1. Introduction
Antimicrobial resistance (AMR) is a serious healthcare problem that is increasing worldwide [
1], and is a threat to global health [
2]. Any use of antimicrobials, whether judicious or non-judicious, creates selection pressure that can lead to AMR emergence [
3]. However, non-judicious use of antimicrobials in human and veterinary medicine is a modifiable driver of AMR, leading to the emergence of bacterial strains resistant to many antimicrobials [
4,
5,
6]. Like in human medicine [
7], initial antimicrobial therapy in veterinary medicine is often empiric and is guided by clinical signs of disease. Under ideal clinical practice situations, empirical use of antimicrobials is discouraged and antimicrobial prescription based on a bacterial culture and antimicrobial susceptibility testing is preferred [
8]. However, high costs and long turnaround times associated with the existing bacterial culture and antimicrobial susceptibility testing methods influence the decision to use them [
9,
10].
In human hospitals, summaries of the patterns of antimicrobial susceptibilities (antibiograms), constructed from laboratory samples that have been submitted institutionally are often used by clinicians. The antibiograms report the percentage of isolates susceptible to a set of antimicrobials, may be stratified (by patient population, medical service or specimen type), and are shared with clinicians, hospital pharmacists and infection-control personnel in the form of web postings and pocket guides [
11]. These antibiograms can complement rapid diagnostic techniques in clinical microbiology [
12], allow clinicians to evaluate the local susceptibility rates to specific antimicrobials and aid them in selection of the most appropriate empiric therapy [
13,
14,
15].
The integration of antibiogram use in veterinary practice to guide empiric antimicrobial therapy is necessary for antimicrobial stewardship [
16]. However, in the United States, the use of cumulative antibiograms to guide empiric antimicrobial therapy in veterinary medicine is still limited to just a few tertiary veterinary hospitals. Even the two tertiary veterinary hospitals that in the past publicly provided cumulative antibiograms to their clinicians via their hospital websites do not have up to date antibiograms posted at their websites. The limited use of cumulative antibiograms in veterinary medicine may be due in part to the fact that there are no specific guidelines or standardized methods available for the construction of veterinary antibiograms. Here, we examined the practicality of constructing cumulative antibiograms for a veterinary tertiary care hospital. This paper describes and documents the approaches used, and the challenges that were encountered in constructing cumulative antibiograms for a veterinary teaching hospital in Indiana, United States.
2. Results
2.1. The isolates
Of the 563 canine antimicrobial susceptibility profiles in the analyzed dataset, 318 (56.5%) isolates were Gram-positive and these included Staphylococcus spp., Enterococcus spp. Streptococcus spp., Corynebacterium spp., Bacillus spp., Aerococcus spp., and Actinomyces spp. Two hundred forty-five isolates (43.5%) were gram-negative and these included Escherichia coli, Pseudomonas spp., Proteus mirabilis, Pasteurella spp., Enterobacter spp., Klebsiella spp., and other Gram-negative organisms.
2.2. General antibiograms
One general antibiogram, with a section for Gram-negative organisms (
Table 1), and another for Gram-positive organisms were prepared (
Table 2). The Gram-negative isolates were 100
Escherichia coli, 27
Pseudomonas spp., and 26
Proteus mirabilis (
Table 1). For the Gram-positive isolates, there were 170
Staphylococcus spp. isolates, 64
Streptococcus spp., 30 C
orynebacterium spp., and 65
Enterococcus spp. isolates (
Table 2).
2.3. Antibiograms stratified by hospital section (medical service), sample type/anatomic region of sample collection
There were enough isolates collected from different hospital sections, the ears, skin, and urine to create individual antibiograms for the different hospital sections and anatomic regions/sample type. Antibiograms with sections for patients admitted to each hospital department were prepared for Gram-positive and Gram-negative organisms (
Table 3 and
Table 4 respectively). Twenty-one
Staphylococcus spp. isolates from the ear and 43 Staphylococcal isolates from the skin were used to create antibiograms for these anatomic regions (
Table 5). One antibiogram, with two parts, one for Gram-negative and the other for Gram-positive bacteria was prepared for isolates from urine (
Table 6).
3. Discussion
This article adds to the literature a description of practical approaches used in making antibiograms for a veterinary teaching hospital. This work shows that creating cumulative antibiograms for dogs seen at a veterinary teaching hospital stratified by area of hospital admission, and sample type is feasible and practical. Similar to ours, another study conducted in the US demonstrated the feasibility of adapting existing guidelines for developing antibiograms in human medicine to the veterinary companion animal private practice setting [
17]. Results of these antibiograms show the tremendous variability in the antimicrobial susceptibilities of the isolates to specific antimicrobials at a veterinary teaching hospital in Indiana e.g., in the general antibiograms, 90% of the staphylococcal isolates were susceptible to amoxicillin-clavulanic acid, only 70% were susceptible to enrofloxacin, 73% of
E. coli were susceptible to amoxicillin-clavulanic acid and 85% were susceptible to enrofloxacin. These susceptibility differences suggest that antibiograms could be very useful in guiding empiric antimicrobial therapy and could guide the framing of antimicrobial stewardship policies at this hospital.
The
E. coli isolates from samples obtained from the Medicine service showed susceptibility profiles similar to those observed in the general Gram-negative antibiogram. Of the 43
Staphylococcus spp. isolates from the skin, only 63% showed susceptibility to clindamycin, a drug of choice for treating drug-resistant skin infections (pyoderma) in dogs. Sixty-nine percent of the 71
E. coli isolated from urine were susceptible to ticarcillin and 73% of the 45
Enterococcus spp. isolated from urine were susceptible to amoxicillin/clavulanic acid and ampicillin. We observe from our antibiograms, that 100% of the canine
E. coli isolated from a veterinary teaching hospital in Indiana in 2015 were susceptible to imipenem. This is similar to a previous report from Marshfield labs where 100% susceptibility of canine
E. coli isolates tested in the years 2017 through 2018 were susceptible to imipenem [
18]. Additionally, a previous study reported 99.9% cumulative susceptibility of
E. coli isolates from human samples to imipenem in Pennsylvania [
15]. Resistance to carbapenems (such as imipenem) among veterinary isolates is rare and imipenem is highly stable against most β-lactamases [
19]. Possibly, the 100% susceptibility observed in our antibiograms is due to the pharmacodynamic properties of imipenem or could be because imipenem is not frequently used and is not a first line drug in the treatment of dogs with bacterial infections. However, it is important to note that the reported proportions of susceptible isolates could be subject to selection bias because selection bias can be an issue in clinical samples submitted to a diagnostic laboratory.
Having an insufficient number of isolates for analysis is a concern when creating antibiograms in smaller human hospitals and for infrequently isolated bacteria [
11]. In our analysis, the number of isolates for other animal species (e.g. cattle), and for some individual bacterial species was very low (fewer than 30 isolates) and therefore antibiograms were not constructed. Depending upon the patient population and how often clinical samples are submitted, we observe that construction of veterinary antibiograms for the various veterinary animal species (except for dogs) and for some bacterial species will be challenging due to the relatively small number of samples submitted for bacterial culture and antimicrobial susceptibility testing. This challenge could be overcome by the use of data collected over a longer period of time.
Periodic preparation and distribution of cumulative antibiograms to clinicians, the hospital pharmacist, and the infection-control personnel at this hospital in the form of pocket guides and website postings will be useful. The impact of the antibiograms on veterinary prescription practices through periodic assessment of hospital pharmacy records and via surveys periodically administered to clinicians should be routinely assessed.
4. Materials and Methods
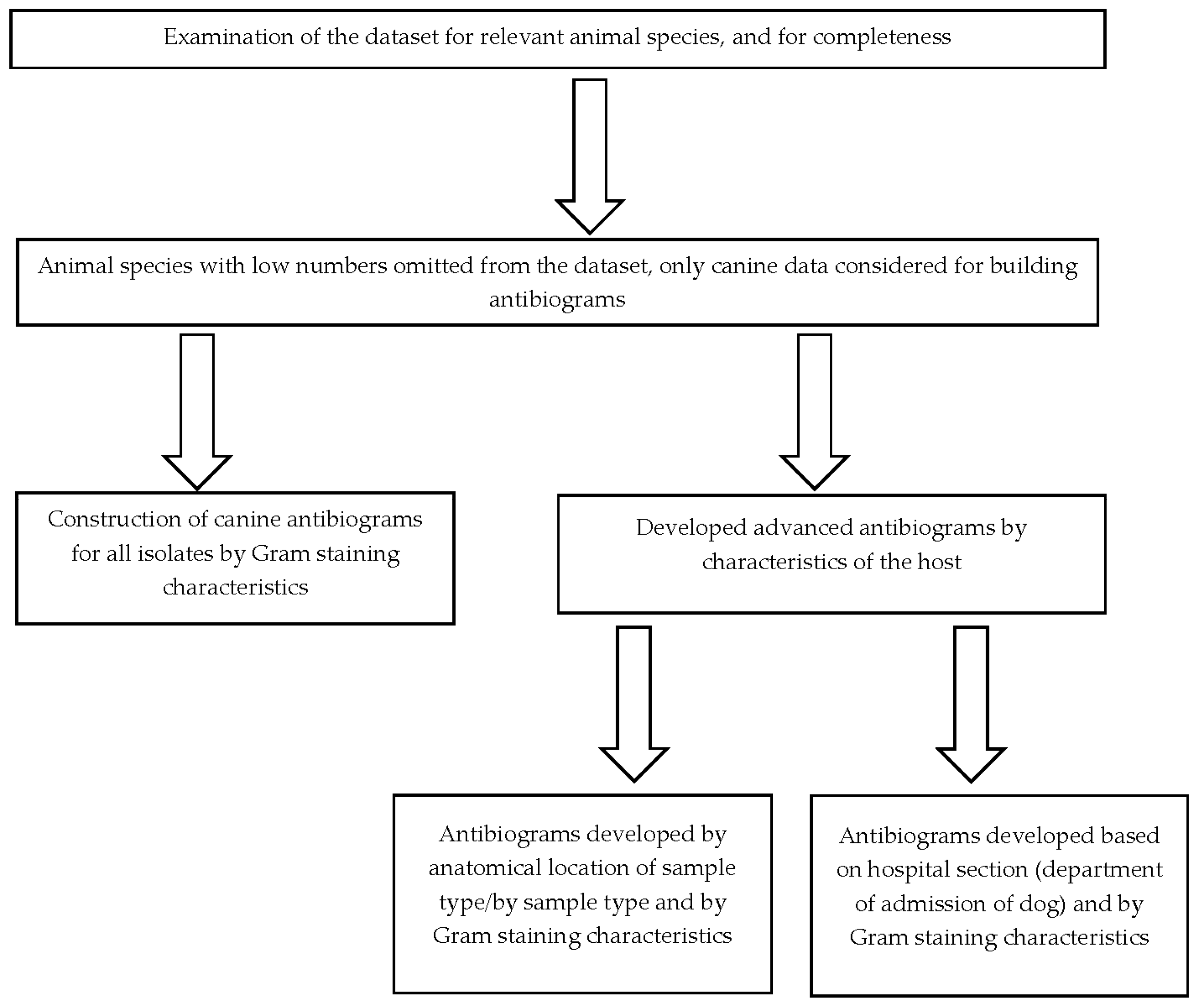
We used laboratory data for the period January 1, 2015 to December 31, 2015 provided by the Animal Disease Diagnostic Lab (ADDL) at a veterinary teaching hospital in Indiana, United States. The data provided did not contain personally identifiable information about the clinician or client and was provided in a Microsoft Excel file. These data consisted of 661 (complete and partial) antimicrobial susceptibility profiles for isolates obtained from dogs (n = 578), cats (n = 68), rats (n = 4), wolves (n = 2), guinea pigs (n = 2), chinchilla (n = 2), fox (n = 1), conure (n = 1), and snake (n = 1). No animal species was recorded for two isolates. The data were verified and examined for completeness. The data captured in the dataset included: the hospital section where the patient was admitted, the location from which the sample was collected, the bacterial isolate identified, and the antimicrobial susceptibility profiles for various antimicrobials. Given the low numbers reported for the various animal species, only dog data was utilized in the construction of the antibiograms.
We used the Clinical and Laboratory Standards Institute (CLSI) guidelines for use in the construction of the antibiograms in human healthcare settings as the basis for the veterinary antibiograms [
20]. In brief, the CLSI recommends that isolates included in antibiogram construction are identified during diagnostic sampling and include only the results from the first isolate recovered from each patient. Information from individual bacterial species should only be presented if there are at least 30 isolates tested. In addition, data should be presented as total percent susceptible for drugs that are routinely tested and should not include percentage of isolates with intermediate susceptibility results. Data included in hospital antibiograms should be updated annually [
11].
In our analysis, we utilized only the first isolate of a given bacterial species recovered from each canine patient during 2015 in the calculation of percent susceptibility for each organism/antimicrobial combination. Antibiograms for drugs that are routinely tested were prepared and presented in tabular form. These drugs include amikacin (AMK), amoxicillin/clavulanic acid (AMC), ampicillin (AMP), cefazolin (CFZ), cefovecin (CFV), cefoxitin (FOX), cefpodoxime (CPD), ceftiofur (CTF), cephalothin (CEF), chloramphenicol (CHL), clindamycin (CLI), doxycycline (DOX), enrofloxacin (ENR), erythromycin (ERY), gentamycin (GEN), imipenem (IPM), marbofloxacin (MBF), oxacillin + 2%NACL (OXA), penicillin (PEN), ticarcillin (TIC), ticarcillin/clavulanic acid (TIM), trimethoprim/ sulfamethoxazole (SXT).
Only the percent susceptible and not those which are of intermediate susceptibility were presented. Organisms with fewer than 30 isolates were only included if their inclusion was deemed essential based on existing knowledge showing that resistance against common antimicrobials is possible [
19]. In some instances, species within a genus with fewer than 30 individual isolates were grouped together. In cases where the number of isolates used to calculate the percent susceptibility to a given drug were fewer compared to other drugs, a note was appended to indicate that the calculation of percent susceptibility was done using fewer isolates. The completed antibiograms were further validated to ensure that only percent susceptibilities for antimicrobial agents that are appropriate for clinical use in the species were reported. However, percent susceptibilities for known intrinsically resistant pathogens like
Pseudomonas aeruginosa and
Enterococcus spp. were reported in the antibiograms to guide the antimicrobial selection. Additionally, percentage susceptibilities for drugs primarily used to treat Gram-positive infections (lincosamides e.g. clindamycin and macrolides e.g. erythromycin) were reported for Gram-negative pathogens with a footnote added stating that these two drug classes are primarily used against Gram-positive organisms. The percentage susceptibilities were calculated using commercial statistical software (SAS, version 9.4, SAS Institute Inc, Cary, NC) and the steps taken in constructing the antibiograms are given in
Figure 1.
5. Conclusions
Creating cumulative antibiograms for dogs seen at a veterinary teaching hospital is feasible and practical. Susceptibility differences to specific antimicrobials were identified among bacteria isolated at a veterinary teaching hospital in Indiana from January 2015 through December 2015. Cumulative antibiograms could be useful in guiding empiric antimicrobial therapy, and for detecting and monitoring AMR trends at this teaching hospital.
Author Contributions
Conceptualization, A.R., L.G., and K.H.; methodology, J.E.E., L.G., M.A., K.H.; software, J.E.E; validation, .E.E., L.G., L.D., K.H.; formal analysis, K.H.; investigation, J.E.E., L.G., M.A., K.H.; resources, A.R., L.G., and K.H.; data curation, J.E.E. and M.A.; writing—original draft preparation, J.E.E.; writing—review and editing, J.E.E., L.G., M.A., K.H; visualization, J.E.E.; supervision, A.R.; project administration, A.R.; funding acquisition, A.R., L.G., and K.H. All authors have read and agreed to the published version of the manuscript.
Funding
This project received funding from the Integrative Data Science Initiative at Purdue University.
Data Availability Statement
The data for this study are available from the corresponding author upon reasonable request.
Acknowledgments
This project was funded by Boehringer Ingelheim and the Integrative Data Science Initiative at Purdue University. We extend our special thanks to Dr. Sharon Erdman of Eskenazi Health for guidance in the antibiograms construction and Amanda Martin of Purdue University for assistance in data preparation. We thank the Indiana Animal Disease Diagnostic Laboratory for providing the data used to construct the antibiograms.
Conflicts of Interest
The authors declare that there is no conflict of interest regarding the publication of this article.
References
- Partridge, S.R.; Kwong, S.M.; Firth, N.; Jensen, S.O. Mobile Genetic Elements Associated with Antimicrobial Resistance. Clinical Microbiology Reviews 2018, 31, e00088-17. [Google Scholar] [CrossRef] [PubMed]
- Baekkeskov, E.; Rubin, O.; Munkholm, L.; Zaman, W. Antimicrobial Resistance as a Global Health Crisis. Available online: https://oxfordre.com/politics/display/10.1093/acrefore/9780190228637.001.0001/acrefore-9780190228637-e-1626 (accessed on 21 April 2023).
- Weese, J.S.; Giguère, S.; Guardabassi, L.; Morley, P.S.; Papich, M.; Ricciuto, D.R.; Sykes, J.E. ACVIM Consensus Statement on Therapeutic Antimicrobial Use in Animals and Antimicrobial Resistance. J Vet Intern Med 2015, 29, 487–498. [Google Scholar] [CrossRef] [PubMed]
- Holmes, A.H.; Moore, L.S.P.; Sundsfjord, A.; Steinbakk, M.; Regmi, S.; Karkey, A.; Guerin, P.J.; Piddock, L.J.V. Understanding the Mechanisms and Drivers of Antimicrobial Resistance. Lancet 2016, 387, 176–187. [Google Scholar] [CrossRef] [PubMed]
- McEwen, S.A.; Collignon, P.J. Antimicrobial Resistance: A One Health Perspective. Microbiol Spectr 2018, 6. [Google Scholar] [CrossRef] [PubMed]
- Hildreth, C.J.; Burke, A.E.; Glass, R.M. Inappropriate Use of Antibiotics. JAMA 2009, 302, 816. [Google Scholar] [CrossRef] [PubMed]
- Leekha, S.; Terrell, C.L.; Edson, R.S. General Principles of Antimicrobial Therapy. Mayo Clin Proc 2011, 86, 156–167. [Google Scholar] [CrossRef] [PubMed]
- Guardabassi, L.; Lars Bøgø, J.; Hilde, K. Guide to Antimicrobial Use in Animals; 1st ed.; John Wiley & Sons, Ltd, 2008.
- De Briyne, N.; Atkinson, J.; Pokludová, L.; Borriello, S.P.; Price, S. Factors Influencing Antibiotic Prescribing Habits and Use of Sensitivity Testing amongst Veterinarians in Europe. Veterinary Record 2013, 173, 475–475. [Google Scholar] [CrossRef] [PubMed]
- Watts, J.L.; Sweeney, M.T.; Lubbers, B.V. Antimicrobial Susceptibility Testing of Bacteria of Veterinary Origin. Microbiology Spectrum 2018, 6, 17–32. [Google Scholar] [CrossRef] [PubMed]
- Hindler, J.F.; Stelling, J. Analysis and Presentation of Cumulative Antibiograms: A New Consensus Guideline from the Clinical and Laboratory Standards Institute. Clin Infect Dis 2007, 44, 867–873. [Google Scholar] [CrossRef] [PubMed]
- Fernández, J.; Vazquez, F. The Importance of Cumulative Antibiograms in Diagnostic Stewardship. Clin Infect Dis 2019, 69, 1086–1087. [Google Scholar] [CrossRef] [PubMed]
- Lautenbach, E.; Nachamkin, I. Analysis and Presentation of Cumulative Antimicrobial Susceptibility Data (Antibiograms): Substantial Variability across Medical Centers in the United States. Infect Control Hosp Epidemiol 2006, 27, 409–412. [Google Scholar] [CrossRef] [PubMed]
- Joshi, S. Hospital Antibiogram: A Necessity. Indian J Med Microbiol 2010, 28, 277–280. [Google Scholar] [CrossRef] [PubMed]
- Guarascio, A.J.; Brickett, L.M.; Porter, T.J.; Lee, N.D.; Gorse, E.E.; Covvey, J.R. Development of a Statewide Antibiogram to Assess Regional Trends in Antibiotic-Resistant ESKAPE Organisms. J Pharm Pract 2019, 32, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Frey, E.; Jacob, M. Commentary: Using Antibiograms to Promote Antimicrobial Stewardship during Treatment of Bacterial Cystitis and Superficial Bacterial Folliculitis in Companion Animal Practice. Journal of the American Veterinary Medical Association 2020, 257, 900–903. [Google Scholar] [CrossRef] [PubMed]
- Frey, E.; Jacob, M. Development of a Method for Creating Antibiograms for Use in Companion Animal Private Practices. J Am Vet Med Assoc 2020, 257, 950–960. [Google Scholar] [CrossRef] [PubMed]
- Marshfield labs. Canine Prevalent Pathogens & Antimicrobial Susceptibility Patterns: January 1, 2017 to December 31, 2018. Available online: Https://Wwwmarshfieldlabsorg/Sites/Ltrm/Vet/Documents/Annual_Cumulative_Antibiogram_Caninepdf.
- Papich, M.G. Antibiotic Treatment of Resistant Infections in Small Animals. Vet Clin North Am Small Anim Pract 2013, 43, 1091–1107. [Google Scholar] [CrossRef] [PubMed]
- M39-A4: Analysis and Presentation of Cumulative Antimicrobial Susceptibility Test Data; Approved Guideline—Fourth Edition.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or prperty resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).