Submitted:
16 May 2023
Posted:
18 May 2023
You are already at the latest version
Abstract
Keywords:
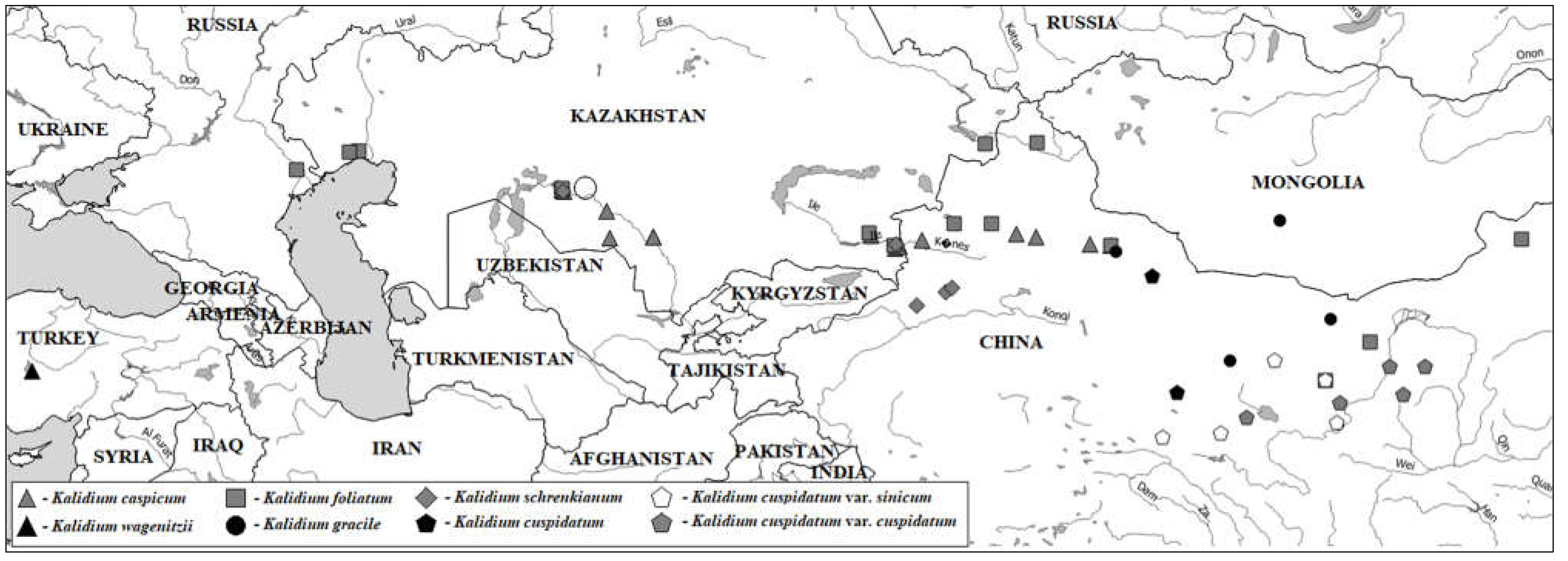
1. Introduction
2. Results
2.1. Flow cytometry
2.2. Molecular phylogeny
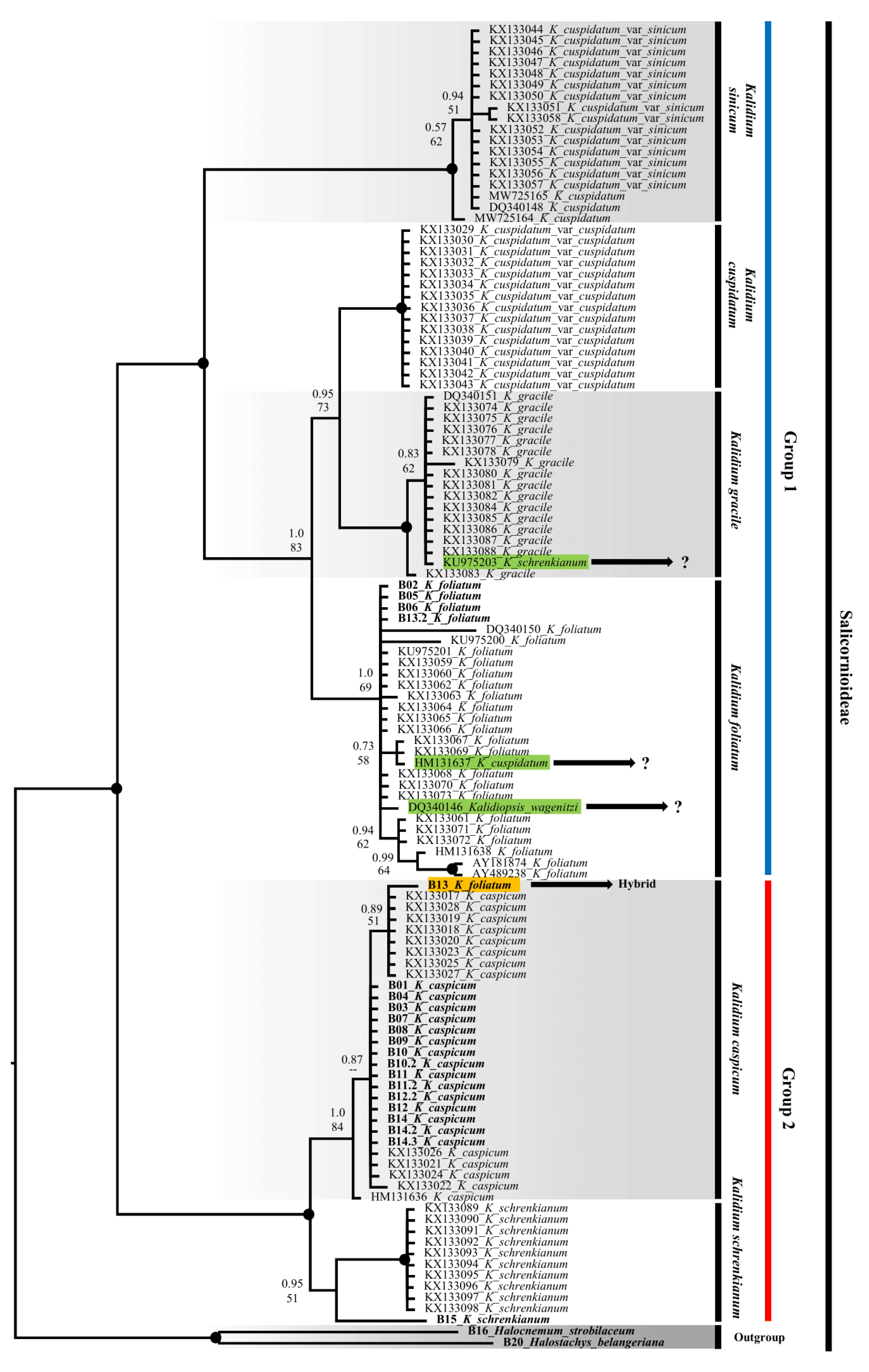
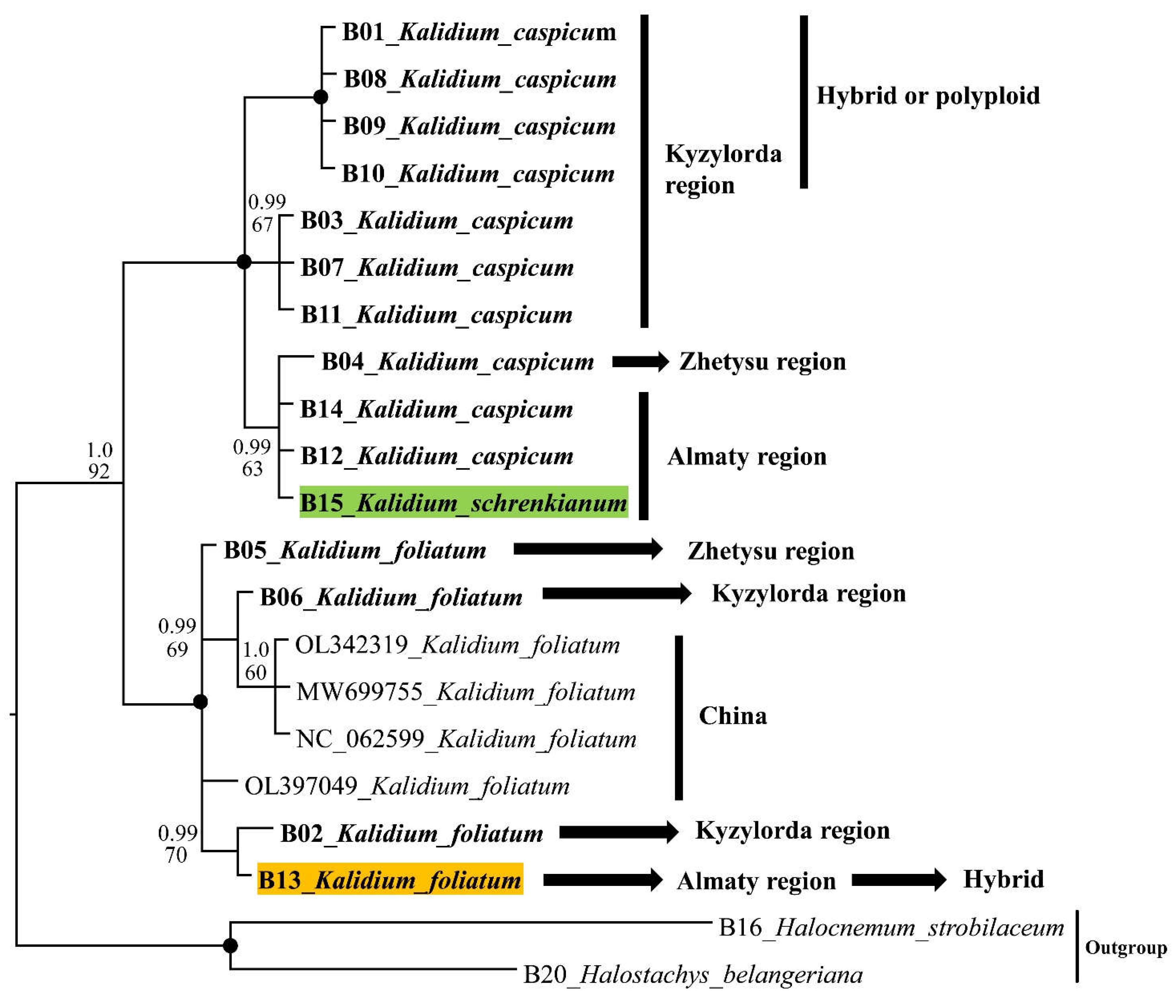
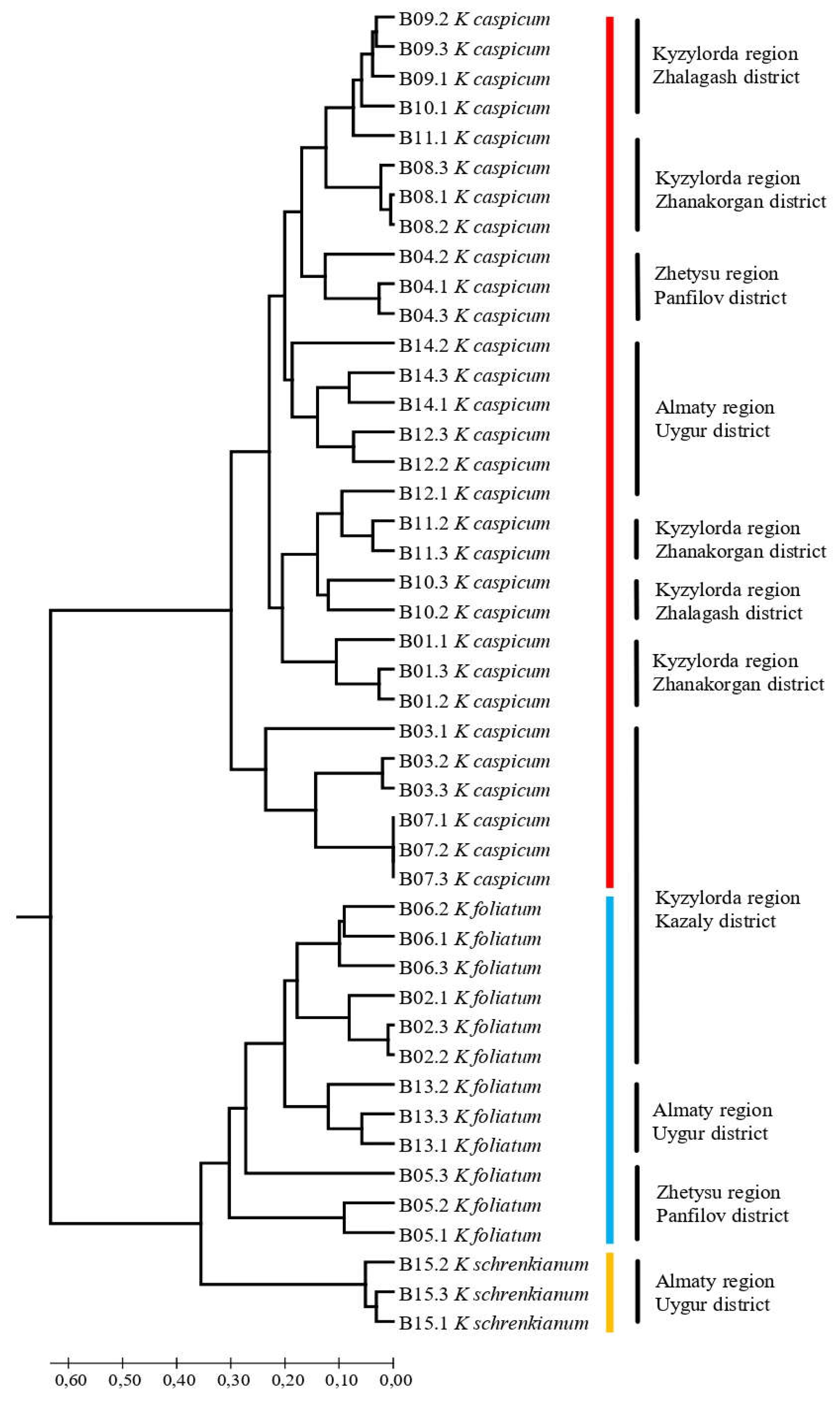
2.3. SCoT results
3. Discussion
4. Materials and Methods
4.1. Flow cytometry
4.2. Molecular genetics methods
4.4. Phylogenetic analyses
4.5. The start codon targeted (SCoT) method
5. Conclusions
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kadereit, G.; Mucina, L.; Freitag, H. Phylogeny of Salicornioideae (Chenopodiaceae): diversification, biogeography, and evolutionary trends in leaf and flower morphology. Taxon 2006, 55, 6. [Google Scholar] [CrossRef]
- Piirainen, M.; Liebisch, O.; Kadereit, G. Phylogeny, biogeography, systematics and taxonomy of Salicornioideae (Amaranthaceae/Chenopodiaceae) – A cosmopolitan, highly specialized hygrohalophyte lineage dating back to the Oligocene. Taxon 2017, 66, 109–132. [Google Scholar] [CrossRef]
- Vesselova, P.V.; Kudabayeva, G.M. Comparative analysis of floristic lists of the desert part of the Syr Darya valley. Ukrainian Journal of Ecology 2018, 8, 427–432. [Google Scholar]
- Chapman, V.J. The New Perspective in the Halophytes. Q. Rev. Biol. 1942, 17, 291–311. [Google Scholar] [CrossRef]
- Shreve, F. Vegetation of the Sonoran Desert. Carnegie Institution of Washington Publication 1951, 591, 1–192. [Google Scholar]
- Wilder, B.T.; Felger, R.S.; Morales, H.R. Succulent Plant Diversity Of the Sonoran Islands, Gulf Of California, Mexico. Haseltonia 2008, 14, 127–160. [Google Scholar] [CrossRef]
- Kung, H.-W.; Chu, G.-L.; Tsien, C.-P.; Li, A.-J.; Ma, C.-G. The Chenopodiaceae in China. Acta Phytotaxonomica Sinica. (In Chinese). 1978, 16, 99–123. [Google Scholar]
- Zhao, Y.-Z.; Zhao, L.-Q.; Cao, R. Flora Intramongolica. 3rd ed. Vol. 1. Huhhot: Typis Intramongolicae Popularis. 2020, 779 pp.
- Sukhorukov, P.; Kushunina, M.A.; Lomonosova, M.N. A new Kalidium species (Amaranthaceae s. l.) from northern Central Asia. Turczaninowia 2022, 25, 24–33. [Google Scholar] [CrossRef]
- Sukhorukov, A.P.; Kushunina, M.A. Taxonomic revision of Chenopodiaceae in Nepal. Phytotaxa 2014, 191, 10–44. [Google Scholar] [CrossRef]
- Zhao, K.F.; Fan, H.; Jiang, X.Y.; et al. Improvement and utiliza- tion of saline soil by planting halophytes. Chinese Journal of Applied & Environmental Biology 2002, 8, 31–35. (in Chinese). [Google Scholar]
- Zhou, Z.Y.; Yan, S.Y.; Qin, Y.; et al. The character of shrub di- versity in arid desert regions in Alashan. Journal of Arid Land Resources and Environment 2009, 23, 146–150. [Google Scholar] [CrossRef]
- Liang, X.H.; Wu, Y.X. Identification of Kalidium species (Chenopodiaceae) by DNA barcoding. Sciences in Cold and Arid Regions 2017, 9, 0089–0096. [Google Scholar] [CrossRef]
- Wang, G.; Jia, J. Effects of NaCl on physiology and leaf ultrastructure in the halophyte Kalidium foliatum. Nordic Journal of Botany 2015, 33, 232–238. [Google Scholar] [CrossRef]
- Tobe, K. Seed Germination and Radicle Growth of a Halophyte, Kalidium capsicum (Chenopodiaceae). Annals of Botany 2000, 85, 391–396. [Google Scholar] [CrossRef]
- Tobe, K.; Li, X.; Omasa, K. Effects of sodium, magnesium and calcium salts on seed germination and radicle survival of a halophyte, Kalidium caspicum (Chenopodiaceae). Aust. J. Bot. 2002, 50, 163–169. [Google Scholar] [CrossRef]
- Song, J.; Feng, G.; Zhang, F. Salinity and Temperature Effects on Germination for Three Salt-resistant Euhalophytes, Halostachys caspica, Kalidium foliatum and Halocnemum strobilaceum. Plant and Soil 2006, 279, 201–207. [Google Scholar] [CrossRef]
- Qu, X.; Baskin, J.M.; Wang, L.; Huang, Z. Effects of cold stratification, temperature, light and salinity on seed germination and radicle growth of the desert halophyte shrub, Kalidium caspicum (Chenopodiaceae). Plant Growth Regulation 2007, 54, 241–248. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, D.Y.; Huang, Z.Y.; Tian, C.Y. Factors influencing seed germination of Kalidium caspicum (Chenopodiaceae), a halophytic desert shrub of Xinjiang, China. Seed Sci. and Technol. 2009, 37, 281–290. [Google Scholar] [CrossRef]
- Gong, D.H.; Wang, G.Z.; Si, W.T.; Zhou, Y.; Liu, Z.; Jia, J. Effects of Salt Stress on Photosynthetic Pigments and Activity of Ribulose-1,5-bisphosphate Carboxylase/Oxygenase in Kalidium foliatum. Russian Journal of Plant Physiology 2018, 65, 98–103. [Google Scholar] [CrossRef]
- Wang, Z.G.; Zhang, P.X.; Shao, Y.T.; Xu, T.T.; Jia, X.Y.; Zhang, X.Q.; Jia, J. Molecular Cloning and the Expression Pattern of a Phospholipid Hydroperoxide Glutathione Peroxidase in Kalidium foliatum under NaCl Treatment. Russian Journal of Plant Physiology 2020, 67, 750–757. [Google Scholar] [CrossRef]
- Wang, J.; Wu, S.; Wang, M. The complete chloroplast genome of halophyte Kalidiumfoliatum (Pall. ) Moq., a dominant species of desert grassland, Mitochondrial DNA Part B. 2022, 7, 544–545. [Google Scholar] [CrossRef] [PubMed]
- Schutze, P.; Freitag, H.; Weising, K. An integrated molecular and morphological study of the subfamily Suaedoideae Ulbr. (Chenopodiaceae). Plant Syst. Evol. 2003, 239, 257–286. [Google Scholar] [CrossRef]
- Wen, Z.-B.; Zhang, M.-L.; Zhu, G.-L.; Sanderson, S.C. Phylogeny of Salsoleae s. l. (Chenopodiaceae) based on DNA sequence data from ITS, psbB–psbH, and rbcL, with emphasis on taxa of northwestern China. Plant Syst. Evol. 2010, 288, 25–42. [Google Scholar] [CrossRef]
- Xue, J.J.; Zhang, M.L. Monophyly and infrageneric variation of Corispermum L. (Chenopodiaceae), evidence from sequence data psbB-psbH, rbcL and ITS. Journal of Arid Land 2011, 3, 240–253. [Google Scholar] [CrossRef]
- Lu, K.-Q.; Li, M.; Wang, G.-H.; Xu, L.-S.; Ferguson, D.K.; Trivedi, A.; Wang, Y.-F. A new pollen classification of Chenopodiaceae for exploring and tracing desert vegetation evolution in the eastern central Asia. Journal of Systematics and Evolution 2018. [CrossRef]
- Ilyin, M.M. The genus Kalidium Moq. Flora of the USSR. - М., 1936, VI, p. 166-168. [in Russian].
- Goloskokov, V.P. and Polyakov, P.P. The genus Kalidium Moq. Alma-Ata. 1960, III, p. 243-246. [in Russian].
- Goloskokov, V.P. and Polyakov, P.P. The genus Kalidium Moq. Illustrated Plant Identifier of Kazakhstan. Alma-Ata. ed. of the Kazakh SSR Academy of Sciences 1969, Т 1, p. 300-301. [in Russian].
- Pratov, U. The genus Kalidium Moq. Plant Identifier of Plants of Central Asia. Tashkent. FAN Publishing House. 1972, III, p. 70-72. [in Russian].
- Abdulina, S.A. Рoд Kalidium Moq. Checklist of vascular plants of Kazakhstan. Almaty. 1998, p. 77. (in Russian).
- Baitenov, M.S. Genus Kalidium Moq. Flora of Kazakhstan. Genus complex flora. Almaty. 2001, 2, p. 72. (in Russian).
- Skaptsov, M.V. and Kutsev, M.G. Possibilities of flow cytometry in modern plant science. "Problems of botany of South Siberia and Mongolia" - XIII International Scientific-Practical Conference. Barnaul. 2014, p. 204-207. (in Russian)
- Friesen, N. Molecular methods used in plant systematics. Barnaul. AzBuka, 64 p. (in Russian). 2007. [Google Scholar]
- Lomonosova, M.N. and Krasnikov, A.A. Chromosome numbers in some members of the Chenopodiaceae. Bot. Zhurn. 1993, 78, 158–159. [Google Scholar]
- Lomonosova, M.N. and Krasnikov, A.A. Chromosome numbers of some Chenopodiaceae representatives of the flora of Russia. Bot. Zhurn. 2006, 91, 1757–1759. [Google Scholar]
- Krasnikov, A. and Schaulo, D. Karyological study of the Tuva Republic flora: a summary. Turczaninowia 2004, 7, 82–95. [Google Scholar]
- Marhold, K. IAPT/IOPB chromosome data 13. Taxon 2012, 61, 892–893. [Google Scholar] [CrossRef]
- Marhold, K. IAPT/IOPB chromosome data 18. Taxon 2014, 63, 1387–1393. [Google Scholar] [CrossRef]
- Marhold, K.; Kucera, J.; Alexeeva, T.V.; Andriyanova, E.A.; Ankova, T.V.; Astashenkov, A.Y.; … Zavgorodnyaya, O.Y. IAPT chromosome data 32. Taxon 2020. [CrossRef]
- Lomonosova, M.N.; Krasnikov, A.A.; Krasnikova, S.A. Chromosome numbers of Chenopodiaceae family members of the Kazakhstan flora. Bot. Zhurn. 2003, 88, 134–135. [Google Scholar]
- Ghaffari, S.M.; Saydrasi, L.; Ebrahimzadeh, H.; Akhani, H. Chromosome numbers and karyotype analyses of species of subfamily Salicornioideae (Chenopodiaceae) from Iran. Iran. J. Bot. 2006, 12, 128–135. [Google Scholar]
- Zhu, G.; Sergei, L.M.; Stevan, E. Chenopodiaceae: Flora of China. 2003, 5, 351–414.
- Skvortsov, A.K. Herbarium: A Manual on Methods and Techniques. Prof. L.I. Prilipko M. Nauka, 1977. 199 p. (in Russian) [Google Scholar]
- Pfosser, М.; Amon, A.; Lelley, Т.; Heberle-Bors, E. Evaluation of sensitivity of flow cytometry in detecting aneuploidy in wheat using disomic and ditelosomic wheat-rye addition lines. Cytometry 1995, 21, 387–393. [Google Scholar] [CrossRef]
- Blattner, F.R. Direct amplification of the entire ITS region from poorly preserved plant material using recombinant PCR. In. Biotechniques 1999, 27, 1180–1186. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Shinsky JJ, White TJ, editors. PCR Protocols: A Guide to Methods and Applications. 1990, 315–322.
- Shaw, J.; Lickey, E.B.; Schilling, E.E.; Small, R.L. Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: the tortoise and the hare III. American Journal of Botany 2007, 94, 275–288. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The clustal X window interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Swofford, D. –L. PAUP*: Phylogenetic analysis using parsimony (* and other methods). version 4 (Sunderland, Massachusetts: Sinauer Associates). 2002.
- Kluge, A.G.; Farris, J.S. Quantitative phyletics and the evolution of anurans. Syst. Zool. 1969, 18, l–32. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, R. and Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef] [PubMed]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 77. [Google Scholar] [CrossRef]
- Bertrand, C.; Collard, Y.; Mackill, D.J. Start Codon Targeted (SCoT) Polymorphism: A Simple, Novel DNA Marker Technique for Generating Gene-Targeted Markers in Plants. Plant Mol. Biol. Rep. 2009, 27, 86–93. [Google Scholar] [CrossRef]

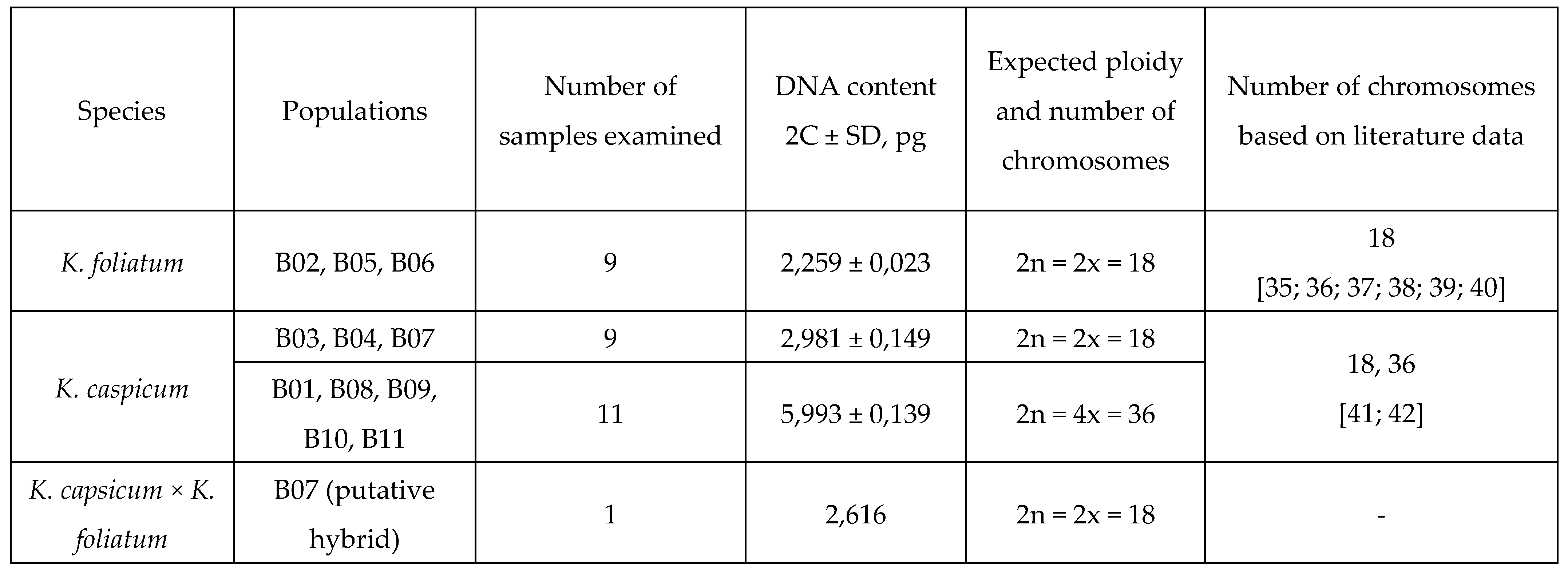 |
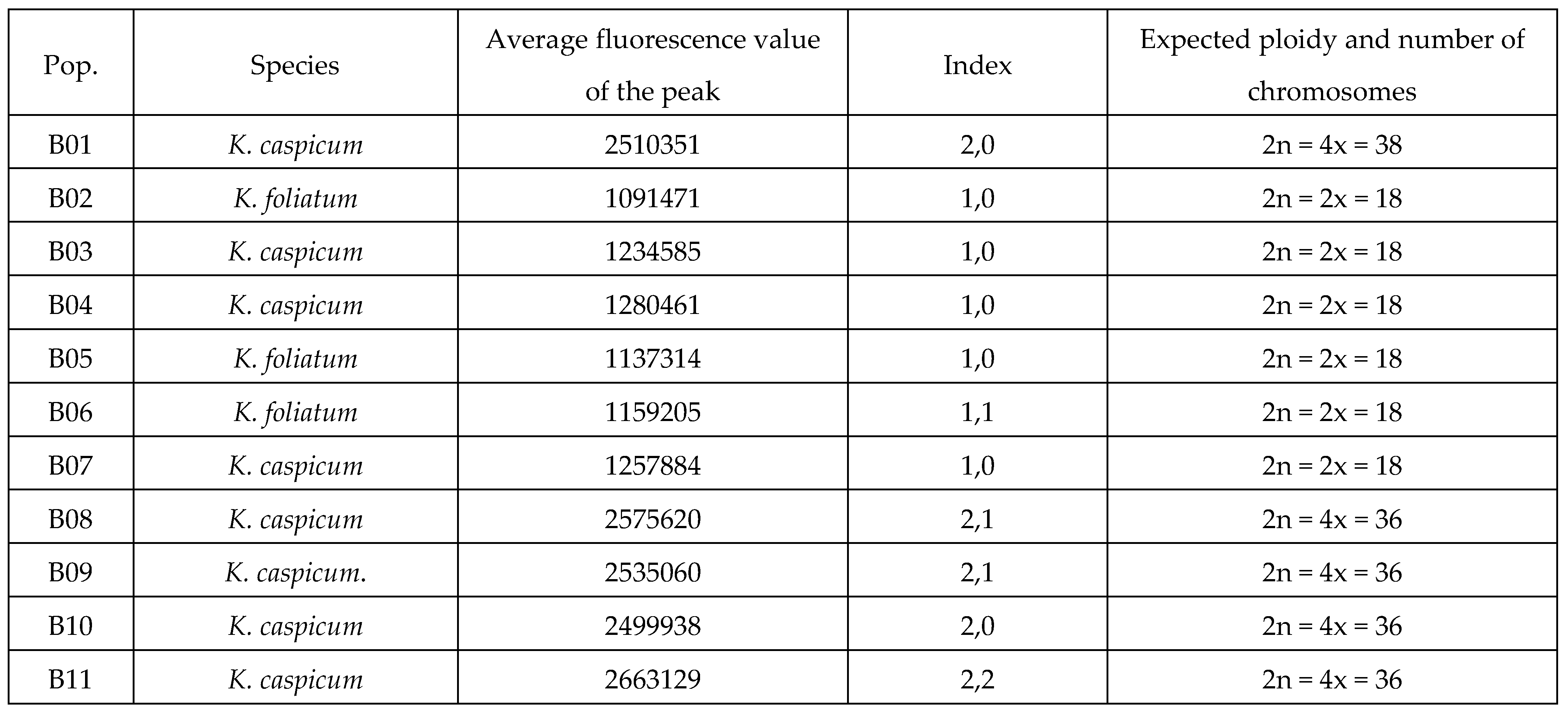 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





