Submitted:
15 May 2023
Posted:
16 May 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
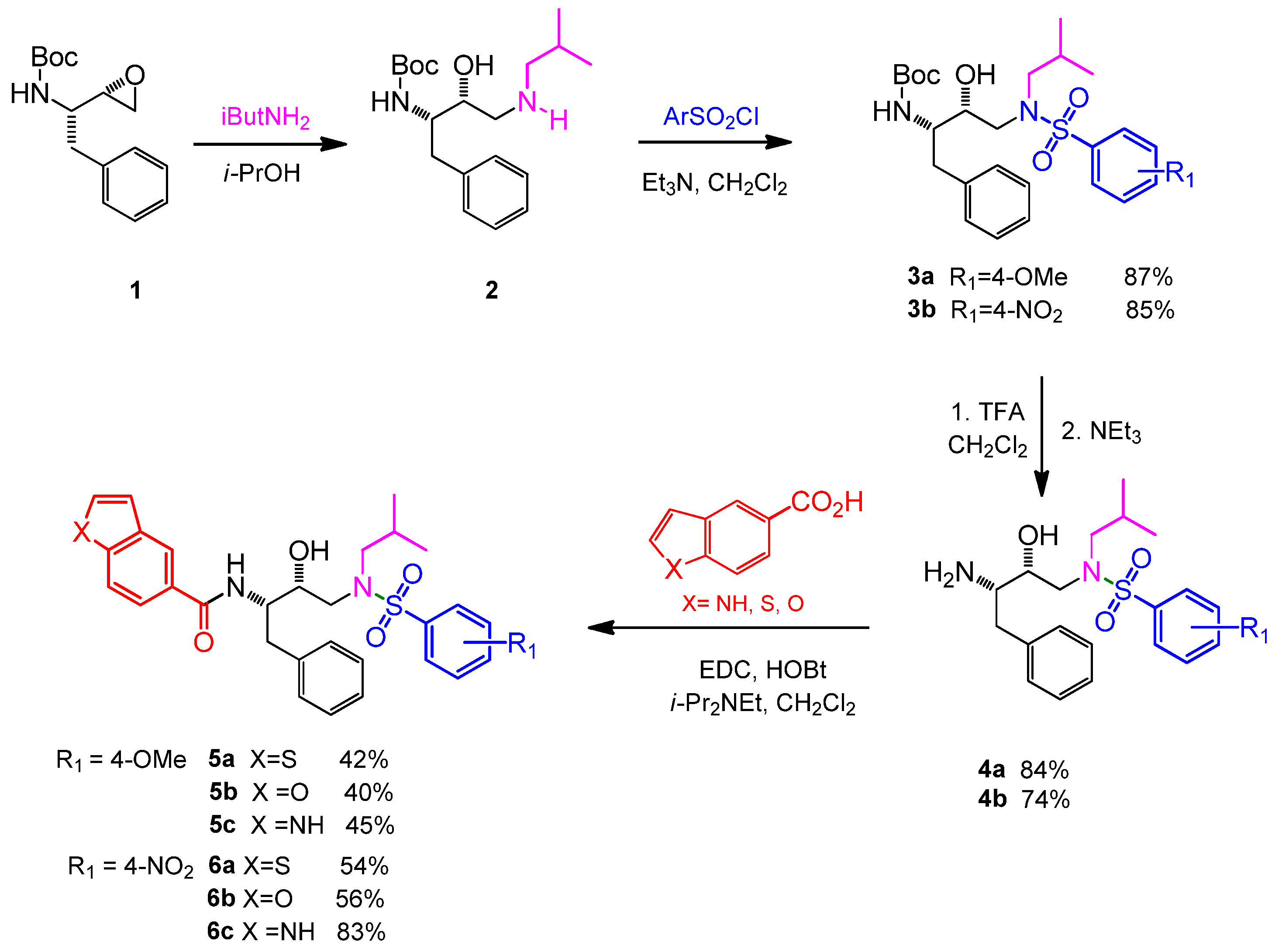
2.1. Chemistry
2.2. Biology
2.2.1. Materials
2.2.2. Cell culture and drug treatment
2.2.3. Cell transfection and FACS analysis
2.2.4. Statistical analysis
3. Results
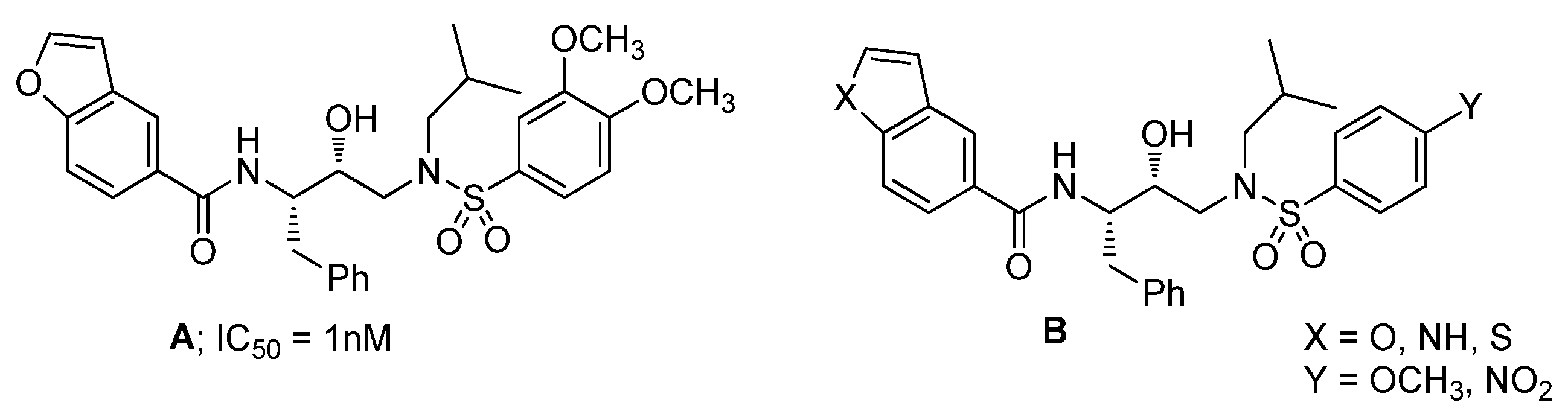
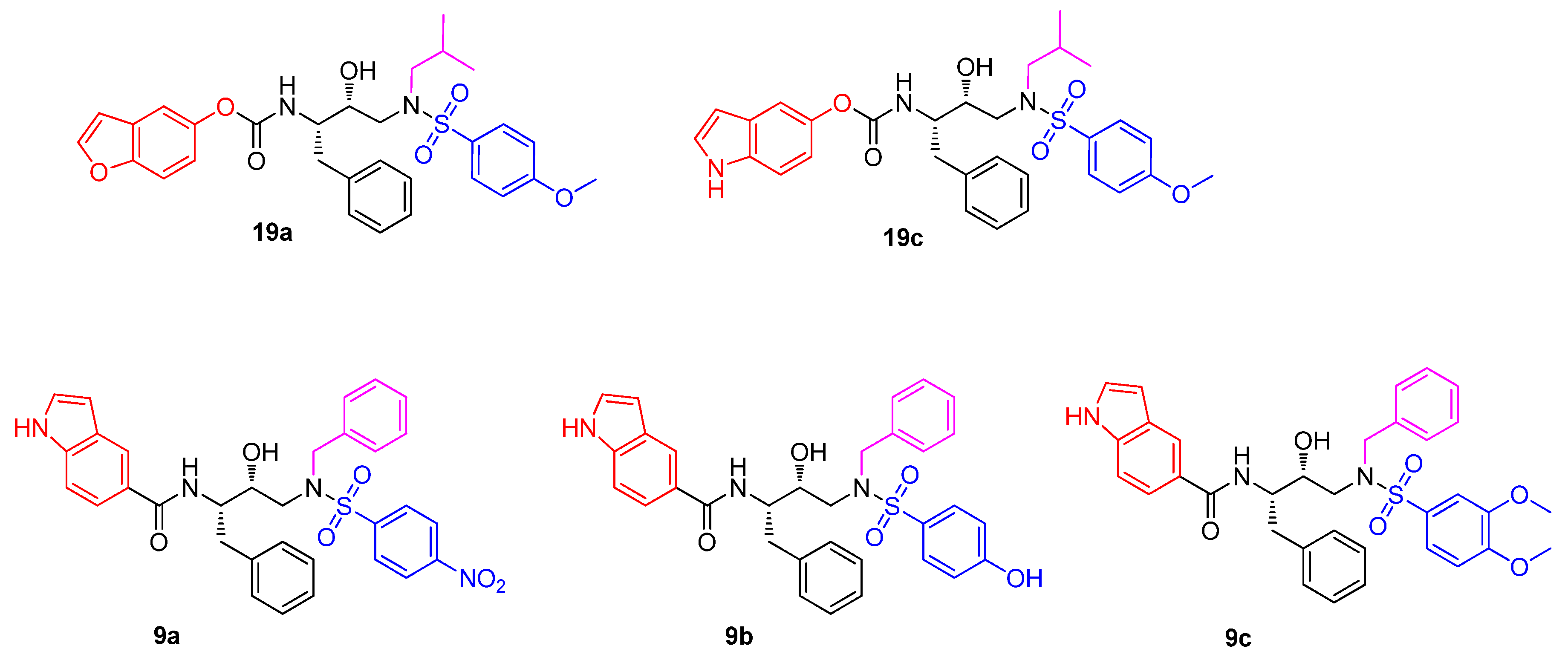
3.1. Chemistry
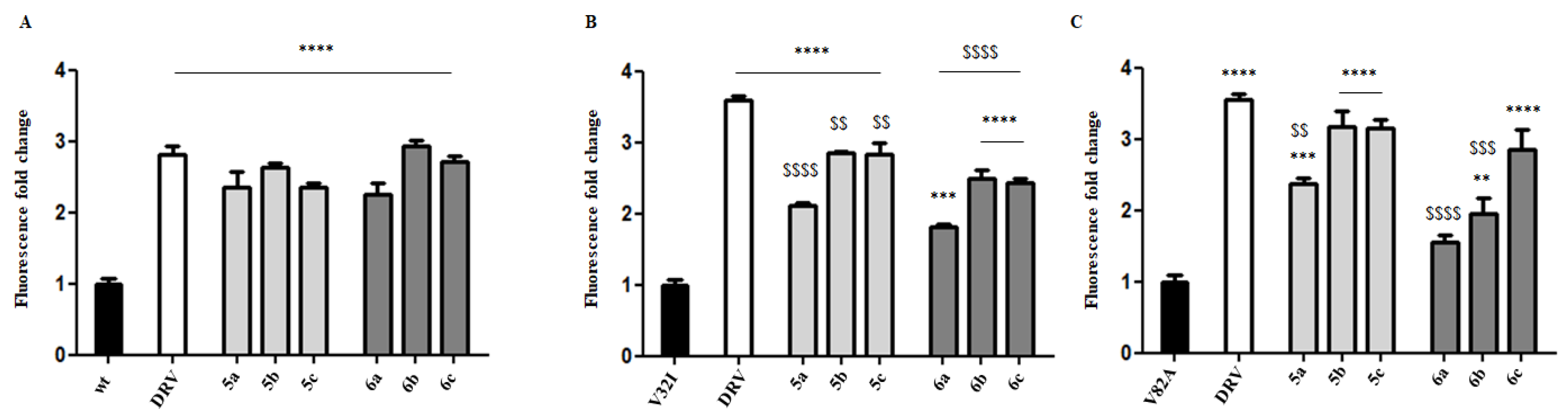
3.2. Biology
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Global Report: UNAIDS report on the global AIDS epidemic 2022. WHO Library Cataloguing-in-Publication Data, Joint United Nations Programme on HIV/AIDS (UNAIDS), 2022.
- World Health Organization. Consolidated guidelines on the use of antiretroviral drugs for treating and preventing HIV infection: recommendations for a public health approach. 2nd ed. Geneva: World Health Organization; 2016.
- Weber, I. T.; Wang, Y-F; Harrison, R. W. HIV Protease: Historical Perspective and Current Research Viruses, 2021, 13, 839. [CrossRef]
- Lv, Z.; Chu, Y.; Wang, Y. HIV protease inhibitors: a review of molecular selectivity and toxicity Research and Palliative Care, 2015, 7, 95-104. [CrossRef]
- Wong-Sam, A.; Wang, Y-F.; Zhang, Y.; Ghosh, A. K.; Harrison, R. W.; Weber, I. T. Drug Resistance Mutation L76V Alters Nonpolar Interactions at the Flap-Core Interface of HIV-1 Protease ACS Omega, 2018, 3, 121132-121140. [CrossRef]
- Wang, B.; He, Y.; Wen, X.; Xi, Z. Prediction and molecular field view of drug resistance in HIV-1 protease mutants, Nature Portfolio Scientific Reports, 2022, 12, 2913. [CrossRef]
- Bonini, C.; Chiummiento, L.; De Bonis, M.; Di Blasio, N.; Funicello, M.; Lupattelli, P.; Pandolfo, R.; Tramutola, F.; Berti, F. Synthesis of New Thienyl Ring Containing HIV-1 Protease Inhibitors: Promising Preliminary Pharmacological Evaluation against Recombinant HIV-1 Proteases. J. Med. Chem. 2010, 53, 1451-1457. [CrossRef]
- Ghosh, A. K.; Shahabi, D.; Kipfmiller, M.; Ghosh, Ajay K.; Johnson, M.; Wang, Y.-F.; Agniswamy, J.; Amano, M.; Weber, I. T.; Mitsuya, H., Evaluation of darunavir-derived HIV-1 protease inhibitors incorporating P2’ amide-derivatives: Synthesis, biological evaluatio and structural studies. Bioorg. Med. Chem. Lett. 2023, 83, 129168. [CrossRef]
- Ghosh, A. K.; Weber, I. T.; Mitsuya, H. Beyond darunavir: recent development of next generation HIV-1 protease inhibitors to combat drug resistance Chem. Commun. 2022, 58, 11762-11782.
- Ghosh, A. K.; Fyvie, W. S.; Brindisi, M.; Steffey, M.; Agniswami, J.; Wang, Y.-F.; Aoki, M.; Amano, M.; Weber, I. T.; Mitsuya, H. Design, Synthesis, Biological Evaluation, and X-ray Studies of HIV-1 Protease Inhibitors with Modified P2’ Ligands of Darunavir ChemMedChem 2017, 12, 1942-1952. [CrossRef]
- Zhang, Y.; Chang, Y.-C. E.; Lousi, J. M.; Wang, Y.-F.; Harrison, R. W.; Weber, I. T. Structures of Darunavir-Resistant HIV-1 Protease Mutant Reveal Atypical Binding of Darunavir to Wide Open Flaps ACS Chem. Biol. 2014, 9, 1351-1358. [CrossRef]
- Ghosh, A. K.; Anderson, D. D.; Weber, I. T.; Mitsuya, H. Enhancing Protein Backbone Binding—A Fruitful Concept for Combating Drug-Resistant HIV. Angew. Chem. Int. Ed. 2012, 51, 1778-1802. [CrossRef]
- Ghosh, A. K.; Chapsal, B. D.; Weber, I. T.; Mitsuya, H, Design of HIV Protease Inhibitors Targeting Protein Backbone: An Effective Strategy for Combating Drug Resistance. Acc. Chem. Res. 2008, 41, 78-86. [CrossRef]
- Bonini, C.; Chiummiento, L.; Di Blasio, N.; Funicello, M.; Lupattelli, P.; Tramutola, F.; Berti, F.; Ostric, A.; Miertus, S.; Frecer, V.; Kong, D.-X. Synthesis and biological evaluation of new simple indolic non peptidic HIV Protease inhibitors: The effect of different substitution patterns. Bioorg. Med. Chem. 2014, 22, 4792-4802. [CrossRef]
- Chiummiento, L.; Funicello, M.; Lupattelli, P.; Tramutola, F.; Berti, F.; Marino-Merlo, F., Synthesis and biological evaluation of novel small non-peptidic HIV-1 PIs: The benzothiophene ring as an effective moiety. Bioorg. Med. Chem. Lett. 2012, 22, 2948-2950. [CrossRef]
- Chiummiento, L.; Funicello, M.; Lupattelli, P.; Tramutola, F.; Campaner, P., New indolic non-peptidic HIV protease inhibitors from (S)-glycidol: synthesis and preliminary biological activity. Tetrahedron, 2009, 65, 5984-5989. [CrossRef]
- Funicello, M.; Chiummiento, L.; Tramutola, F.; Armentano, M. F.; Bisaccia, F.; Miglionico, R.; Milella, L.; Benedetti, F.; Berti, F.; Lupattelli, P. Synthesis and biological evaluation in vitro and in mammalian cells of new heteroaryl carboxyamides as HIV-protease inhibitors Bioorg. Med. Chem. 2017, 25, 4715-4722. [CrossRef]
- Tramutola, F.; Armentano, M. F.; Berti, F.; Chiummiento, L.; Lupattelli, P.; D’Orsi, R.; Miglionico, R.; Milella, L.; Bisaccia, F.; Funicello, M., New heteroaryl carbamates: Synthesis and biological screening in vitro and in mammalian cells of wild-type and mutant HIV protease inhibitors. Bioorg. Med. Chem. 2019, 27, 1863-1870. [CrossRef]
- D’Orsi, R.; Funicello, M.; Laurita, T.; Lupattelli, P.; Berti, F.; Chiummiento, L. The Pseudo-Symmetric N-benzyl Hydroxyethylamine Core in a New Series of Heteroarylcarboxyamide HIV-1 Pr Inhibitors: Synthesis, Molecular Modelling and Biological Evaluation. Biomolecules, 2021, 11, 1584. [CrossRef]
- Rinaldi, R.; Miglionico, R.; Nigro, I.; D’Orsi, R.; Chiummiento, L.; Funicello, M.; Lupattelli, P.; Laurenzana, I.; Sgambato, A.; Monné, M.; Bisaccia, F.; Armentano, M. F., Two Novel Precursors of the HIV-1 Protease Inhibitor Darunavir Target the UPR/Proteasome System in Human Hepatocellular Carcinoma Cell Line HepG2. Cells, 2021, 10, 3052. [CrossRef]
- Kanemitsu, T.; Inoue, M.; Yoshimura, N.; Yoneyama, K.; Watarai, R.; Miyazaki, M.; Odanaka, Y.; Nagata, K.; Itoh, T. A Concise One-Pot Organo- and Biocatalyzed Preparation of Enantiopure Hexahydrofuro[2,3-b]furan-3-ol: An Approach to the Synthesis of HIV Protease Inhibitors. Eur. J. Org. Chem. 2016, 1874-1880. [CrossRef]
- Lindsten, K.; Uhlikova, T.; Konvalinka, J.; Masucci, M.G.; Dantuma, N.P. Cell-based fluorescence assay for human immunodeficiency virus type 1 protease activity. Antimicrob Agents Chemother. 2001, 45, 2616–2622. [CrossRef]
- Chiummiento, L.; Funicello, M.; Lupattelli, P.; Tramutola, F.; Berti, F.; Marino-Merlo, F. Synthesis and biological evaluation of novel small non-peptidic HIV-1 PIs: the benzothiophene ring as an effective moiety. Bioorg. Med. Chem. Lett. 2012, 22, 2948-2950. [CrossRef]
- Cerminara, I.; Chiummiento, L.; Funicello, M.; Guarnaccio, A.; Lupattelli, P. Heterocycles in Peptidomimetics and Pseudopeptides: Design and Synthesis. Pharmaceutical 2012, 5, 297. [CrossRef]
- Chiummiento, L.; Funicello, M.; Lupattelli, P.; Tramutola, F. Ligand-Free Suzuki Coupling of Arylboronic Acids with Methyl (E)-4-Bromobut-2-enoate: Synthesis of Unconventional Cores of HIV-1 Protease Inhibitors. Org. Lett. 2012, 14, 3928-3931. [CrossRef]
- Tramutola, F.; Chiummiento, L.; Funicello, M.; Lupattelli, P. Practical and efficient ipso-iodination of arylboronic acids via KF/I2 system. Tetrahedron Lett. 2015, 56, 1122-1123. [CrossRef]
- Bonini, C.; Chiummiento, L.; De Bonis, M.; Funicello, M.; Lupattelli, P.; Suanno, G.; Berti, F.; Campaner, P. Synthesis, biological activity and modelling studies of two novel anti HIV PR inhibitors with a thiophene containing hydroxyethylamino core, Tetrahedron 2005, 61, 6580-6589. [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules.Sci. Rep. 2017, 7, 42717. [CrossRef]
| Entry | Inhibitor | IC50 (nM) | Entry | Inhibitor | IC50 (nM) |
|---|---|---|---|---|---|
| 1 |
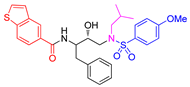 5a |
< 0.6 | 4 |
 6a |
< 0.6 |
| 2 |
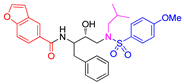 5b |
< 0.6 n | 5 |
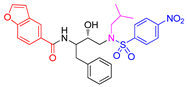 6b |
4.0 ± 0.5 |
| 3 |
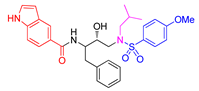 5c |
95 ± 7 | 6 |
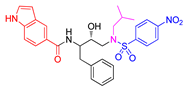 6c |
0.60 ± 0.03 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





