1. Introduction
Croplands are important not only for global food security, but also for water security in a world two-thirds of its population are estimated to experience severe water scarcity at least once a month [
1,
2]. According to the United Nations’ Food and Agriculture Organization (FAO) cropland is “land used for the cultivation of crops, both temporary (annuals) and permanent (perennials), and may include areas periodically left fallow or used as temporary pasture” [
3]. Cropland mapping and global agriculture monitoring are therefore very essential for food security, agricultural resources management, agricultural economics, vegetation studies, and land use/land cover mapping purposes [
4,
5,
6]. Croplands mapping and monitoring is a widely used application of remote sensing given its evident advantages of spatial coverage, cost effectiveness, monitoring ability for its revisiting frequency, and the ability to detect crops stress [
7,
8,
9,
10,
11,
12,
13].
Among vegetation indices, Normalized Difference Vegetation Index (NDVI) data is very popular remote sensing-based information for cropland coverage mapping and crops conditions assessment. NDVI is widely used for estimating the extent and vigor of vegetation on earth. NDVI data have proven its robustness for many cropland extent mapping [
14,
15], classification of agricultural lands [
16], land suitability analysis for crop cultivation [
17], crop type mapping [
18], assessment of leaf area index (LAI) and vegetation fraction identification [
19], to mention a few.
NDVI is typically estimated using the percentage surface reflectance values of the red and near-infrared (NIR) bands acquired through remote sensing. NDVI is a unit-less index that ranges from -1 to +1, with values close to -1 indicating areas with no vegetation or water, values close to 0 indicating areas with sparse vegetation or bare soil, and values close to +1 indicating areas with dense and healthy vegetation.
The availability of remote sensing data from different satellite programs and the ease of access to such data have led to the availability of time series NDVI datasets at global, regional, and local scales. Some of satellite programs that make data freely available to the end users include MODIS, Landsat, and Sentinel, along with limited commercial satellite data with higher spatial and temporal resolutions from programs such as WorldView-3,-4 and Planet [
20].
The NDVI time series data however typically contain some errors due to atmosphere conditions, cloud cover, and sensor capacity. It is therefore very important to analyze, smooth and reconstruct NDVI data before the use for cropland mapping [
21,
22,
23]. The common NDVI data reconstruction techniques for local and regional studies are spatial and based on proximity analysis. These methods consider the correlation between pixels and their neighborhood in order to restore the missing values. Some of the common spatial methods are based on linear, bilinear and kriging interpolation. Some of the popular methods for reconstructing NDVI time series data are temporal- or spatiotemporal- based, which can generally be categorized into temporal interpolation, temporal filtering, temporal function-fitting, temporal deep learning, frequency-based methods, and hybrid techniques [
24,
25,
26].
Given their high computational speed and good classification results, many machine learning and AI techniques have been used with remote sensing data for mapping and monitoring of cropland. Some of the popular methods include random forest, neural networks, and support vector machines [
27,
28,
29,
30]. These algorithms typically learn about the target class characteristics from training dataset and identify the class in the input dataset. Some of the recent literature on this topic includes a study by Ketchum et al. (2020) in which a method for large-scale mapping of irrigated agricultural lands in the western U.S. using random forest machine learning was developed [
31]. Chao et al. (2022) have mapped croplands in China with a machine learning classifier on Google Earth Engine [
32].
Conducted as part of the International Telecommunication Union (ITU) competition, the main objective of this study was to develop a robust, cost-effective, machine learning and AI based method for cropland mapping in four study areas; Iran, Mozambique, Sri Lanka, and Sudan. The competition tasks included training dataset creation, NDVI data time series reconstruction, and cropland extent mapping as well as crop intensity mapping. Our method achieved the highest accuracy in cropland extent (98%) using Bidirectional LSTM on the Iran study region and (95%) using Random Forest on the Sri-Lanka study area. The contributions of this paper are summarized with the following points:
Provide a detailed methodology of data points collection of Sentinel-2 satellite assets though google earth engine for cropland extent and intensity applications for machine learning.
perform NDVI time-series reconstruction on the data-points collected to patch gaps and errors within the series using Savitsky-Golay filter and linear interpolation technique.
Developing an adaptive threshold approach for crop intensity cycles detection based on the obtained NDVI time-series.
Apply different machine learning techniques on the dataset at hand to generate cropland extent and intensity maps.
Compare between context aware to regular machine learning techniques in terms of efficiency and speed.
This paper will go as follows; The materials and methods section discusses the study areas and dataset at hand along with the samples collection process, time-series reconstruction methodology, and machine learning algorithms used. Later, the results and discussion section illustrates the obtained results with a comparison between different machine learning models. The conclusions provides an overall summary of the work discussed and a glance on the possible future work in this field.
2. Materials and Methods
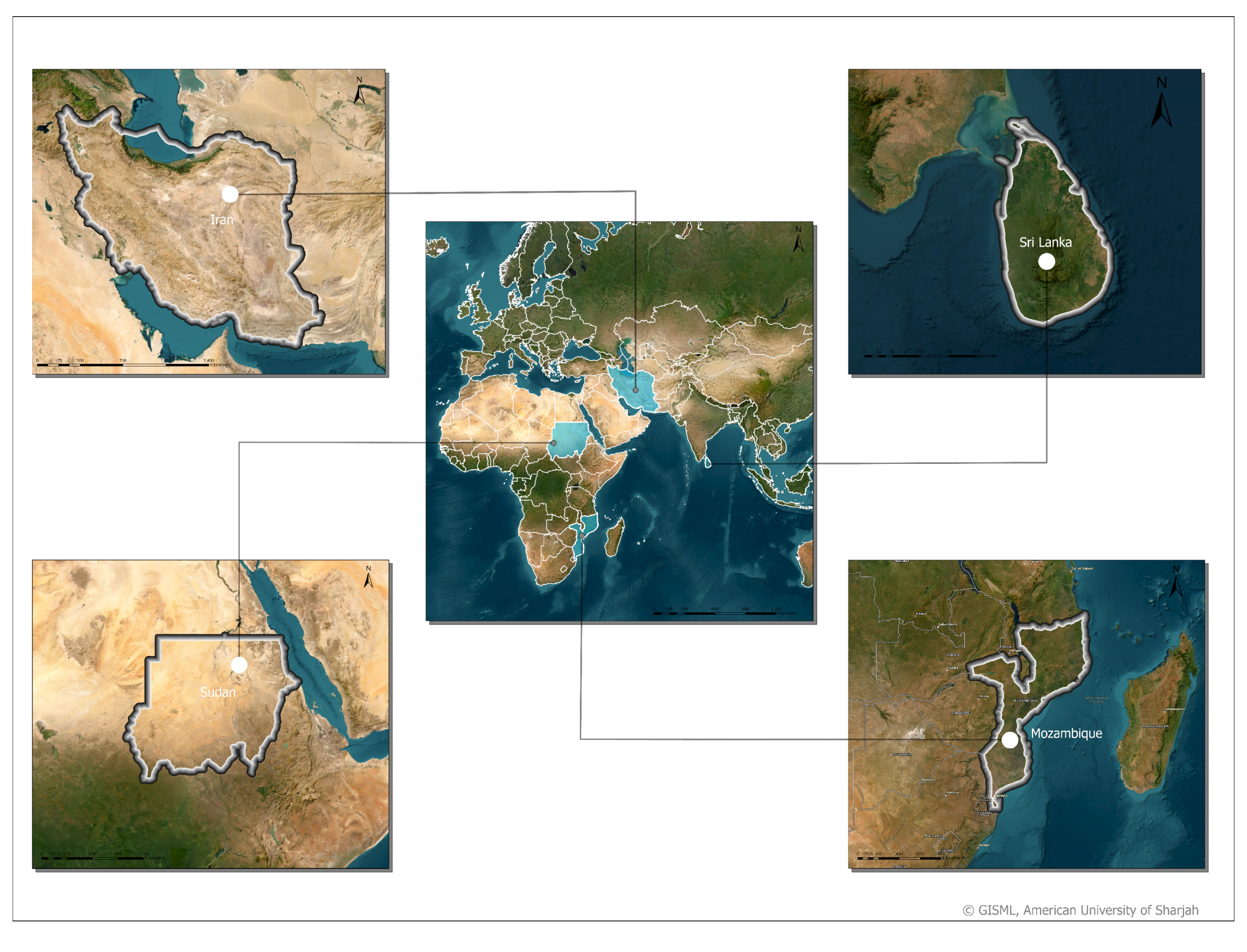
2.1. Study Area and Dataset
The study region of this paper covers four different countries: Iran, Sudan, Mozambique, and Sri Lanka. The regions covered are fairly diverse in land cover types, including grassland, forests, water, bare soil, and asphalt, but most importantly, they include ample agricultural land. The four regions differ in climate, with the region in Iran being of a semi-arid climate, the region in Sudan being of a hot desert climate, and lastly, the region of both Mozambique and Sri Lanka having a tropical climate. Consequently, the cropland captured in the study area’s collected data is diverse in the type of crops included.
The datasets used in this paper consist of Normalized Difference Vegetation Index (NDVI) time series data of 10-meter spatial resolution collected from a 15-day Sentinel-2 composite, which covers a region of size 0.5 degrees by 0.5 degrees for each of the four studied countries. NDVI is a remote sensing index used to assess vegetation health and quantity [
33,
34]. NDVI also helps differentiate vegetated land from other land-cover types in imagery, as it tends to be of a positive value for pixels related to vegetated land and zero or of a negative value for pixels pertaining to other types of land-cover, such as water [
35]. Hence, it was used in this work in order to perform both cropland and crop intensity mapping. NDVI relies on the red band and the Near-infrared band and is calculated as follows where
NIR and
red are the surface reflectance values of the Near-infrared band and the Red band, respectively.:
The geo-spatial assets were provided by the international AI for good ITU Cropland mapping competition organizers and were used to generate data points using Google earth engine by applying two different data collection approaches based on the problem requirement. For the problem of cropland extent, we collected 75 geometric points of cropland and 75 points of non-cropland using eye inspection for each of the four study regions. The geometric points were then appointed a binary label according to their class (cropland, non-cropland). As for the problem of crop intensity mapping, we have used geometric polygons of water surfaces, cropland, and non-cropland areas to form our datasets. The polygons allowed us to collect a large number of data points, ensuring that we capture data pertaining to all crop intensity classes, of which we only used 200 points. These data points were then appointed a label of zero crop cycles, one crop cycle, two crop cycles, or three crop cycles through the crop cycle counting algorithm described later in this paper. After appointing the labels, we dealt with any imbalances found in the datasets caused by under-representation of certain classes by oversampling these underrepresented classes. It is worth mentioning that the size of the datasets was restricted to a maximum of 500 training data points per dataset as per competition rules, with smaller datasets being preferred, hence the small size of the collected datasets for this paper. Each of these data-points represented a pixel-wise NDVI value recorded every two weeks which resulted in a time-series of 24 points in total.
2.2. Sentinel-2 Data
Sentinel-2 data was used as input for our cropland extent and intensity models. The European Space Agency oversaw the development of the Sentinel-2 which is a constellation of two Earth observation satellites, as a part of the Copernicus Earth observation program of the European Commission. Sentinel-2 satellites’ wide-swath, multi-spectral imaging capabilities offer an unprecedented perspective of the Earth, encompassing all of its landmasses, sizable islands, and waterways. Applications in forestry, agriculture, and other land management fields benefit greatly from Sentinel-2 data. For instance, it can be used to map forest cover and soils, investigate leaf area together with chlorophyll and water content, and monitor inland waterways and coastal regions [
36]. Two identical satellites make up the Sentinel-2 mission: Sentinel-2A, which was launched on June 23, 2015, and Sentinel-2B, which was launched in 2017. The constellation may revisit each location on the surface of the Planet once every five days with both satellites launched. Each satellite is equipped with a Multi-Spectral Instrument (MSI), which creates photographs of the Earth with a resolution of ten meters per pixel, a field of view of 290 km, and thirteen bands spanning the visible and infrared spectrum [
36].
2.3. Data Pre-Processing
This section details the dataset pre-processing and preparation procedure followed in this paper. The
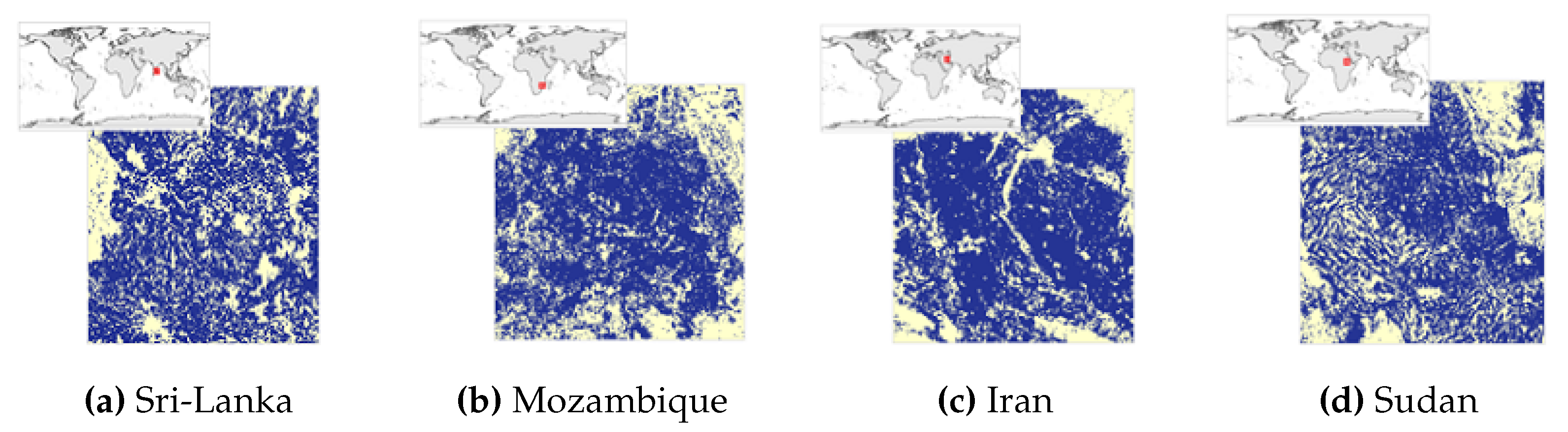
Figure 1 shows the study regions we are working on.
2.3.1. Cropland Extent Samples
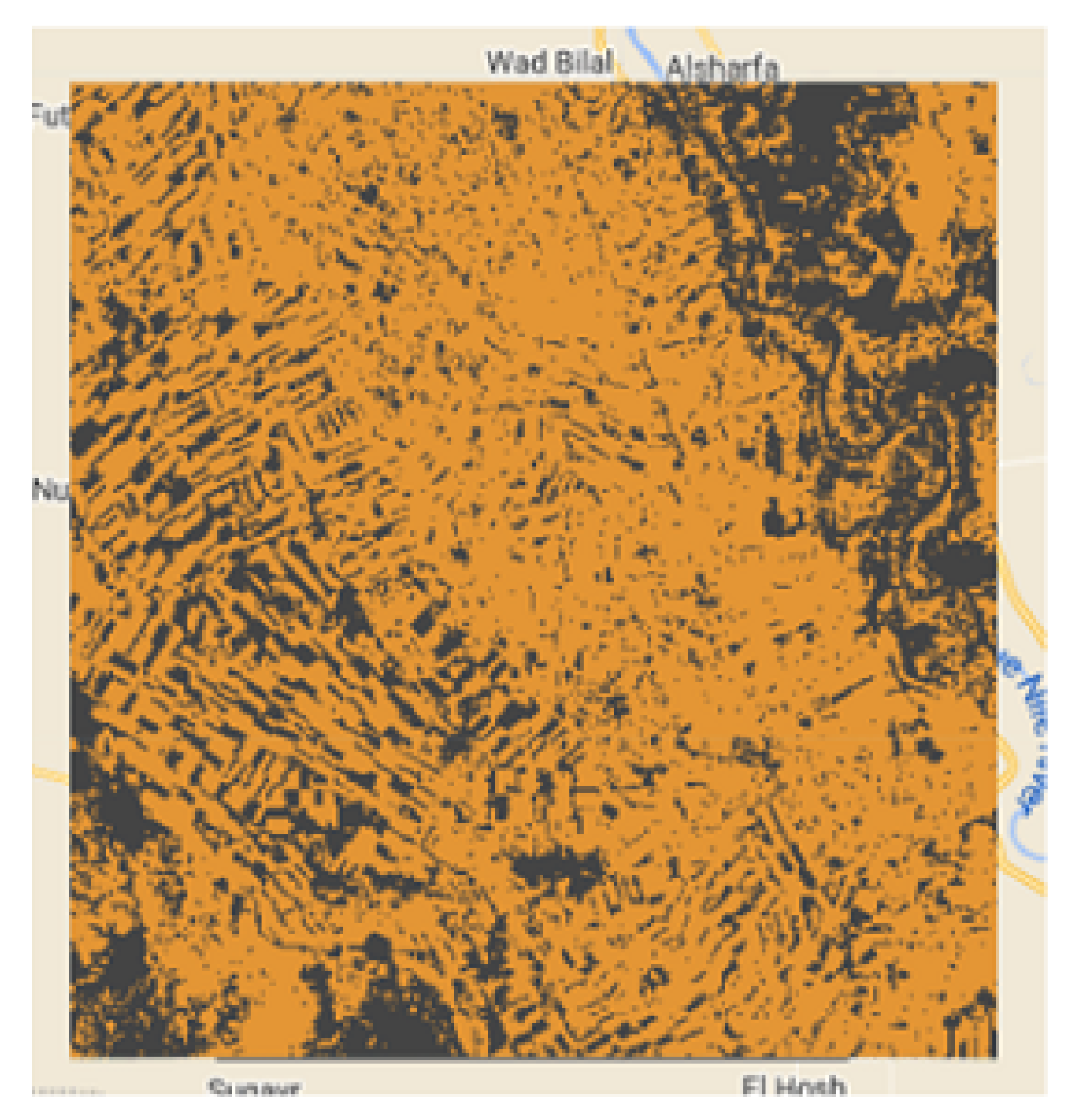
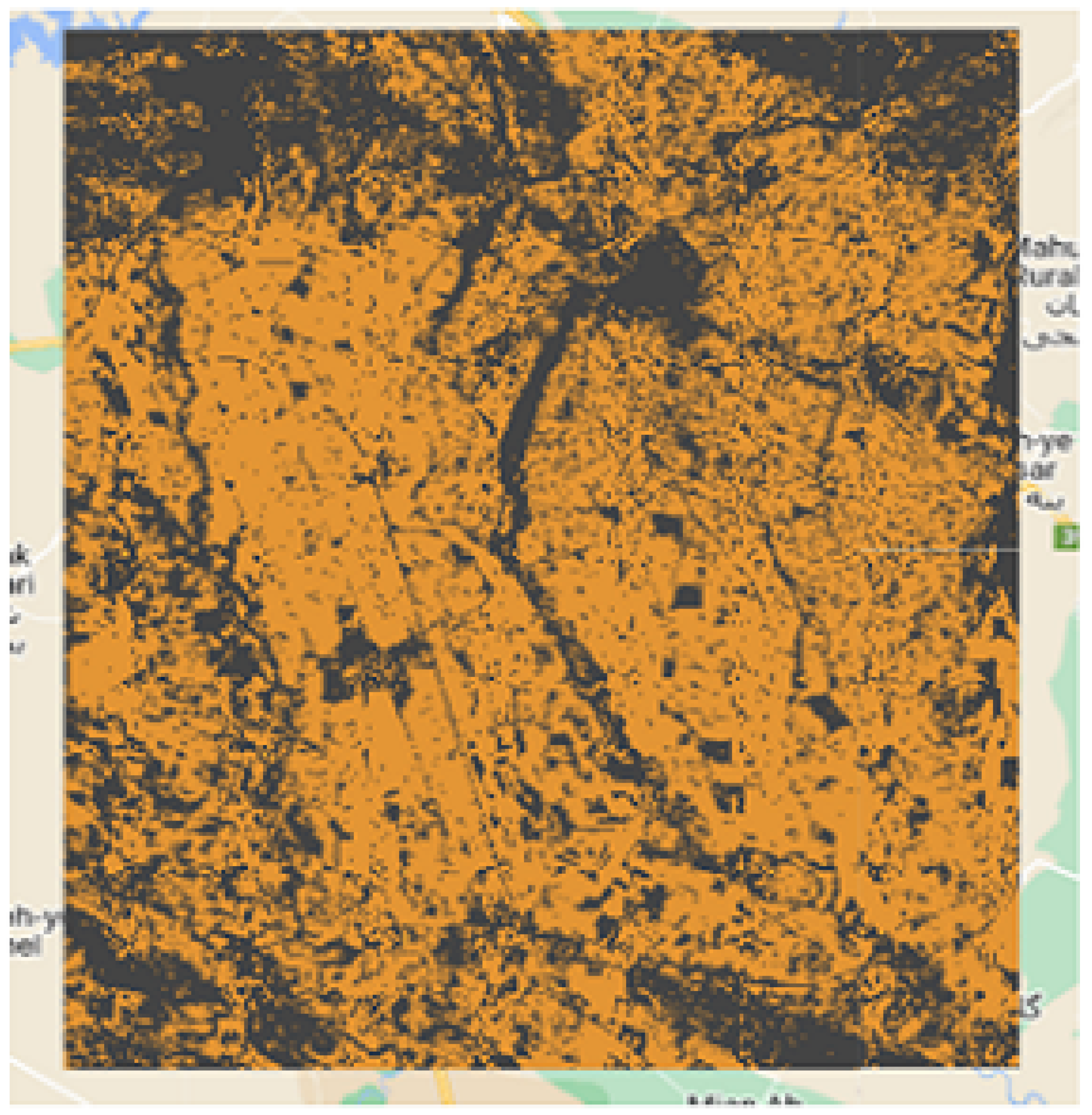
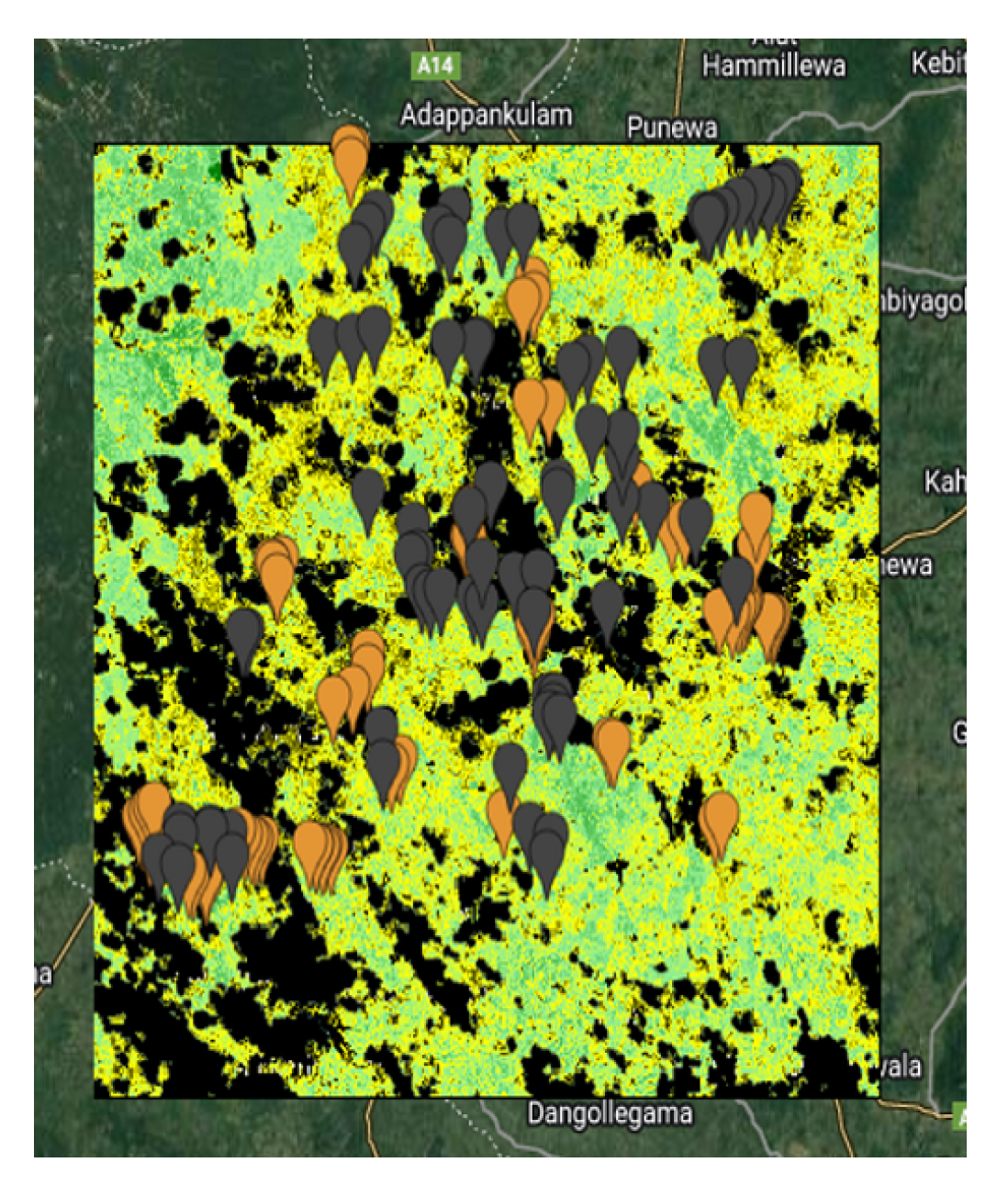
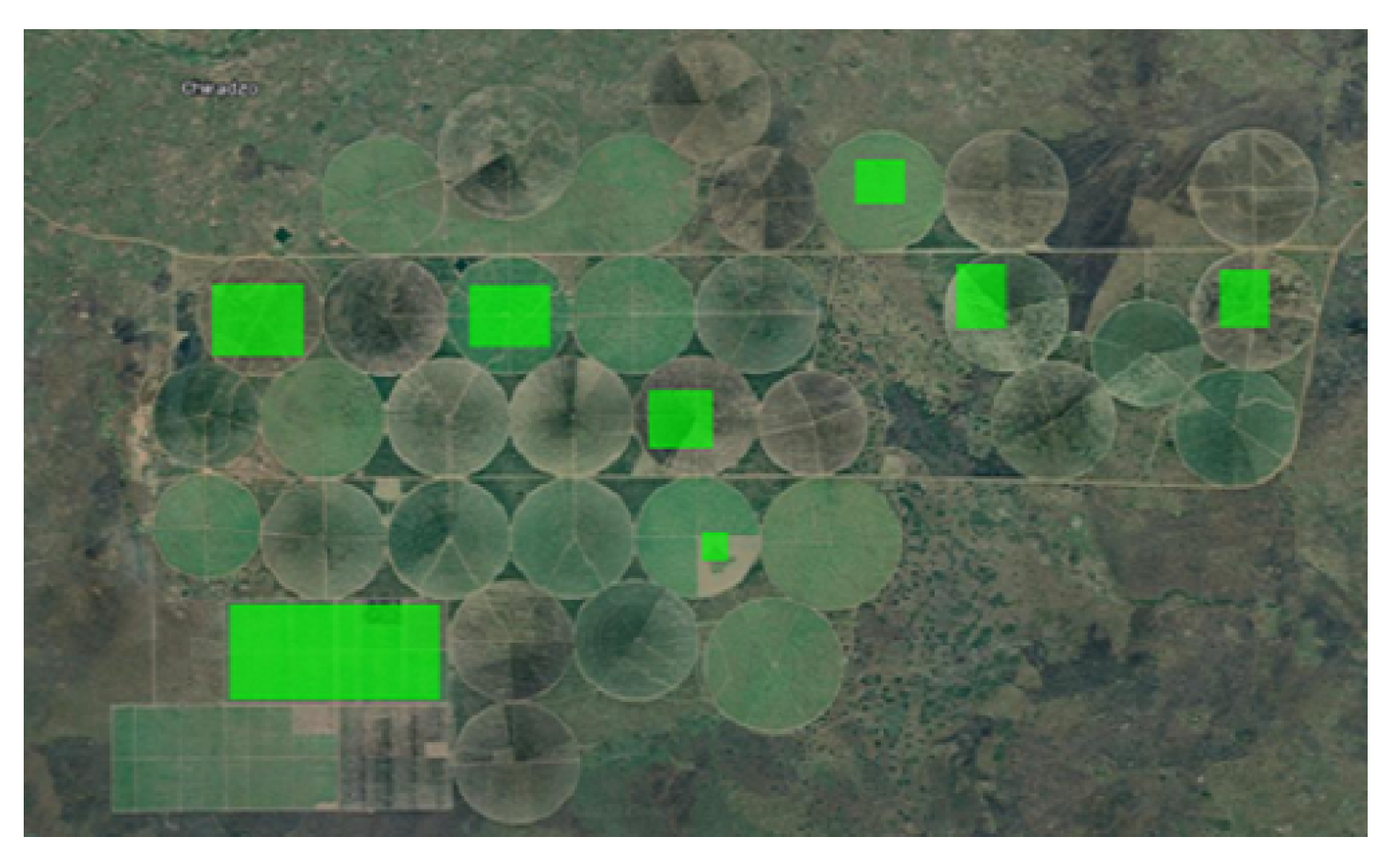
The
Figure 2,
Figure 3,
Figure 4 and
Figure 5 show the 4 study areas data collection for the cropland extent problem with 150 samples per country. Those 150 samples were divided into 75 cropland and 75 non cropland. The shape that contains different colors are the NDVI values for each pixel in that region of interest for a specific period in time.
2.3.2. Crop Intensity Samples
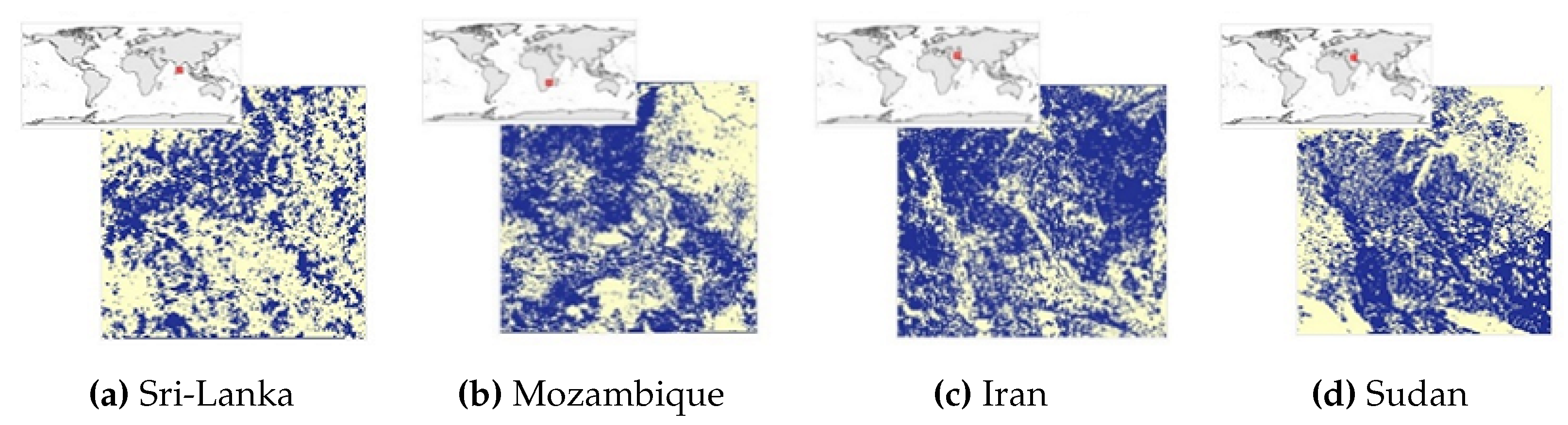
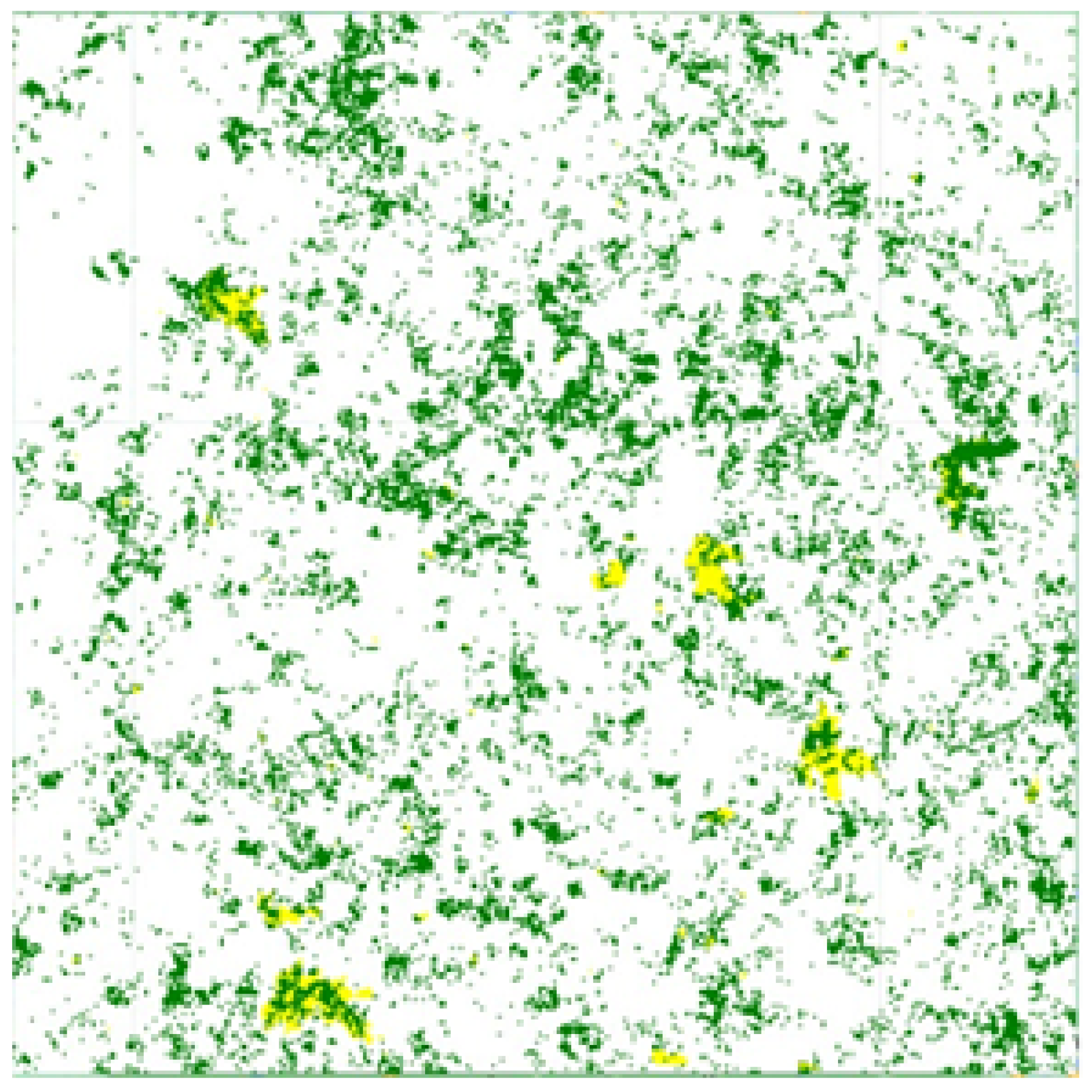
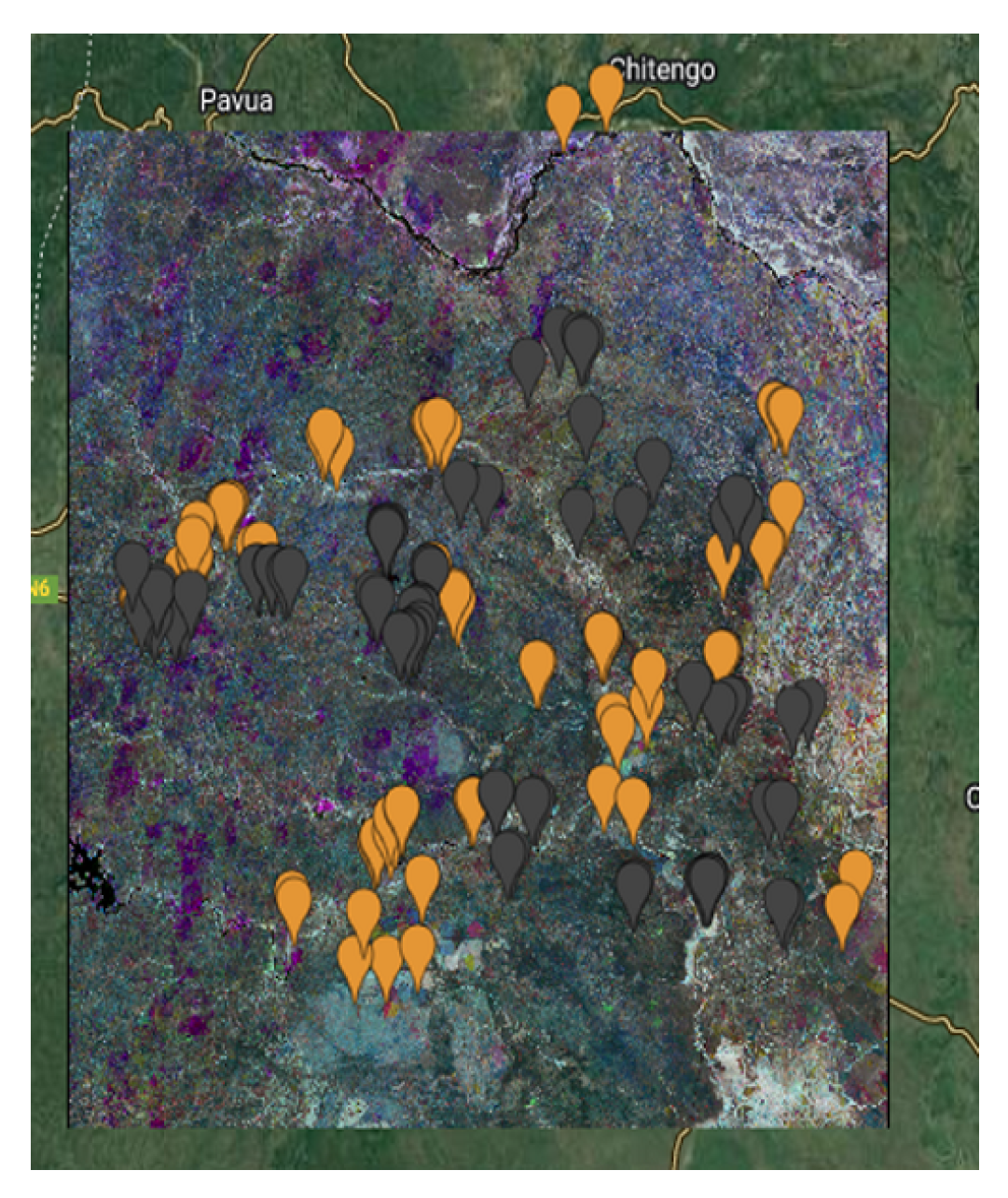
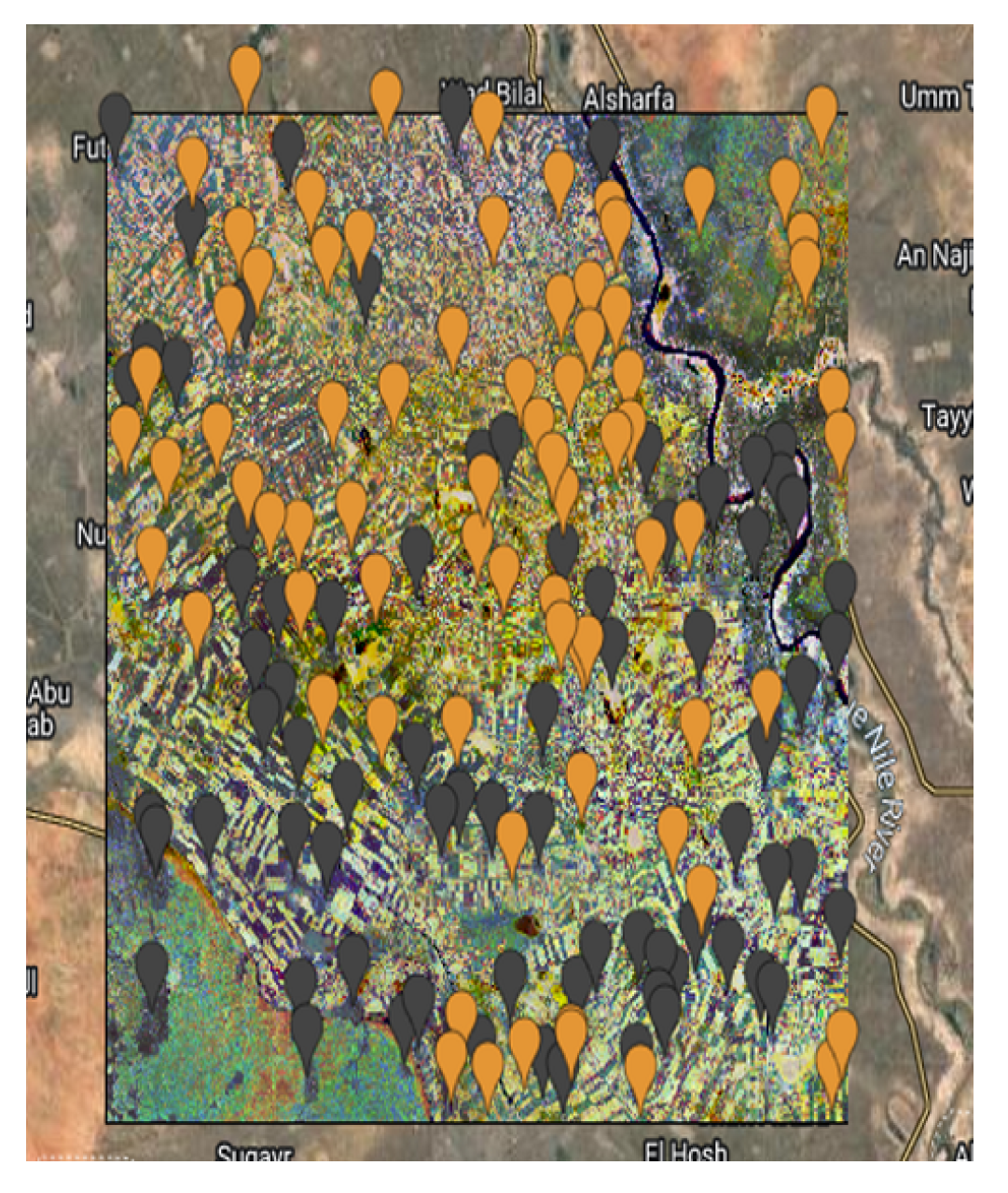
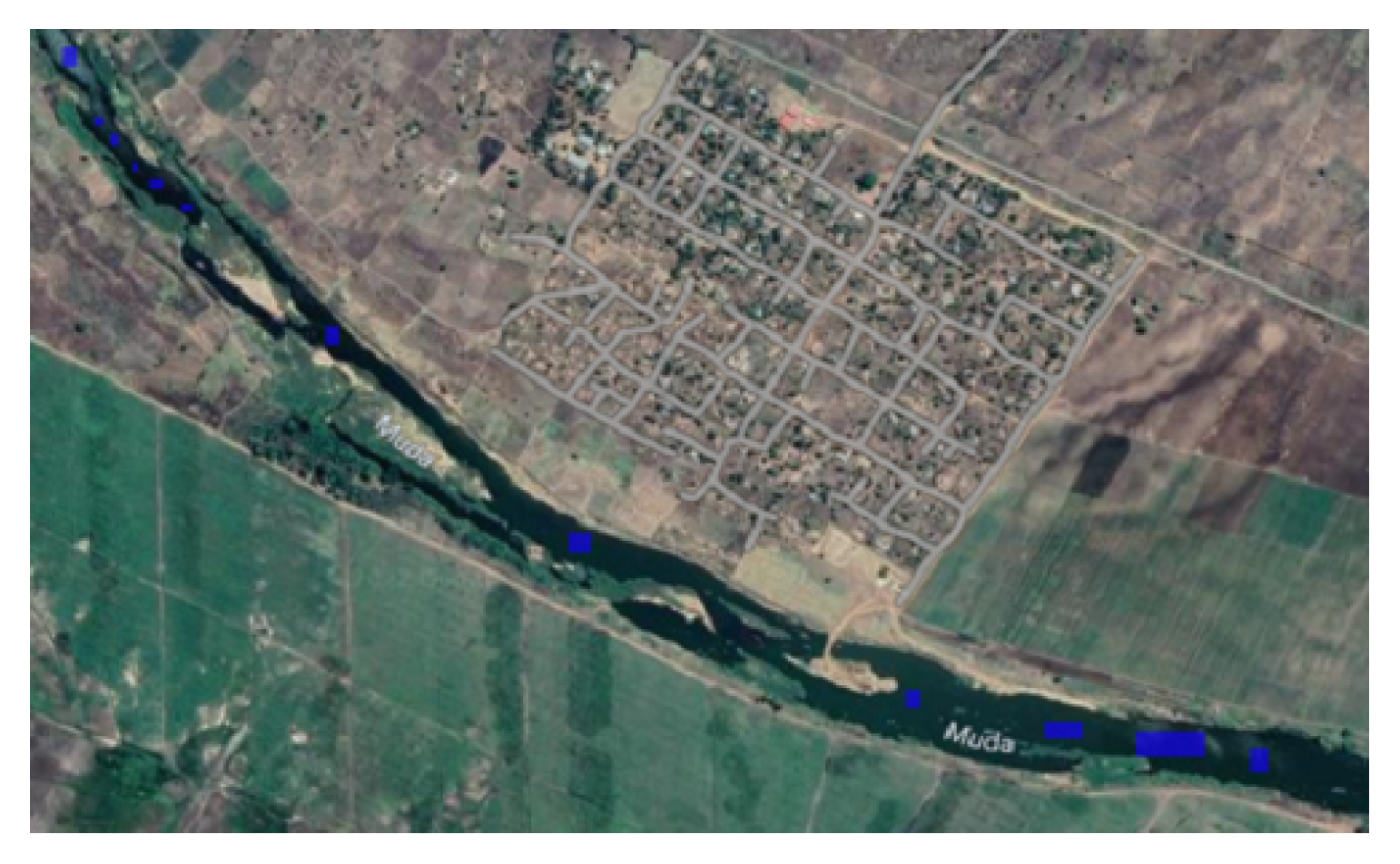
The
Figure 6,
Figure 7,
Figure 8 and
Figure 9 show the Mozambique data collection for the cropland intensity problem. This was done for the four study areas. The samples were taken as polygons in order to collect as many samples as possible to cover the three classes for crops. Polygons were taken for water samples and non cropland samples to make it random when taking the sample points.
2.3.3. Cropland Extent and Intensity Samples Taken Procedure
The dataset used for the two problems of cropland extent and intensity mapping was collected from a gap-filled NDVI time series provided by the competition organizers. Since training classifiers on an incomplete time-series negatively impacts their accuracy, it was necessary to first reconstruct the dataset’s time series data, filling in the gaps where data was missing, before proceeding to use the dataset to train any classifier. Hence, to fill in the gaps, iterative interpolation for data reconstruction (IDR) and was used and missing data points were simply replaced with the average value of their adjacent points where sudden and drastic changes in NDVI values were noted. Further, linear interpolation was also applied when an NDVI value difference greater than 0.4 -around half of the greatest NDVI value found in the collected dataset - was noted between two adjacent points where the second point was replaced to achieve a more natural rate of change in the NDVI scores of the time series data. This method was selected as the gap-filling procedure amongst other more complex options such as Fourier-based approach (Fourier), the double logistic model (DL), the the Whittaker smoother (Whit), and the locally adjusted cubic spline capping approach (LACC) [
37] given that the missing NDVI values were not expected to drastically deviate from their neighboring values, that is, following a typical vegetation growth cycle, and that such a replacement was not going to affect the mapping for which the classifiers are training. The time-series was further cleaned by applying a quadratic-polynomial-based Savitsky-Golay filter to the collected time series data, resulting in smoother NDVI time series curves.Given that the dataset’s time series curves contained 24 points only, the filter was configured to have a sliding window of 3 points so as to not cause significant loss in the time series’ curve characteristics.
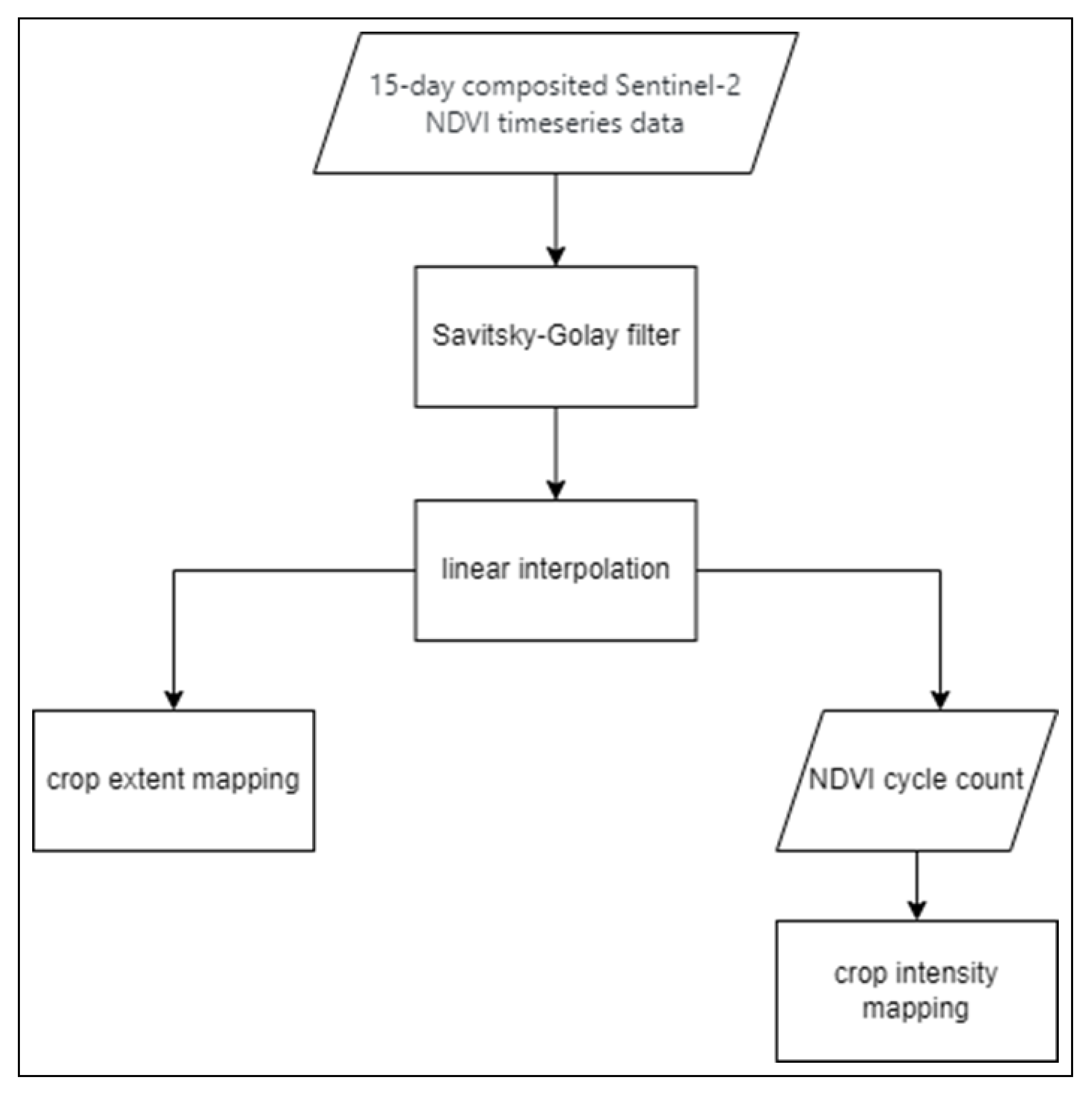
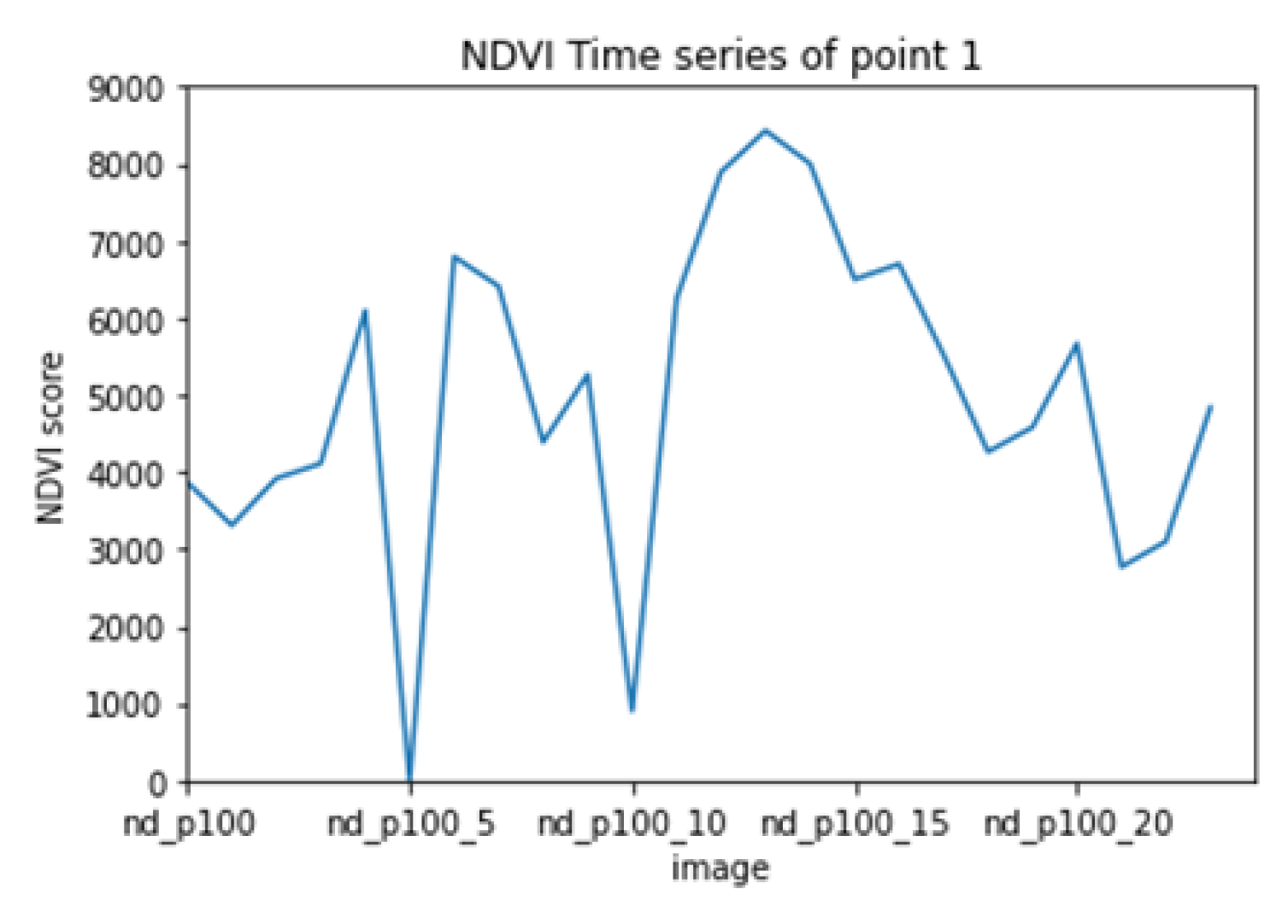
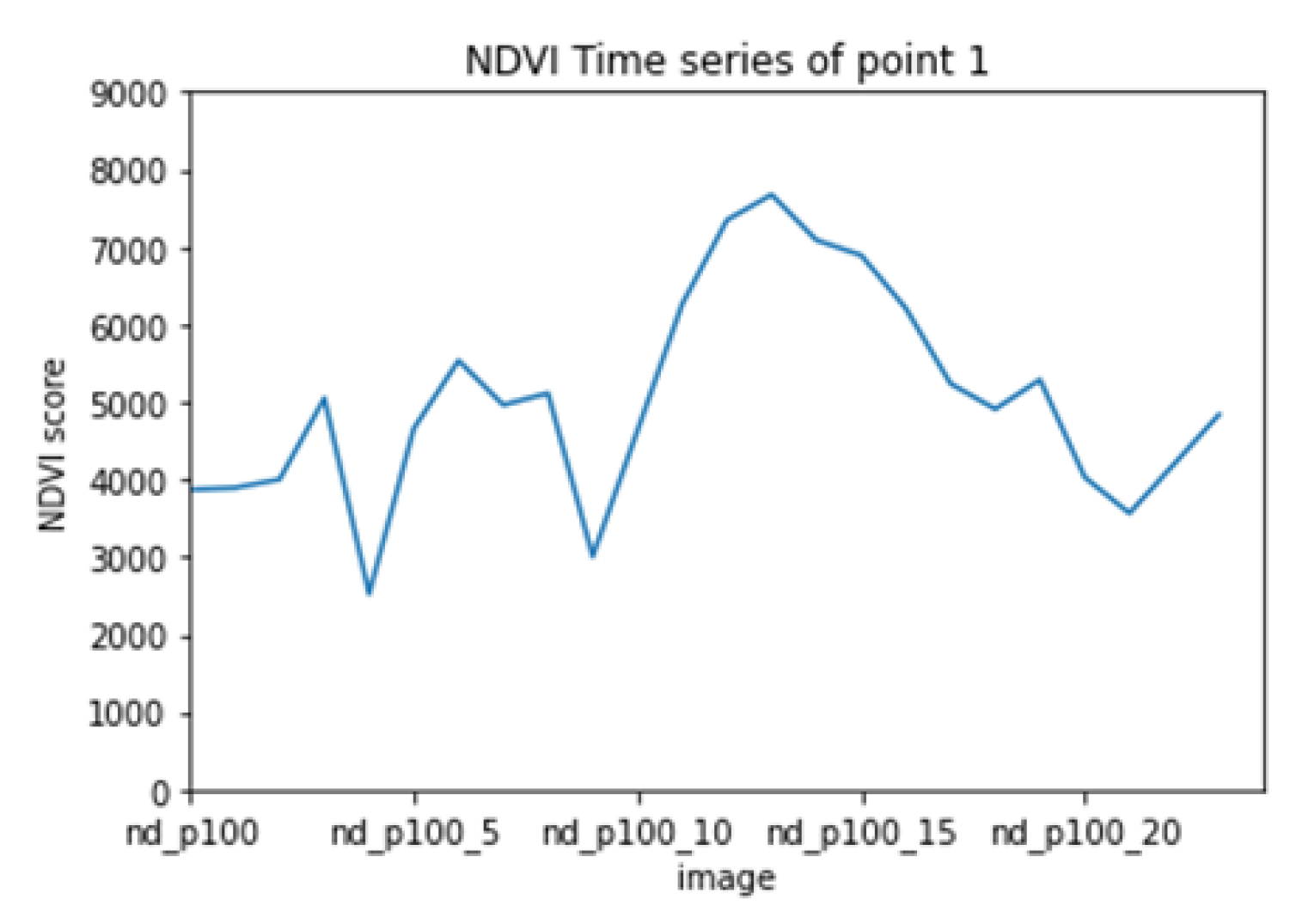
Figure 10 and
Figure 11 show a sample time series from the collected dataset before (
Figure 10) and after (
Figure 11) applying the detailed reconstruction method. the flowchart in
Figure 12 is depicting said procedure
2.4. Adaptive Threshold Approach for Crop Intensity Cycles Detection
While labelling for the cropland extent dataset was done through visual interpretation, crop intensity labels were not determined as easily. Crop intensity was instead determined through an algorithm developed for the purpose of crop cycle counting. The developed algorithm traversed each and every time series in the dataset and found the total number of peaks in the time series curves. Based on the number of peaks found, a time series was assigned a label: A class 0 label when no crop cycles were detected, a class 1 label when only one crop cycle was detected, a class 2 label when two crop cycles were detected, and lastly, a class 3 label when three crop cycles were detected.
The developed algorithm paid particular attention to eliminating spurious peaks from the crop cycle count. Spurious peaks mainly occur in time series data due to the growth of weeds in fields, which cause spikes in NDVI scores.These spikes are presented in NDVI time series curves as small peaks with values, much lower than that of cropland peaks which have NDVI scores typically above 0.5 [
38]. When these peaks are not dealt with, the crop cycle count in a given time series is overestimated and hence the resulting crop intensity estimate made is inaccurate. The algorithm first traverses a given time series and finds the maximum NDVI score. If the highest NDVI score in the time series is found to be less than 0.3, the time series is immediately given the class 0 label, meaning it does not contain any crop cycle. This is because NDVI timeseries of such low NDVI scores cannot correspond with actively used cropland, which typically reflect higher NDVI scores, instead they are more likely to pertain to barren land, rocks, and water or sparse vegetation. Otherwise, two thresholds are created based on this value: a minimum value for thresholding peaks and a maximum value for valleys. These thresholds are used to find the number of crop cycles in a given time series while making sure that spurious peaks caused by weeds are not included in the count. In previous literature relating to crop intensity mapping, spurious peaks were removed from the crop cycle count by simply using a static threshold for the peak values, where peaks that fell below a specific NDVI or EVI score were considered spurious [
38,
39,
40]. However, we have found that such an algorithm is insensitive to the fact that different types of crops differ in the NDVI scores they are capable of attaining during a crop cycle, assuming one threshold for all types of crops. This insensitivity can result in underestimating crop intensity. In order to deal with this problem, we have introduced the concept of adaptive thresholds, where peaks and valleys in a given time series are accepted based on thresholding values tailored to that time series. In our algorithm’s case, we have set the thresholding value for peaks in any time series to be 0.70 of the NDVI score of the absolute maxima in that time series, whereas the valley threshold was set to be 0.20 of that value. After finding the thresholds, the time series is traversed once more in order to find the number of crop cycles. A crop cycle is then considered to have occurred when a peak value that is encompassed between two valley values is found. In the special case that the peak value is found at an endpoint, a crop cycle is counted if and only if the peak is either lead or followed by a valley.
2.5. Classification Methods
There are primarily five machine learning algorithms deployed for the purpose of the binary classification of our sample points for the cropland extent problem as well as the categorical classification of our reconstructed NDVI time series for the crop intensity problem. The models used for this purpose are as follows:
2.5.1. Random Forest
Random forest is a classification algorithm that is built up of multiple decision tress. Firstly, n records at random are selected from a data set with k records. Next, each sample’s decision tree is built separately and consequently each decision tree produces an output. Finally, the model’s output is based on the Majority Voting ensemble that combines the results from all the individual decision trees [
41].
2.5.2. XGBoost Classifier
XGBoost is short for Extreme Gradient Boosting. It is a distributed gradient-boosted decision tree. The trees are boosted by parallel trees and is usually the leading machine learning algorithm for classification problems. Similar to random forest, XGBoost also employs decision trees as base learners. However, the trees used by XGBoost are CART (Classification and Regression trees) that contain real-value scores in each leaf node instead of a single decision [
42].
2.5.3. LSTM
LSTM is short for Long Short-Term Memory and are a special type of RNN (Recurrent Neural Networks) capable of handling the vanishing gradient issue faced by RNN. It can process the entire sequence of data and remember long term dependencies. LSTM has feedback and is one input flow which can either be backwards or forwards [
43]. Equations (
2) and (
3) describe the encoder and decoder of an LSTM. where
is the encoders hidden state at time step
t with input token embedding
. On the decoder side,
denotes the hidden state at time step
t with input embedding token
.
2.5.4. Bidirectional LSTM
Bidirectional LSTM works similar to LSTM but the main difference is that bidirectional can make input flow in both directions, backwards and forward since it consists of two LSTMs: one which takes the input in the forward direction and the other taking it in the backward direction. With the help of BiLSTMs, the network has access to more information, which improves the context available to the algorithm [
44].
2.5.5. KNN DTW
KNN is the K-nearest neighbors algorithm. It is well known for classification and works by finding the distances between a value and other examples in the data and then chooses the most frequent label for classification. DTW Stands for Dynamic Time Warping and is used in time series analysis. It measures the similarity between two temporal sequences.This approach is popular for time series classification given the algorithm’s speed and scalability. Since we are dealing with the crop intensity problem involving NDVI time series classification, KNN with DTW seemed to be the more appropriate choice of an algorithm for this specific problem [
45].
2.5.6. Computational Complexity
As we are looking into developing a light-weight solution to cropland mapping, accuracy is not the only metric that should be taken into consideration but also the computational complexity of the algorithms implemented. In general, traditional machine learning algorithms are faster than ensemble methods and recurrent neural networks. The complexity of Random forest is the lowest among tested algorithms with a training time complexity of
and a space complexity as small as
where
T is the number of trees,
n is the number of training samples,
m is the number of features,
d is the depth of tree and
k is the number of neighbours [
46]. On the other hand, KNN-DTW follows a slightly more expensive algorithm with a training time complexity of
and a space complexity of
[
47] while XGBoost follows a lower efficiency level of
[
48]. Finally the time complexity of LSTM and Bidirectional LSTM relies on the number of edges in the network
W with a time complexity of
and
respectively [
49]. In general,literature suggests that there is a trade off between accuracy and speed when it comes to generating cropland maps. traditional machine learning algorithms are not as computationally expensive as RNNs and thereby faster but at the cost of slightly less accurate results.
3. Results and Discussion
Our results showed that traditional machine learning algorithms such as Random Forest and XGBoost performed well across all four test regions for cropland extent. XGBoost model achieved the highest average accuracy (88.5%) overall outperforming more computationally expensive recurrent neural network models such as LSTM and Bidirectional LSTM. In general, the test region of Iran resulted in the best performance where Bi-LSTM achieved 98% accuracy. On the other hand, Mozambique’s test region was the most challenging as the highest accuracy achieved was 82% through both XGBoost and LSTM models. The results obtained for cropland intensity mapping problem were consistent with those of cropland extent mapping as XGBoost scored the highest average accuracy (87.3%) across all test regions and outperformed random forest model and KNN DTW model. Context aware models such as LSTM, Bidirectional LSTM and KNN DTW seemed to perform better than context unaware algorithms.
3.1. Maps Generated by Google Earth Engine as Assets
Figure 13.
Cropland extent resulted map.
Figure 13.
Cropland extent resulted map.
Figure 14.
Cropland intensity resulted map.
Figure 14.
Cropland intensity resulted map.
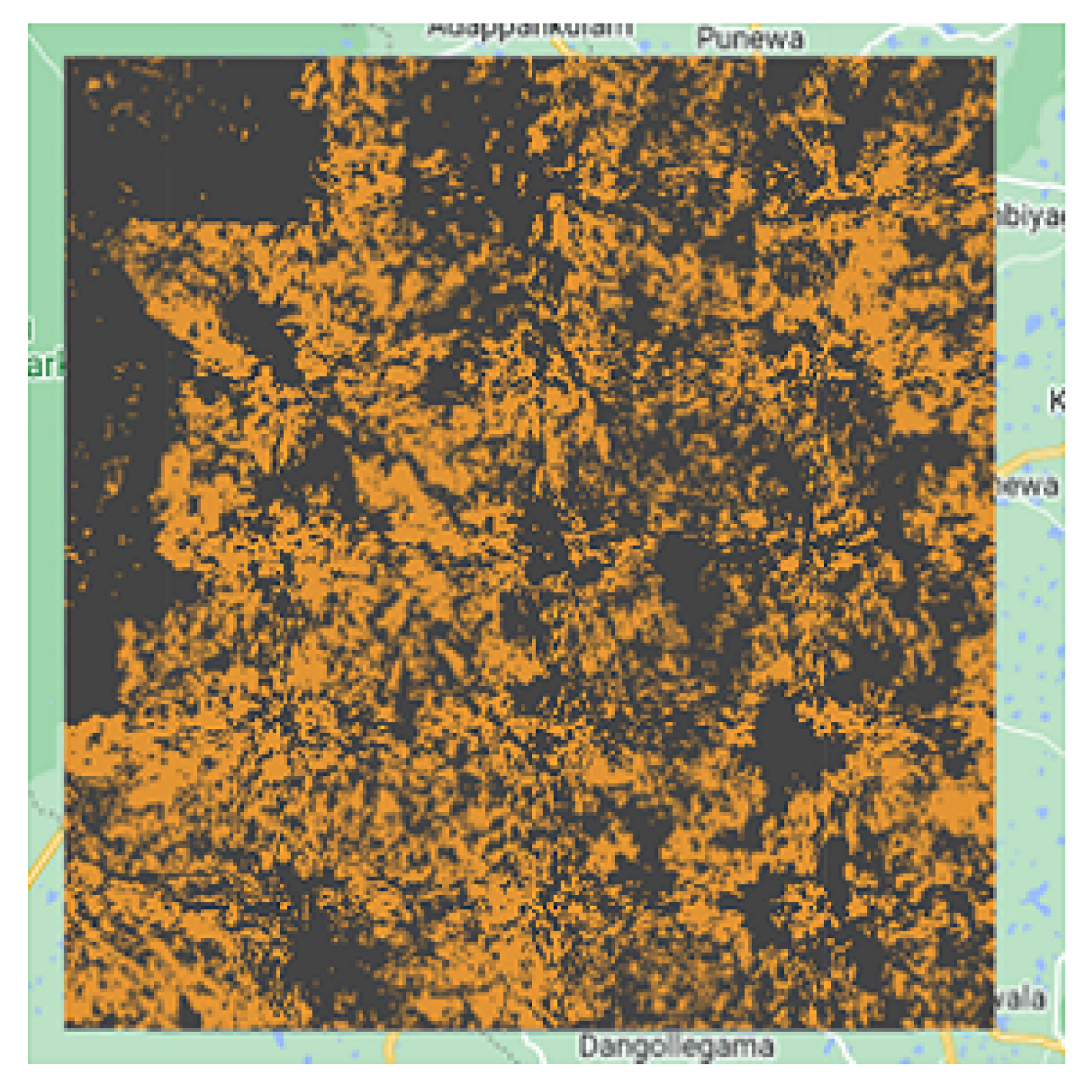
These results can be explained by the geographical nature of the four test regions. We believe that the presence of many forests in the region of Sri-Lanka (
Figure 15) reduced the model’s performance as the corresponding time series of forests samples were like those of cropland areas. The region of Mozambique (
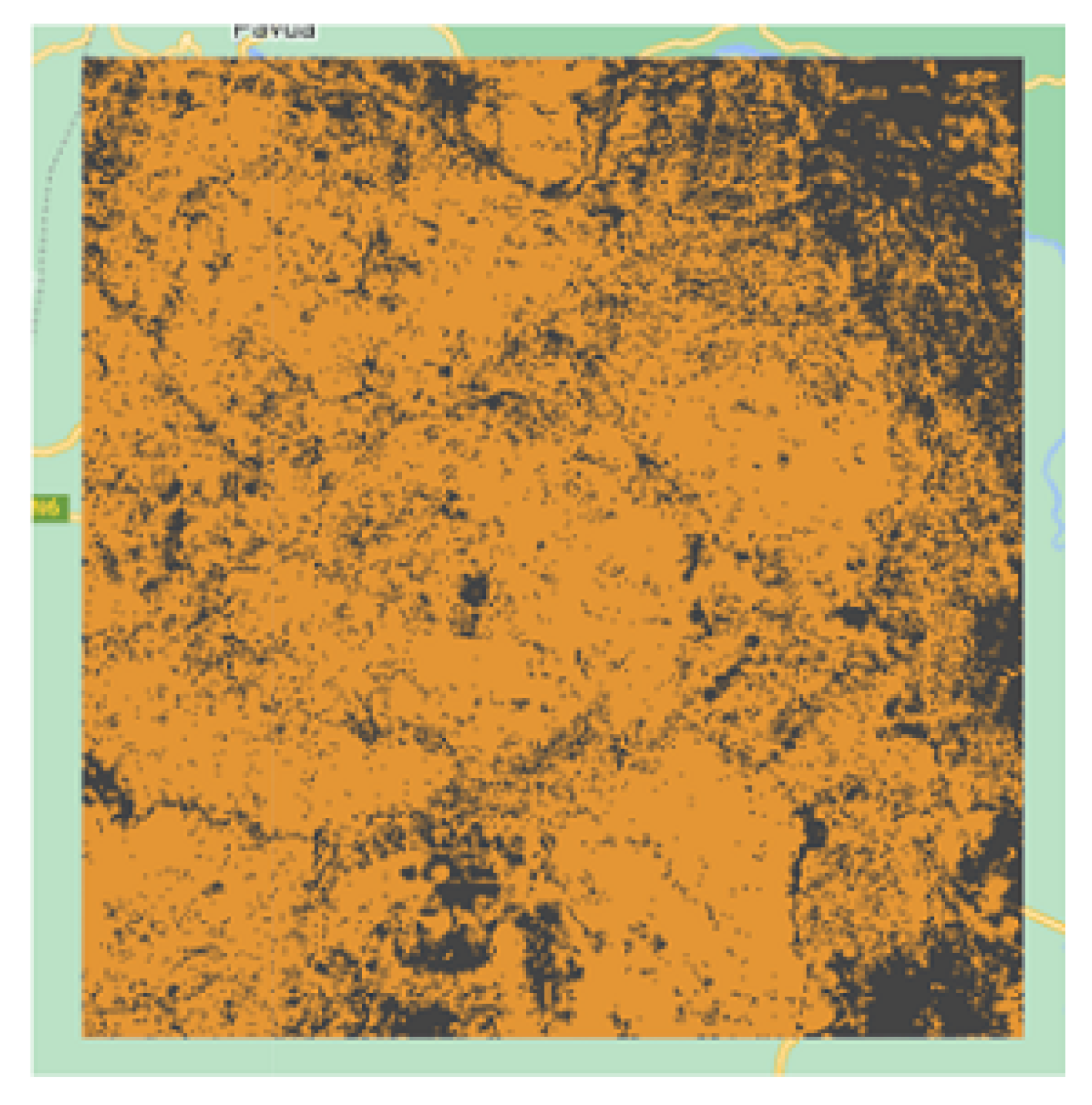
Figure 16) contained tedium samples that were difficult to distinguish even through eye-inspection, while the region of Iran (
Figure 17) contained easily distinguished cropland or non-cropland areas with a high variance present in the corresponding time series. Besides, the presence of a body of water such as the Nile River in the test region of Sudan (
Figure 18) could decrease the performance because it is fringed by gallery forests and herbaceous vegetation which can be misleading.
Table 1.
Cropland extent accuracy.
Table 1.
Cropland extent accuracy.
| Model |
Mozambique |
Sudan |
Iran |
SriLanka |
| Random Forest |
0.74 |
0.84 |
0.92 |
0.86 |
| XGBoost |
0.82 |
0.92 |
0.92 |
0.88 |
| LSTM |
0.82 |
0.84 |
0.96 |
0.90 |
| Bidirectional LSTM |
0.80 |
0.90 |
0.98 |
0.80 |
Figure 15.
Sri-Lanka cropland extent resulted map.
Figure 15.
Sri-Lanka cropland extent resulted map.
Figure 16.
Mozambique cropland extent resulted map.
Figure 16.
Mozambique cropland extent resulted map.
Figure 17.
Iran cropland extent resulted map.
Figure 17.
Iran cropland extent resulted map.
Figure 18.
Sudan cropland extent resulted map.
Figure 18.
Sudan cropland extent resulted map.
Table 2.
Crop intensity accuracy.
Table 2.
Crop intensity accuracy.
| Model |
Mozambique |
Sudan |
Iran |
SriLanka |
| Random Forest |
0.87 |
0.86 |
0.803 |
0.95 |
| XGBoost |
0.92 |
0.83 |
0.803 |
0.94 |
| KNN DTW |
0.75 |
0.92 |
0.80 |
0.92 |
Figure 19.
Iran cropland extent resulted map.
Figure 19.
Iran cropland extent resulted map.
Figure 20.
Sri-Lanka cropland intensity resulted map.
Figure 20.
Sri-Lanka cropland intensity resulted map.
Figure 21.
Mozambique cropland intensity resulted map.
Figure 21.
Mozambique cropland intensity resulted map.
Figure 22.
Iran cropland intensity resulted map.
Figure 22.
Iran cropland intensity resulted map.
Figure 23.
Sudan cropland intensity resulted map.
Figure 23.
Sudan cropland intensity resulted map.
To test he inference time of the algorithms at hand, we measured the time needed to generalize the results on a 0.5° by 0.5° grid which corresponds to about 55km squared in the four test regions. Traditional machine learning methods seemed to be more time-efficient when applied on the full map with an average inference time of 4 minutes for Random Forest and and less than a minute using XGBoost using a Python 3 Google Compute Engine backend. On the other hand, context aware models took longer time to perform the same task with an approximately 1h 30 minutes for the LSTM model using the same computational resources. These results are consistent with the theoretical computational complexity analysis of the algorithms discussed earlier. We believe that traditional machine learning algorithms are the optimum solution to operate on large grid maps given its relatively high accuracy, low computational cost, and fast inference time.
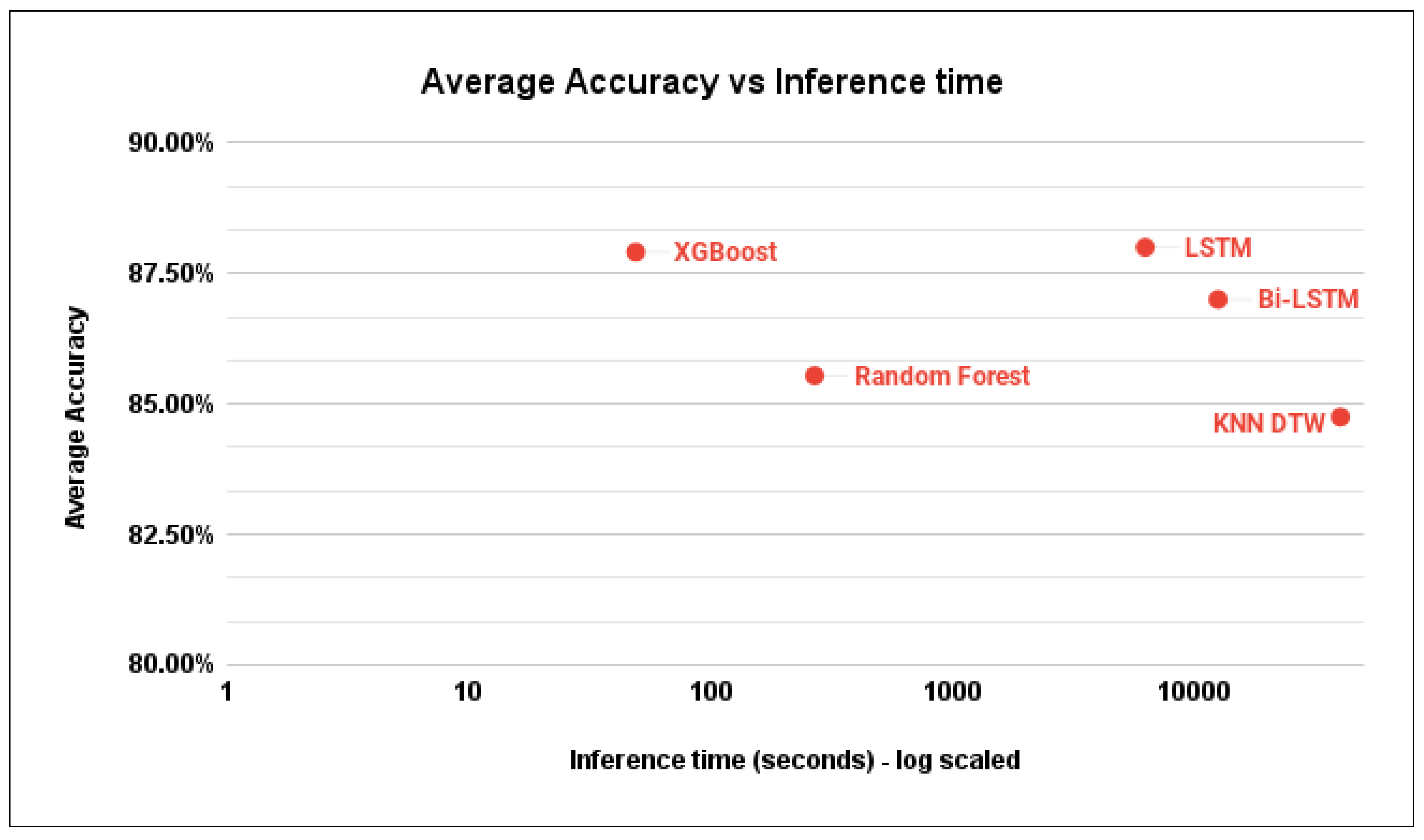
Figure 23 illustrates the average inference time of each model with respect to the average accuracy. A more detailed summary is provided in
Table 3.
Figure 24.
Inference time of different models on a 0.5° by 0.5° grid.
Figure 24.
Inference time of different models on a 0.5° by 0.5° grid.
Table 3.
Average inference time using different Algorithms.
Table 3.
Average inference time using different Algorithms.
| model |
avg inference time (seconds) |
| XGBoost |
49 |
| Random Forest |
269 |
| LSTM |
6259 |
| Bi-LSTM |
12518 |
| KNN DTW |
40065 |
4. Conclusions
In conclusion, we have discussed a full pipeline to perform cropland mapping to obtain cropland extent and intensity assets using a light weight machine learning based framework. Samples were collected from Sentienal-2 data using google earth engine before performing time-series reconstruction involving the utilization of Sevitsky-Golay filters and linear interpolation to fill the gaps within the NDVI time series. Later, the reconstructed series was fed into two different types of algorithms in which the first consists of traditional machine learning algorithms such as Random Forest and XGBoost while the second included context-aware methods such as KNN-DTW and Recurrent Neural Networks like LSTMs and Bi-LSTMs achieving high accuracy results ranging between and up to across four test regions including Iran, Sri-lanka, Sudan, and Mozambique. The obtained results were finally generalized over a full map of a 0.5° by 0.5° grid size captured from 10-meters spatial resolution and the inference time of each algorithm was measured. We concluded that traditional machine learning algorithms provide a more efficient and light weight solution for cropland mapping given its relatively high accuracy and fast inference time.
References
- Summary Progress Update 2021: SDG 6—water and sanitation for all.
- Remote Sensing—Free Full-Text—Global Croplands and their Importance for Water and Food Security in the Twenty-first Century: Towards an Ever Green Revolution that Combines a Second Green Revolution with a Blue Revolution.
- Detail.
- Matton, N.; Canto, G.S.; Waldner, F.; Valero, S.; Morin, D.; Inglada, J.; Arias, M.; Bontemps, S.; Koetz, B.; Defourny, P. An Automated Method for Annual Cropland Mapping along the Season for Various Globally-Distributed Agrosystems Using High Spatial and Temporal Resolution Time Series. Remote. Sens. 2015, 7, 13208–13232. [Google Scholar] [CrossRef]
- Mtibaa, S.; Irie, M. Land cover mapping in cropland dominated area using information on vegetation phenology and multi-seasonal Landsat 8 images. Euro-Mediterr. J. Environ. Integr. 2016, 1, 6. [Google Scholar] [CrossRef]
- Boryan, C.; Yang, Z.; Mueller, R.; Craig, M. Monitoring US agriculture: The US Department of Agriculture, National Agricultural Statistics Service, Cropland Data Layer Program. Geocarto Int. 2011, 26, 341–358. [Google Scholar] [CrossRef]
- Oliphant, A.J.; Thenkabail, P.S.; Teluguntla, P.; Xiong, J.; Gumma, M.K.; Congalton, R.G.; Yadav, K. Mapping cropland extent of Southeast and Northeast Asia using multi-year time-series Landsat 30-m data using a random forest classifier on the Google Earth Engine Cloud. Int. J. Appl. Earth Obs. Geoinf. 2019, 81, 110–124. [Google Scholar] [CrossRef]
- Belgiu, M.; Csillik, O. Sentinel-2 cropland mapping using pixel-based and object-based time-weighted dynamic time warping analysis. Remote. Sens. Environ. 2018, 204, 509–523. [Google Scholar] [CrossRef]
- Chakrabarti, S.; Cormier, T.; Malizia, N.; Potere, D.; Sulla-Menashe, D.; Zmijewski, K.; Friedl, M. Mapping Cropland Extent by Asynchronous Fusion of Optical and Active Microwave Imagery. IGARSS 2018 - 2018 IEEE International Geoscience and Remote Sensing Symposium, 2018, pp. 5319–5321. ISSN 2153-7003. [CrossRef]
- Potapov, P.; Turubanova, S.; Hansen, M.C.; Tyukavina, A.; Zalles, V.; Khan, A.; Song, X.P.; Pickens, A.; Shen, Q.; Cortez, J. Global maps of cropland extent and change show accelerated cropland expansion in the twenty-first century. Nat. Food 2022, 3, 19–28. [Google Scholar] [CrossRef]
- See, L.; Fritz, S.; You, L.; Ramankutty, N.; Herrero, M.; Justice, C.; Becker-Reshef, I.; Thornton, P.; Erb, K.; Gong, P.; Tang, H.; van der Velde, M.; Ericksen, P.; McCallum, I.; Kraxner, F.; Obersteiner, M. Improved global cropland data as an essential ingredient for food security. Glob. Food Secur. 2015, 4, 37–45. [Google Scholar] [CrossRef]
- Xiong, J.; Thenkabail, P.S.; Tilton, J.C.; Gumma, M.K.; Teluguntla, P.; Oliphant, A.; Congalton, R.G.; Yadav, K.; Gorelick, N. Nominal 30-m Cropland Extent Map of Continental Africa by Integrating Pixel-Based and Object-Based Algorithms Using Sentinel-2 and Landsat-8 Data on Google Earth Engine. Remote. Sens. 2017, 9, 1065. [Google Scholar] [CrossRef]
- Hendricks, N.P.; Er, E. Changes in cropland area in the United States and the role of CRP. Food Policy 2018, 75, 15–23. [Google Scholar] [CrossRef]
- Gumma, M.K.; Thenkabail, P.S.; Maunahan, A.; Islam, S.; Nelson, A. Mapping seasonal rice cropland extent and area in the high cropping intensity environment of Bangladesh using MODIS 500m data for the year 2010. ISPRS J. Photogramm. Remote. Sens. 2014, 91, 98–113. [Google Scholar] [CrossRef]
- Gumma, M.K.; Thenkabail, P.S.; Maunahan, A.; Islam, S.; Nelson, A. Mapping seasonal rice cropland extent and area in the high cropping intensity environment of Bangladesh using MODIS 500m data for the year 2010. ISPRS J. Photogramm. Remote. Sens. 2014, 91, 98–113. [Google Scholar] [CrossRef]
- Gumma, M.K.; Thenkabail, P.S.; Teluguntla, P.G.; Oliphant, A.; Xiong, J.; Giri, C.; Pyla, V.; Dixit, S.; Whitbread, A.M. Agricultural cropland extent and areas of South Asia derived using Landsat satellite 30-m time-series big-data using random forest machine learning algorithms on the Google Earth Engine cloud. GIScience Remote. Sens. 2020, 57, 302–322. [Google Scholar] [CrossRef]
- Halder, J. Land Suitability Assessment for Crop Cultivation by Using Remote Sensing and GIS. J. Geogr. Geol. 2013, 5, p65. [Google Scholar] [CrossRef]
- Tran, K.H.; Zhang, H.K.; McMaine, J.T.; Zhang, X.; Luo, D. 10 m crop type mapping using Sentinel-2 reflectance and 30 m cropland data layer product. Int. J. Appl. Earth Obs. Geoinf. 2022, 107, 102692. [Google Scholar] [CrossRef]
- Helman, D.; Lensky, I.M.; Tessler, N.; Osem, Y. A Phenology-Based Method for Monitoring Woody and Herbaceous Vegetation in Mediterranean Forests from NDVI Time Series. Remote. Sens. 2015, 7, 12314–12335. [Google Scholar] [CrossRef]
- The International Encyclopedia of Geography: People, The Earth, Environment and Technology. Blackwell publishing: 2017.
- Kong, D.; Zhang, Y.; Gu, X.; Wang, D. A robust method for reconstructing global MODIS EVI time series on the Google Earth Engine. ISPRS J. Photogramm. Remote. Sens. 2019, 155, 13–24. [Google Scholar] [CrossRef]
- Li, X.; Shen, R.; Chen, R. Improving Time Series Reconstruction by Fixing Invalid Values and its Fidelity Evaluation. IEEE Access 2019, 1. [Google Scholar] [CrossRef]
- Li, X.; Wang, L.; Cheng, Q.; Wu, P.; Gan, W.; Fang, L. Cloud removal in remote sensing images using nonnegative matrix factorization and error correction. ISPRS J. Photogramm. Remote. Sens. 2019, 148, 103–113. [Google Scholar] [CrossRef]
- Xu, L.; Li, B.; Yuan, Y.; Gao, X.; Zhang, T. A Temporal-Spatial Iteration Method to Reconstruct NDVI Time Series Datasets. Remote Sensing 2015, 7, 8906–8924. [Google Scholar] [CrossRef]
- Padhee, S.K.; Dutta, S. Spatio-Temporal Reconstruction of MODIS NDVI by Regional Land Surface Phenology and Harmonic Analysis of Time-Series. GIScience Remote. Sens. 2019, 56, 1261–1288. [Google Scholar] [CrossRef]
- Spatiotemporal reconstruction of MODIS land surface temperature with the help of GLDAS product using kernel-based nonparametric data assimilation.
- You, N.; Dong, J.; Huang, J.; Du, G.; Zhang, G.; He, Y.; Yang, T.; Di, Y.; Xiao, X. The 10-m crop type maps in Northeast China during 2017–2019. Sci. Data 2021, 8, 41. [Google Scholar] [CrossRef]
- d’Andrimont, R.; Verhegghen, A.; Lemoine, G.; Kempeneers, P.; Meroni, M.; van der Velde, M. From parcel to continental scale – A first European crop type map based on Sentinel-1 and LUCAS Copernicus in-situ observations. Remote. Sens. Environ. 2021, 266, 112708. [Google Scholar] [CrossRef]
- Yaramasu, R.; Bandaru, V.; Pnvr, K. Pre-season crop type mapping using deep neural networks. Comput. Electron. Agric. 2020, 176, 105664. [Google Scholar] [CrossRef]
- Zheng, B.; Myint, S.W.; Thenkabail, P.S.; Aggarwal, R.M. A support vector machine to identify irrigated crop types using time-series Landsat NDVI data. Int. J. Appl. Earth Obs. Geoinf. 2015, 34, 103–112. [Google Scholar] [CrossRef]
- Ketchum, D.; Jencso, K.; Maneta, M.P.; Melton, F.; Jones, M.O.; Huntington, J. IrrMapper: A Machine Learning Approach for High Resolution Mapping of Irrigated Agriculture Across the Western U.S. Remote. Sens. 2020, 12, 2328. [Google Scholar] [CrossRef]
- Zhang, C.; Dong, J.; Xie, Y.; Zhang, X.; Ge, Q. Mapping irrigated croplands in China using a synergetic training sample generating method, machine learning classifier, and Google Earth Engine. Int. J. Appl. Earth Obs. Geoinf. 2022, 112, 102888. [Google Scholar] [CrossRef]
- Gupta, V.D.; Areendran, G.; Raj, K.; Ghosh, S.; Dutta, S.; Sahana, M. Assessing habitat suitability of leopards (Panthera pardus) in unprotected scrublands of Bera, Rajasthan, India. In Forest Resources Resilience and Conflicts; Elsevier, 2021; pp. 329–342.
- Dutta, S.; Rehman, S.; Chatterjee, S.; Sajjad, H. Analyzing seasonal variation in the vegetation cover using NDVI and rainfall in the dry deciduous forest region of Eastern India. In Forest Resources Resilience and Conflicts; Elsevier, 2021; pp. 33–48.
- Viana, C.M.; Oliveira, S.; Oliveira, S.C.; Rocha, J. Land use/land cover change detection and urban sprawl analysis. In Spatial modeling in GIS and R for earth and environmental sciences; Elsevier: 2019; pp. 621–651.
- Sentinel-2 – Marketplace – Google Cloud console.
- Liu, R.; Shang, R.; Liu, Y.; Lu, X. Global evaluation of gap-filling approaches for seasonal NDVI with considering vegetation growth trajectory, protection of key point, noise resistance and curve stability. Remote Sensing of Environment 2017, 189, 164–179. [Google Scholar] [CrossRef]
- HAO, P.y.; Tang, H.j.; CHEN, Z.x.; Le, Y.; Wu, M.q. High resolution crop intensity mapping using harmonized Landsat-8 and Sentinel-2 data. J. Integr. Agric. 2019, 18, 2883–2897. [Google Scholar] [CrossRef]
- Pan, L.; Xia, H.; Yang, J.; Niu, W.; Wang, R.; Song, H.; Guo, Y.; Qin, Y. Mapping cropping intensity in Huaihe basin using phenology algorithm, all Sentinel-2 and Landsat images in Google Earth Engine. Int. J. Appl. Earth Obs. Geoinf. 2021, 102, 102376. [Google Scholar] [CrossRef]
- Li, L.; Friedl, M.A.; Xin, Q.; Gray, J.; Pan, Y.; Frolking, S. Mapping crop cycles in China using MODIS-EVI time series. Remote Sensing 2014, 6, 2473–2493. [Google Scholar] [CrossRef]
- Sruthi, E. Understanding Random Forest. In Data Science Bloagathon; Analytics Vidhya, 2021.
- Seif, G. A Beginner’s guide to XGBoost. Towards Data Science 2019. [Google Scholar]
- Saxena, S. Introduction to long short term memory (LSTM). Analytics Vidhya 2021. [Google Scholar]
- Cornegruta, S.; Bakewell, R.; Withey, S.; Montana, G. Modelling radiological language with bidirectional long short-term memory networks. arXiv 2016, arXiv:1609.08409. [Google Scholar]
- Pawar, R. k-NN based Time Series Classification An overview of the different approaches from the literature.
- Sole, X.; Ramisa, A.; Torras, C. Evaluation of Random Forests on large-scale classification problems using a Bag-of-Visual-Words representation.
- Dynamic Time Warping and Geometric Edit Distance: Breaking the Quadratic Barrier: ACM Transactions on Algorithms: Vol 14, No 4.
- Chen, T.; Guestrin, C. XGBoost: A Scalable Tree Boosting System. Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, 2016, pp. 785–794. arXiv:1603.02754. [CrossRef]
- Hochreiter, S.; Schmidhuber, J. Long Short-Term Memory. Neural Comput. 1997, 9, 1735–1780. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).