ||
| Transporter and transport processes | |||||||
| Strongly down-accumulated proteins in Sama in media with 10 mM B | |||||||
| A0A1D6H2R4 | Zm00001d015569 | H+-exporting diphosphatase | 0.33 | 0.0169 | Ion transport. Pyrophosphate hydrolysis-driven proton transmembrane transporter activity | ||
| A0A1D6K7N5 | Zm00001d029762 | Hexose transporter | 0.20 | 0.0439 | Hexose transporter | ||
| Unknown or not well determined | |||||||
| Strongly down-accumulated proteins in Sama in media with 10 mM B | |||||||
| A0A1D6KKK1 | Zm00001d031677 | MtN19-like protein | 0.23 | 0.0121 | Not well determined | ||
| Protein ID1 | Gene Name/ID2 | Protein name/Annotation | Function/Biological process3 |
|---|---|---|---|
| DNA AND CHROMATIN ORGANIZATION AND DNA REPAIR | |||
| Protein exclusively detected in Pachía in 10 mM B | |||
| A0A1D6KX75 | Zm00001d033247 | Nfc103a - nucleosome/chromatin assembly factor C | Nucleosome/chromatin assembly. DNA repair. Chromatin remodeling, regulation of DNA-templated transcription |
| OTHERS | |||
| Protein exclusively detected in Sama in both B treatments | |||
| K7VAT7 | Zm00001d046569 | Protein kinase superfamily protein with octicosapeptide/Phox/Bem1p domain | Protein serine/threonine kinase activity. Protein phosphorylation |
| REACTIVE OXYGEN SPECIES (ROS) SCAVENGING PATHWAYS / RESPONSE TO OXIDATIVE STRESS | |||
| Protein exclusively detected in Pachía in both B treatments | |||
| B4FKV6 | Zm00001d014341 | Peroxidase 54 | Response to oxidative stress. Peroxidase activity |
| TRANSCRIPTION AND TRANSLATION | |||
| Protein exclusively detected in Pachía in 10 mM B | |||
| A0A096RFR6 | Zm00001d039518 | Eukaryotic translation initiation factor 3 subunit A (eIF3a) | Translation initiation factor activity. Protein synthesis. Formation of cytoplasmic translation initiation complex |
| Transporter and transport processes | |||
| Proteins exclusively detected in Pachía in both B treatments | |||
| A0A1D6EU13 | Zm00001d006238 | Calcium lipid binding protein-like | Lipid transport |
| A0A1D6JN64 | Zm00001d027580 | Outer mitochondrial membrane porin1 (ommp1) | Voltage-gated anion channel activity. Inorganic anion transport, transmembrane transport, anion transmembrane transport |
| Protein exclusively detected proteins in Sama in both B treatments | |||
| Q7Y1W6 | Zm00001d018693 | Pentatricopeptide repeat 2 (PPR2) | Chloroplast translation |
| Unknown or not well determined | |||
| Protein exclusively detected proteins in Sama in both B treatments | |||
| A0A1D6DWG9 | Zm00001d002089 | Tetratricopeptide repeat (TPR)-like superfamily protein | Unknown |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





