Submitted:
05 May 2023
Posted:
08 May 2023
You are already at the latest version
Abstract
Keywords:
Introduction
Methods
Sources of aircraft wastewater samples and virus concentration
Nucleic acid extraction
PCR inhibition assessment
RT-PCR, qPCR and RT-qPCR analysis
qPCR and RT-qPCR assay limit of detection
Quality control
Data analysis
Results
qPCR and RT-qPCR assay performance and relevant QA/QC
Prevalence of indicator, enteric and respiratory viruses in aircraft wastewater samples
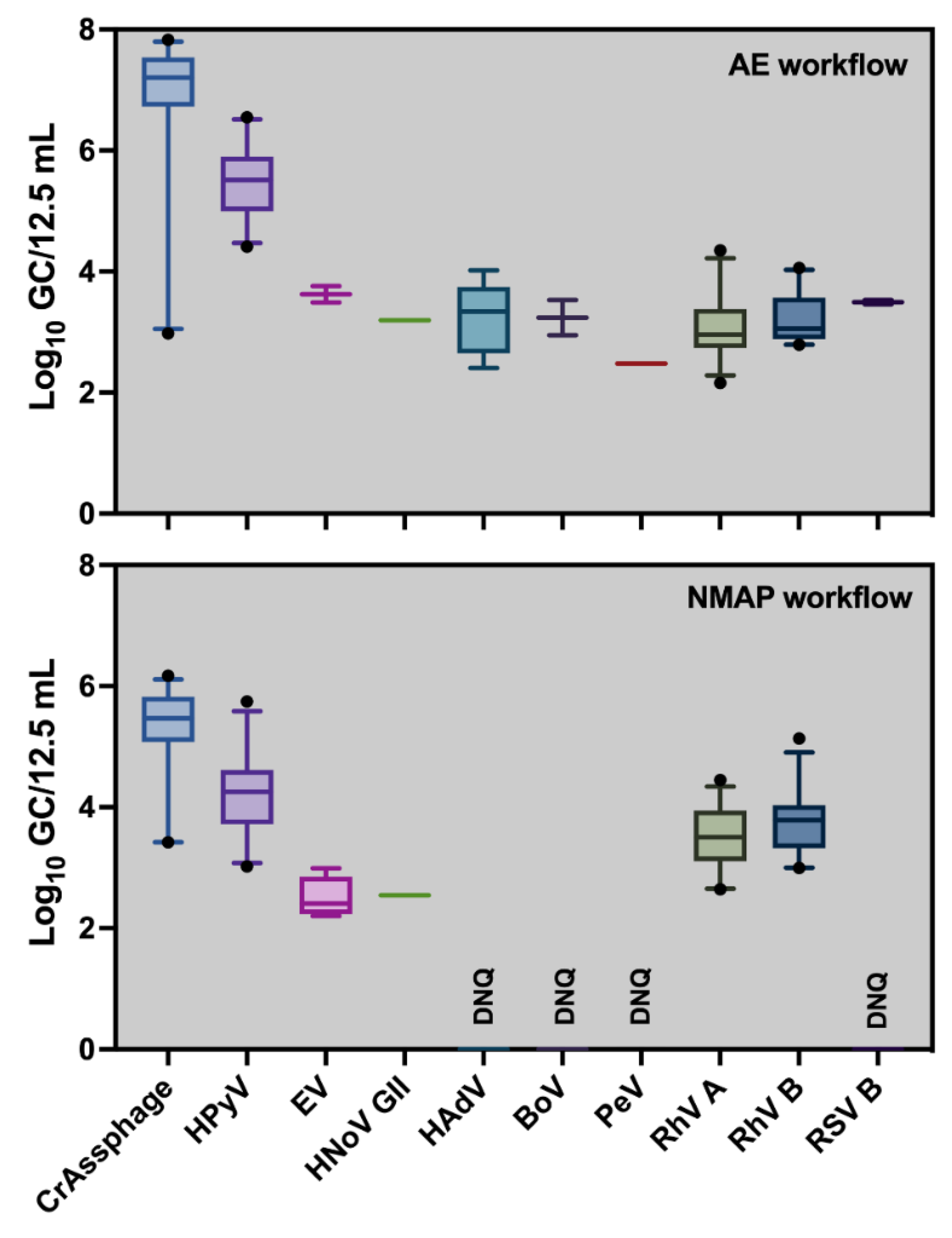
Concentrations of indicator, enteric and respiratory viruses in aircraft wastewater samples
Discussion
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Hovi, T.; Stenvik, M.; Partanen, H.; Kangas, A. Poliovirus surveillance by examining sewage specimens. Quantitative recovery of virus after introduction into sewerage at remote upstream location/Epidemiol Infect 2001, 127, 101–106. [Google Scholar] [CrossRef] [PubMed]
- O’Reilly, K.M.; Allen, D.J.; Fine, P.; Asghar, H. The challenges of informative wastewater sampling for SARS-CoV-2 must be met: Lessons from polio eradication. The Lancet Microbe 2020, 1, e189–e190. [Google Scholar] [CrossRef]
- Fong, T.-T.; Phanikumar, M.S.; Xagoraraki, I.; Rose, J.B. Quantitative detection of human adenoviruses in wastewater and combined sewer overflows influencing a Michigan river. Appl Environm Microbiol 2010, 76, 715–723. [Google Scholar] [CrossRef]
- Prevost, B.; Lucas, F.; Goncalves, A.; Richard, F.; Moulin, L.; Wurtzer, S. Large scale survey of enteric viruses in river and waste water underlines the health status of the local population. Environ Int 2015, 79, 42–50. [Google Scholar] [CrossRef]
- Levican, J.; Levican, A.; Ampuero, M.; Gaggero, A. JC polyomavirus circulation in one-year surveillance in wastewater in Santiago, Chile. Infect Genet Evol 2019, 71, 151–158. [Google Scholar] [CrossRef]
- Wolfe, M.K.; Duong, D.; Bakker, K.M.; et al. Wastewater-based detection of two influenza outbreaks. Environ Sci Technol Lett 2022, 9, 687–692. [Google Scholar] [CrossRef]
- Faleye, T.O.C.; Bowes, D.A.; Driver, E.M.; Adhikari, S.; Adams, D.; Varsani, A.; Halden, R.U.; Scotch, M. Wastewater-based epidemiology and long-read sequencing to identify enterovirus circulation in three municipalities in Maricopa County, Arizona, Southwest United States between June and October 2020. Viruses 2021, 13, 1803. [Google Scholar] [CrossRef]
- Ahmed, W.; Bertsch, P.M.; Angel, N.; et al. Detection of SARS-CoV-2 RNA in commercial passenger aircraft and cruise ship wastewater: A surveillance tool for assessing the presence of COVID-19 infected travellers. J Travel Med. 2020, 27, taaa116. [Google Scholar] [CrossRef]
- Weidhaas, J.; Aanderud, Z.T.; Roper, D.K.; et al. Correlation of SARS-CoV-2 RNA in wastewater with COVID-19 disease burden in sewersheds. Sci Total Environ 2021, 775, 145790. [Google Scholar] [CrossRef]
- Ahmed, W.; Bivins, A.; Simpson, S.L.; Bertsch, P.M.; Ehret, J.; Hosegood, I.; Metcalfe, S.S.; Smith, W.J.; Thomas, K.V.; Tynan, J.; et al. Wastewater surveillance demonstrates high predictive value for COVID-19 infection on board repatriation flights to Australia. Environ Int. 2020, 158, 106938. [Google Scholar] [CrossRef]
- Jones, D.L.; Rhymes, J.M.; Wade, M.J.; Kevill, J.L.; Malham, S.K.; Grimsley, J.M.; Rimmer, C.; Weightman, A.J.; Farkas, K. Suitability of aircraft wastewater for pathogen detection and public health surveillance. Sci Total Environ 2023, 856, 159162. [Google Scholar] [CrossRef]
- Morfino, R.C.; Bart, S.M.; Franklin, A.; et al. Notes from the Field: Aircraft wastewater surveillance for early detection of SARS-CoV-2 variants—John, F. Kennedy International Airport, New York City, August–September 2022. MMWR Morb Mortal Wkly Rep 2023, 72, 210–211. [Google Scholar] [CrossRef]
- Hjelmso, M.H.; Mollerup, S.; Jensen, R.H.; Pietroni, C.; Lukjancenko, O.; Schultz, A.C.; Aarestrup, F.M.; Hansen, A.J. Metagenomic analysis of viruses in toilet waste from long distance flights-A new procedure for global infectious disease surveillance. PLoS ONE 2019, 14, e0210368. [Google Scholar] [CrossRef]
- Albastaki, A.; Naji, M.; Lootah, R.; et al. First confirmed detection of SARS-COV-2 in untreated municipal and aircraft wastewater in Dubai, UAE: The use of wastewater-based epidemiology as an early warning tool to monitor the prevalence of COVID-19. Sci Total Environ 2021, 760, 143350. [Google Scholar] [CrossRef]
- Le Targa, L.; Wurtz, N.; Lacoste, A.; et al. SARS-CoV-2 testing of aircraft wastewater shows that mandatory tests and vaccination pass before boarding did not prevent massive importation of omicron variant into Europe. Viruses 2022, 14, 1511. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, W.; Bivins, A.; Smith, W.J.M.; et al. Detection of the Omicron (B.1.1.529) variant of SARS-CoV-2 in aircraft wastewater. Sci Total Environ 2022, 820, 153171. [Google Scholar] [CrossRef]
- Ahmed, W.; Bertsch, P.M.; Bivins, A.; et al. Comparison of virus concentration methods for the RT-qPCR-based recovery of murine hepatitis virus, a surrogate for SARS-CoV-2 from untreated wastewater. Sci Total Environ 2020, 739, 139960. [Google Scholar] [CrossRef]
- Ahmed, W.; Bivins, A.; Korajkic, A.; Metcalfe, S.; Smith, W.J.; Simpson, S.L. Comparative analysis of Adsorption-Extraction (AE) and Nanotrap® Magnetic Virus Particles (NMVP) workflows for the recovery of endogenous enveloped and non-enveloped viruses in wastewater. Sci Total Environ 2023, 859, 160072. [Google Scholar] [CrossRef]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular cloning: a laboratory manual COLD Spring Harbor. NY Cold Spring Harbor Laboratory press (1989).
- Haugland, R.A.; Siefring, S.C.; Wymer, L.J.; Brenner, K.P.; Dufour, A.P. Comparison of Enterococcus measurements in freshwater at two recreational beaches by quantitative polymerase chain reaction and membrane filter culture analysis. Water Res 2005, 39, 559–568. [Google Scholar] [CrossRef]
- Besselsen, D.G.; Wagner, A.M.; Loganbill, J.K. Detection of rodent coronaviruses by use of fluorogenic reverse transcriptase-polymerase chain reaction analysis. Comp Med 2002, 52, 111–116. [Google Scholar] [PubMed]
- Staley, C.; Gordon, K.V.; Schoen, M.E.; Harwood, V.J. Performance of two quantitative PCR methods for microbial source tracking of human sewage and implications for microbial risk assessment in recreational waters. Appl Environ Microbiol 2012, 78, 7317–7326. [Google Scholar] [CrossRef] [PubMed]
- Heim, A.; Ebnet, C.; Harste, G.; Pring-Akerblom, P. Rapid and quantitative detection of human adenovirus DNA by real-time PCR. J Med Virol 2003, 70, 228–239. [Google Scholar] [CrossRef] [PubMed]
- Jothikumar, N.; Lowther, J.A.; Henshilwood, K.; et al. Rapid and sensitive detection of noroviruses by using TaqMan-based one-step reverse transcriptio-PCR assays and application to naturally contaminated shellfish samples. Appl Environ Microbiol 2005, 71, 1870–1875. [Google Scholar] [CrossRef]
- Costafreda, M.I.; Bosch, A.; Pinto, R.M. Development, evaluation, and standardization of a real-time TaqMan reverse Transcription-PCR assay for quantification of hepatitis A virus in clinical and shellfish samples. Appl Environ Microbiol 2006, 72, 3846–3855. [Google Scholar] [CrossRef] [PubMed]
- McQuaig, S.M.; Scott, T.M.; Lukasik, J.O.; Paul, J.H.; Harwood, V.J. Quantification of human polyomaviruses JC Virus and BK Virus by TaqMan quantitative PCR and comparison to other water quality indicators in water and fecal samples. Appl Environ Microbiol 2009, 75, 3379–3388. [Google Scholar] [CrossRef] [PubMed]
- Cashdollar, J.L.; Brinkman, N.E.; Griffin, S.M.; McMinn, B.R.; Rhodes, E.R.; Varughese, E.A.; Grimm, A.C.; Parshionikar, S.U.; Wymer, L.; Fout, G.S. Development and evaluation of EPA method 1615 for detection of enterovirus and norovirus in water. Appl Environ Microbiol 2013, 79, 215–223. [Google Scholar] [CrossRef] [PubMed]
- Stachler, E.; Kelty, C.; Sivaganesan, M.; Li, X.; Bibby, K.; Shanks, O.C. Quantitative CrAssphage PCR Assays for human fecal pollution measurement. Environ Sci Technol 2017, 51, 9146–9154. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, W.; Bivins, A.; Stephens, M.; Metcalfe, S.; Smith, W.J.; Sirikanchana, K.; Kitajima, M.; Simpson, S.L. Occurrence of multiple respiratory viruses in wastewater in Queensland, Australia: Potential for community disease surveillance. Sci Total Environ 2023, 864, 161023. [Google Scholar] [CrossRef]
- Ahmed, W.; Payyappat, S.; Cassidy, M.; Harrison, N.; Besley, C. Microbial source tracking of untreated human wastewater and animal scats in urbanized estuarine waters. Sci. Total Environ 2023, 877, 162764. [Google Scholar] [CrossRef]
- Verbyla, M.E.; Symonds, E.M.; Kafle, R.C.; Cairns, M.R.; Iriarte, M.; Guzmán, A.M.; Coronado, O.; Breitbart, M.; Ledo, C.; Mihelcic, J.R. Managing microbial risks from indirect wastewater reuse for irrigation in urbanizing watersheds. Environ Sci Technol 2016, 50, 6803–6813. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin Chem 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Ciannella, S.; González-Fernández, C.; Gomez-Pastora, J. Recent progress on wastewater-based epidemiology for COVID-19 surveillance: A systematic review of analytical procedures and epidemiological modeling. Sci Total Environ 2023, 878, 162953. [Google Scholar] [CrossRef]
- Shah, S.; Gwee, S.X.W.; Ng, J.Q.X.; Lau, N.; Koh, J.; Pang, J. Wastewater surveillance to infer COVID-19 transmission: A systematic review. Sci Total Environ 2022, 804, 150060. [Google Scholar] [CrossRef]
- Servetas, S.L.; Parratt, K.H.; Brinkman, N.E.; et al. Standards to support an enduring capability in wastewater surveillance for public health: Where are we? Case Stud Chem Environ Eng. 2002, 6, 100247. [Google Scholar] [CrossRef]
- Rachmadi, A.T.; Torrey, J.R.; Kitajima, M. Human polyomavirus: Advantages and limitations as a human-specific viral marker in aquatic environments. Water Res. 2016, 105, 456–469. [Google Scholar] [CrossRef]
- Stachler, E.; Akyon, B.; de Carvalho, N.A.; Ference, C.; Bibby, K. Correlation of crAssphage qPCR markers with culturable and molecular indicators of human fecal pollution in an impacted urban watershed. Enviro Sci Technol 2018, 52, 7505–7512. [Google Scholar] [CrossRef]
- Wilder, M.L.; Middleton, F.; Larsen, D.A.; et al. Co-quantification of crAssphage increases confidence in wastewater-based epidemiology for SARS-CoV-2 in low prevalence areas. Water Res. X 2021, 11, 100100. [Google Scholar] [CrossRef] [PubMed]
- Greenwald, H.D.; Kennedy, L.C.; Hinkle, A.; et al. Tools for interpretation of wastewater SARS-CoV-2 temporal and spatial trends demonstrated with data collected in the San Francisco Bay Area. Water Res. X 2021, 12, 100111. [Google Scholar] [CrossRef] [PubMed]
- Holm, R.H.; Nagarkar, M.; Yeager, R.A. Surveillance of RNase P, PMMoV, and CrAssphage in wastewater as indicators of human fecal concentration across urban sewer neighborhoods, Kentucky. FEMS Microbes 2022, 3, xtac003. [Google Scholar] [CrossRef]
- Crank, K.; Li, X.; North, D.; Ferraro, G.B.; Iaconelli, M.; Mancini, P.; La Rosa, G.; Bibby, K. CrAssphage abundance and correlation with molecular viral markers in Italian wastewater. Water Res. 2020, 184, 116161. [Google Scholar] [CrossRef] [PubMed]
- Bofill-Mas, S.; Albinana-Gimenez, N.; Clemente-Casares, P.; Hundesa, A.; Rodriguez-Manzano, J.; Allard, A.; Calvo, M.; Girones, R. Quantification and stability of human adenoviruses and polyomavirus JCPyV in wastewater matrices. Appl Environ Microbiol 2006, 72, 7894–7896. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Kim, E.T.; Huh, J.-S. Virus in the urine of healthy people and patients with infectious diseases. Urogenit Tract Infect 2021, 16, 44–48. [Google Scholar] [CrossRef]
- Kazama, S.; Masago, Y.; Tohma, K.; Souma, N.; Imagawa, T.; Suzuki, A.; Liu, X.; Saito, M.; Oshitani, H. ; Omura, T Temporal dynamics of norovirus determined through monitoring of municipal wastewater by pyrosequencing and virological surveillance of gastroenteritis cases. Water Res 2016, 92, 244–253. [Google Scholar] [CrossRef] [PubMed]
- Farkas, K.; Marshall, M.; Cooper, D.; McDonald, J.E.; Malham, S.K.; Peters, D.E.; Maloney, J.D.; Jones, D.L. Seasonal and diurnal surveillance of treated and untreated wastewater for human enteric viruses. Environ Sci Pollut Res Int 2018, 25, 33391–33401. [Google Scholar] [CrossRef] [PubMed]
- Kilaru, P.; Hill, D.; Anderson, K.; et al. Wastewater Surveillance for Infectious Disease: A Systematic Revie. Am. J. Epidemiol. 2023, 192, 305–322. [Google Scholar] [CrossRef] [PubMed]
- Crank, K.; Chen, W.; Bivins, A.; et al. Contribution of SARS-CoV-2 RNA shedding routes to RNA loads in wastewater. Sci Total Environ 2022, 806, 150376. [Google Scholar] [CrossRef]
- Boehm, A.B.; Hughes, B.; Duong, D.; Chan-Herur, V.; Buchman, A.; Wolfe, M.K.; White, B.J. Wastewater concentrations of human influenza, metapneumovirus, parainfluenza, respiratory syncytial virus, rhinovirus, and seasonal coronavirus nucleic-acids during the COVID-19 pandemic: A surveillance study. Lancet Microbe 2023, 4, E340–E348. [Google Scholar] [CrossRef]
| Viruses | No. of positive/no. of samples tested (%) | ||
|---|---|---|---|
| AE | NMAP | HA and NMAP combined | |
| CrAssphage | 24/24 (100) | 24/24 (100) | 24/24 (100) |
| HPyV | 24/24 (100) | 24/24 (100) | 24/24 (100) |
| HAV | 0/24 (0) | 0/24 (0) | 0/24 (0) |
| EV | 9/24 (37.5) | 10/24 (41.7) | 14/24 (58.3) |
| HNoV GII | 1/24 (4.20) | 1/24 (4.20) | 1/24 (4.20) |
| HAdV | 13/24 (54.2) | 4/24 (16.7) | 13/24 (54.2) |
| BoV | 7/24 (29.2) | 2/24 (8.30) | 8/24 (33.3) |
| PeV | 2/24 (8.30) | 3/24 (12.5) | 4/24 (16.7) |
| RhV A | 24/24 (100) | 24/24 (100) | 24/24 (100) |
| RhV B | 24/24 (100) | 24/24 (100) | 24/24 (100) |
| EBV | 5/24 (20.8) | 4/24 (16.7) | 7/24 (29.2) |
| IAV | 2/24 (8.30) | 2/24 (8.30) | 4/24 (16.7) |
| IBV | 0/24 (0) | 0/24 (0) | 0/24 (0) |
| RsV A | 0/24 (0) | 0/24 (0) | 0/24 (0) |
| RsV B | 4/24 (16.7) | 6/24 (25.0) | 9/24 (37.5) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





