Submitted:
19 May 2023
Posted:
19 May 2023
You are already at the latest version
Abstract
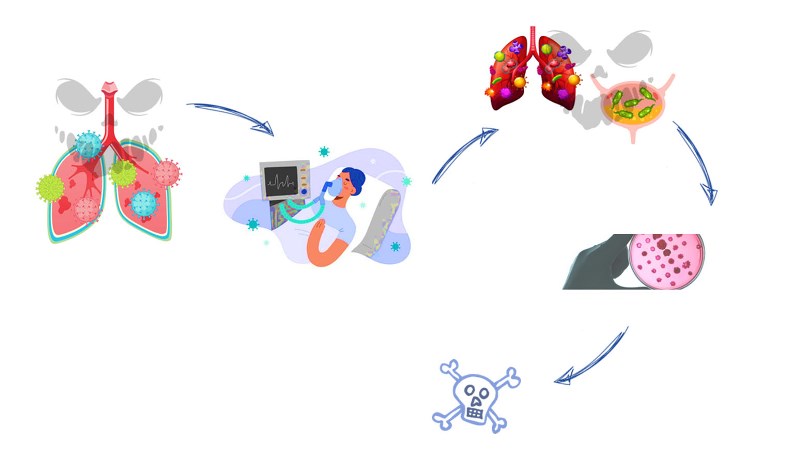
Keywords:
1. Introduction
2. Methods
2.1. The impact of the coronavirus disease 2019 (COVID-19)
2.2. The role of coinfections in patients with COVID-19
2.3. Multidrug-resistant bacterial infections in COVID-19 patients admitted to intensive care units
3. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Liu, Y.; Ling, L.; Wong, S.H.; Wang, M.H.; Fitzgerald, J.R.; Zou, X.; Fang, S.; Liu, X.; Wang, X.; Hu, W.; et al. Outcomes of respiratory viral-bacterial co-infection in adult hospitalized patients. EClinicalMedicine 2021, 37, 100955. [CrossRef]
- Arnold, F.W.; Fuqua, J.L. Viral respiratory infections: a cause of community-acquired pneumonia or a predisposing factor? Current opinion in pulmonary medicine 2020, 26, 208-214. [CrossRef]
- Klugman, K.P.; Astley, C.M.; Lipsitch, M. Time from illness onset to death, 1918 influenza and pneumococcal pneumonia. Emerging infectious diseases 2009, 15, 346-347. [CrossRef]
- Hussell, T.; Wissinger, E.; Goulding, J. Bacterial complications during pandemic influenza infection. Future microbiology 2009, 4, 269-272. [CrossRef]
- Feldman, C.; Anderson, R. The role of co-infections and secondary infections in patients with COVID-19. Pneumonia 2021, 13, 5. [CrossRef]
- Kilbourne, E.D. Influenza pandemics of the 20th century. Emerging infectious diseases 2006, 12, 9-14. [CrossRef]
- McFee, R.B. Nosocomial or hospital-acquired infections: an overview. Disease-a-month : DM 2009, 55, 422-438. [CrossRef]
- Raoofi, S.; Pashazadeh Kan, F.; Rafiei, S.; Hosseinipalangi, Z.; Noorani Mejareh, Z.; Khani, S.; Abdollahi, B.; Seyghalani Talab, F.; Sanaei, M.; Zarabi, F.; et al. Global prevalence of nosocomial infection: A systematic review and meta-analysis. PloS one 2023, 18, e0274248. [CrossRef]
- Fattorini, L.; Creti, R.; Palma, C.; Pantosti, A.; Unit of Antibiotic, R.; Special, P.; Unit of Antibiotic, R.; Special Pathogens of the Department of Infectious Diseases, I.S.d.S.R. Bacterial coinfections in COVID-19: an underestimated adversary. Annali dell'Istituto superiore di sanita 2020, 56, 359-364. [CrossRef]
- Wong, K.C.; Leung, K.S. Transmission and prevention of occupational infections in orthopaedic surgeons. The Journal of bone and joint surgery. American volume 2004, 86, 1065-1076. [CrossRef]
- Szabo, S.; Feier, B.; Capatina, D.; Tertis, M.; Cristea, C.; Popa, A. An Overview of Healthcare Associated Infections and Their Detection Methods Caused by Pathogen Bacteria in Romania and Europe. Journal of clinical medicine 2022, 11. [CrossRef]
- Hassan, R.; El-Gilany, A.H.; Abd Elaal, A.M.; El-Mashad, N.; Azim, D.A. An overview of healthcare-associated infections in a tertiary care hospital in Egypt. Infection prevention in practice 2020, 2, 100059. [CrossRef]
- Alrebish, S.A.; Yusufoglu, H.S.; Alotibi, R.F.; Abdulkhalik, N.S.; Ahmed, N.J.; Khan, A.H. Epidemiology of Healthcare-Associated Infections and Adherence to the HAI Prevention Strategies. Healthcare 2022, 11. [CrossRef]
- Sahu, T.; Verma, H.K.; Bhaskar, L. Bacterial and fungal co-infection is a major barrier in COVID-19 patients: A specific management and therapeutic strategy is required. World journal of virology 2022, 11, 107-110. [CrossRef]
- Suetens, C.; Latour, K.; Karki, T.; Ricchizzi, E.; Kinross, P.; Moro, M.L.; Jans, B.; Hopkins, S.; Hansen, S.; Lyytikainen, O.; et al. Prevalence of healthcare-associated infections, estimated incidence and composite antimicrobial resistance index in acute care hospitals and long-term care facilities: results from two European point prevalence surveys, 2016 to 2017. Euro surveillance : bulletin Europeen sur les maladies transmissibles = European communicable disease bulletin 2018, 23. [CrossRef]
- Hassan Ahmed Khan, F.K.B., Riffat Mehboob. Nosocomial infections: Epidemiology, prevention, control and surveillance. Asian Pacific Journal of Tropical Biomedicine 2017, 7, 4. [CrossRef]
- Munyeshyaka, E.; Cyuzuzo, P.; Yadufashije, C.; Karemera, J. Contribution of Medical Wards Contamination to Wound Infection among Patients Attending Ruhengeri Referral Hospital. International journal of microbiology 2021, 2021, 7838763. [CrossRef]
- Klevens, R.M.; Edwards, J.R.; Richards, C.L., Jr.; Horan, T.C.; Gaynes, R.P.; Pollock, D.A.; Cardo, D.M. Estimating health care-associated infections and deaths in U.S. hospitals, 2002. Public health reports 2007, 122, 160-166. [CrossRef]
- Biondo, C. New Insights into Bacterial Pathogenesis. Pathogens 2022, 12. [CrossRef]
- Biondo, C. Bacterial Antibiotic Resistance: The Most Critical Pathogens. Pathogens 2023, 12. [CrossRef]
- Biondo, C.; Midiri, A.; Gerace, E.; Zummo, S.; Mancuso, G. SARS-CoV-2 Infection in Patients with Cystic Fibrosis: What We Know So Far. Life 2022, 12. [CrossRef]
- Dhama, K.; Khan, S.; Tiwari, R.; Sircar, S.; Bhat, S.; Malik, Y.S.; Singh, K.P.; Chaicumpa, W.; Bonilla-Aldana, D.K.; Rodriguez-Morales, A.J. Coronavirus Disease 2019-COVID-19. Clinical microbiology reviews 2020, 33. [CrossRef]
- Baker, M.A.; Sands, K.E.; Huang, S.S.; Kleinman, K.; Septimus, E.J.; Varma, N.; Blanchard, J.; Poland, R.E.; Coady, M.H.; Yokoe, D.S.; et al. The Impact of Coronavirus Disease 2019 (COVID-19) on Healthcare-Associated Infections. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America 2022, 74, 1748-1754. [CrossRef]
- Dobrovic, K.; Skrobo, T.; Selec, K.; Jelic, M.; Civljak, R.; Persec, J.; Sakan, S.; Busic, N.; Mihelcic, A.; Hleb, S.; et al. Healthcare-Associated Bloodstream Infections Due to Multidrug-Resistant Acinetobacter baumannii in COVID-19 Intensive Care Unit: A Single-Center Retrospective Study. Microorganisms 2023, 11. [CrossRef]
- Bazaid, A.S.; Barnawi, H.; Qanash, H.; Alsaif, G.; Aldarhami, A.; Gattan, H.; Alharbi, B.; Alrashidi, A.; Al-Soud, W.A.; Moussa, S.; et al. Bacterial Coinfection and Antibiotic Resistance Profiles among Hospitalised COVID-19 Patients. Microorganisms 2022, 10. [CrossRef]
- Grasselli, G.; Cattaneo, E.; Florio, G. Secondary infections in critically ill patients with COVID-19. Critical care 2021, 25, 317. [CrossRef]
- Westblade, L.F.; Simon, M.S.; Satlin, M.J. Bacterial Coinfections in Coronavirus Disease 2019. Trends in microbiology 2021, 29, 930-941. [CrossRef]
- World Health Organization (WHO) Coronavirus (COVID-19) Dashboard. 2023.
- Zeng, H.; Ma, Y.; Zhou, Z.; Liu, W.; Huang, P.; Jiang, M.; Liu, Q.; Chen, P.; Luo, H.; Chen, Y. Spectrum and Clinical Characteristics of Symptomatic and Asymptomatic Coronavirus Disease 2019 (COVID-19) With and Without Pneumonia. Frontiers in medicine 2021, 8, 645651. [CrossRef]
- Gain, C.; Song, S.; Angtuaco, T.; Satta, S.; Kelesidis, T. The role of oxidative stress in the pathogenesis of infections with coronaviruses. Frontiers in microbiology 2022, 13, 1111930. [CrossRef]
- Paidas, M.J.; Sampath, N.; Schindler, E.A.; Cosio, D.S.; Ndubizu, C.O.; Shamaladevi, N.; Kwal, J.; Rodriguez, S.; Ahmad, A.; Kenyon, N.S.; et al. Mechanism of Multi-Organ Injury in Experimental COVID-19 and Its Inhibition by a Small Molecule Peptide. Frontiers in pharmacology 2022, 13, 864798. [CrossRef]
- Samadizadeh, S.; Masoudi, M.; Rastegar, M.; Salimi, V.; Shahbaz, M.B.; Tahamtan, A. COVID-19: Why does disease severity vary among individuals? Respiratory medicine 2021, 180, 106356. [CrossRef]
- Yildirim, Z.; Sahin, O.S.; Yazar, S.; Bozok Cetintas, V. Genetic and epigenetic factors associated with increased severity of Covid-19. Cell biology international 2021, 45, 1158-1174. [CrossRef]
- Ganguli, S.; Howlader, S.; Dey, K.; Barua, S.; Islam, M.N.; Aquib, T.I.; Partho, P.B.; Chakraborty, R.R.; Barua, B.; Hawlader, M.D.H.; et al. Association of comorbidities with the COVID-19 severity and hospitalization: A study among the recovered individuals in Bangladesh. International journal of health sciences 2022, 16, 30-45.
- Brodin, P. Immune determinants of COVID-19 disease presentation and severity. Nature medicine 2021, 27, 28-33. [CrossRef]
- Mustafa, Z.U.; Tariq, S.; Iftikhar, Z.; Meyer, J.C.; Salman, M.; Mallhi, T.H.; Khan, Y.H.; Godman, B.; Seaton, R.A. Predictors and Outcomes of Healthcare-Associated Infections among Patients with COVID-19 Admitted to Intensive Care Units in Punjab, Pakistan; Findings and Implications. Antibiotics 2022, 11. [CrossRef]
- Bardi, T.; Pintado, V.; Gomez-Rojo, M.; Escudero-Sanchez, R.; Azzam Lopez, A.; Diez-Remesal, Y.; Martinez Castro, N.; Ruiz-Garbajosa, P.; Pestana, D. Nosocomial infections associated to COVID-19 in the intensive care unit: clinical characteristics and outcome. European journal of clinical microbiology & infectious diseases : official publication of the European Society of Clinical Microbiology 2021, 40, 495-502. [CrossRef]
- Ak, O.; Batirel, A.; Ozer, S.; Colakoglu, S. Nosocomial infections and risk factors in the intensive care unit of a teaching and research hospital: a prospective cohort study. Medical science monitor : international medical journal of experimental and clinical research 2011, 17, PH29-34. [CrossRef]
- Wang, Y.; Ren, J.; Yao, Z.; Wang, W.; Wang, S.; Duan, J.; Li, Z.; Zhang, H.; Zhang, R.; Wang, X. Clinical Impact and Risk Factors of Intensive Care Unit-Acquired Nosocomial Infection: A Propensity Score-Matching Study from 2018 to 2020 in a Teaching Hospital in China. Infection and drug resistance 2023, 16, 569-579. [CrossRef]
- Suleyman, G.; Alangaden, G.; Bardossy, A.C. The Role of Environmental Contamination in the Transmission of Nosocomial Pathogens and Healthcare-Associated Infections. Current infectious disease reports 2018, 20, 12. [CrossRef]
- Yin, X.; Xu, X.; Li, H.; Jiang, N.; Wang, J.; Lu, Z.; Xiong, N.; Gong, Y. Evaluation of early antibiotic use in patients with non-severe COVID-19 without bacterial infection. International journal of antimicrobial agents 2022, 59, 106462. [CrossRef]
- Zhang, H.; Zhang, Y.; Wu, J.; Li, Y.; Zhou, X.; Li, X.; Chen, H.; Guo, M.; Chen, S.; Sun, F.; et al. Risks and features of secondary infections in severe and critical ill COVID-19 patients. Emerging microbes & infections 2020, 9, 1958-1964. [CrossRef]
- Vijay, S.; Bansal, N.; Rao, B.K.; Veeraraghavan, B.; Rodrigues, C.; Wattal, C.; Goyal, J.P.; Tadepalli, K.; Mathur, P.; Venkateswaran, R.; et al. Secondary Infections in Hospitalized COVID-19 Patients: Indian Experience. Infection and drug resistance 2021, 14, 1893-1903. [CrossRef]
- De Bruyn, A.; Verellen, S.; Bruckers, L.; Geebelen, L.; Callebaut, I.; De Pauw, I.; Stessel, B.; Dubois, J. Secondary infection in COVID-19 critically ill patients: a retrospective single-center evaluation. BMC infectious diseases 2022, 22, 207. [CrossRef]
- Ripa, M.; Galli, L.; Poli, A.; Oltolini, C.; Spagnuolo, V.; Mastrangelo, A.; Muccini, C.; Monti, G.; De Luca, G.; Landoni, G.; et al. Secondary infections in patients hospitalized with COVID-19: incidence and predictive factors. Clinical microbiology and infection : the official publication of the European Society of Clinical Microbiology and Infectious Diseases 2021, 27, 451-457. [CrossRef]
- Adler, H.; Ball, R.; Fisher, M.; Mortimer, K.; Vardhan, M.S. Low rate of bacterial co-infection in patients with COVID-19. The Lancet. Microbe 2020, 1, e62. [CrossRef]
- Gupta, R.K.; George, R.; Nguyen-Van-Tam, J.S. Bacterial pneumonia and pandemic influenza planning. Emerging infectious diseases 2008, 14, 1187-1192. [CrossRef]
- Garg, S.K. Antibiotic misuse during COVID-19 Pandemic: A Recipe for Disaster. Indian journal of critical care medicine : peer-reviewed, official publication of Indian Society of Critical Care Medicine 2021, 25, 617-619. [CrossRef]
- Papst, L.; Luzzati, R.; Carevic, B.; Tascini, C.; Gorisek Miksic, N.; Vlahovic Palcevski, V.; Djordjevic, Z.M.; Simonetti, O.; Sozio, E.; Lukic, M.; et al. Antimicrobial Use in Hospitalised Patients with COVID-19: An International Multicentre Point-Prevalence Study. Antibiotics 2022, 11. [CrossRef]
- Liang, S.T.; Liang, L.T.; Rosen, J.M. COVID-19: a comparison to the 1918 influenza and how we can defeat it. Postgraduate medical journal 2021, 97, 273-274. [CrossRef]
- Owolabi, M.; Ali, R.; Dacosta, J.; Muhanna, A.; Slim, J. When Influenza, Bacterial Pneumonia, and COVID-19 Co-exist. Cureus 2022, 14, e32686. [CrossRef]
- Huttner, B.D.; Catho, G.; Pano-Pardo, J.R.; Pulcini, C.; Schouten, J. COVID-19: don't neglect antimicrobial stewardship principles! Clinical microbiology and infection : the official publication of the European Society of Clinical Microbiology and Infectious Diseases 2020, 26, 808-810. [CrossRef]
- Bendala Estrada, A.D.; Calderon Parra, J.; Fernandez Carracedo, E.; Muino Miguez, A.; Ramos Martinez, A.; Munez Rubio, E.; Rubio-Rivas, M.; Agudo, P.; Arnalich Fernandez, F.; Estrada Perez, V.; et al. Inadequate use of antibiotics in the covid-19 era: effectiveness of antibiotic therapy. BMC infectious diseases 2021, 21, 1144. [CrossRef]
- Bergmann, F.; Gabler, C.; Nussbaumer-Proll, A.; Wolfl-Duchek, M.; Blaschke, A.; Radtke, C.; Zeitlinger, M.; Jorda, A. Early Bacterial Coinfections in Patients Admitted to the ICU With COVID-19 or Influenza: A Retrospective Cohort Study. Critical care explorations 2023, 5, e0895. [CrossRef]
- Vaughn, V.M.; Gandhi, T.N.; Petty, L.A.; Patel, P.K.; Prescott, H.C.; Malani, A.N.; Ratz, D.; McLaughlin, E.; Chopra, V.; Flanders, S.A. Empiric Antibacterial Therapy and Community-onset Bacterial Coinfection in Patients Hospitalized With Coronavirus Disease 2019 (COVID-19): A Multi-hospital Cohort Study. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America 2021, 72, e533-e541. [CrossRef]
- Parasher, A. COVID-19: Current understanding of its Pathophysiology, Clinical presentation and Treatment. Postgraduate medical journal 2021, 97, 312-320. [CrossRef]
- Bigdelou, B.; Sepand, M.R.; Najafikhoshnoo, S.; Negrete, J.A.T.; Sharaf, M.; Ho, J.Q.; Sullivan, I.; Chauhan, P.; Etter, M.; Shekarian, T.; et al. COVID-19 and Preexisting Comorbidities: Risks, Synergies, and Clinical Outcomes. Frontiers in immunology 2022, 13, 890517. [CrossRef]
- Russell, C.D.; Lone, N.I.; Baillie, J.K. Comorbidities, multimorbidity and COVID-19. Nature medicine 2023, 29, 334-343. [CrossRef]
- Pourajam, S.; Kalantari, E.; Talebzadeh, H.; Mellali, H.; Sami, R.; Soltaninejad, F.; Amra, B.; Sajadi, M.; Alenaseri, M.; Kalantari, F.; et al. Secondary Bacterial Infection and Clinical Characteristics in Patients With COVID-19 Admitted to Two Intensive Care Units of an Academic Hospital in Iran During the First Wave of the Pandemic. Frontiers in cellular and infection microbiology 2022, 12, 784130. [CrossRef]
- Xu, Y.; Xu, Z.; Liu, X.; Cai, L.; Zheng, H.; Huang, Y.; Zhou, L.; Huang, L.; Ling, Y.; Deng, L.; et al. Clinical Findings of COVID-19 Patients Admitted to Intensive Care Units in Guangdong Province, China: A Multicenter, Retrospective, Observational Study. Frontiers in medicine 2020, 7, 576457. [CrossRef]
- Riera, J.; Barbeta, E.; Tormos, A.; Mellado-Artigas, R.; Ceccato, A.; Motos, A.; Fernandez-Barat, L.; Ferrer, R.; Garcia-Gasulla, D.; Penuelas, O.; et al. Effects of intubation timing in patients with COVID-19 throughout the four waves of the pandemic: a matched analysis. The European respiratory journal 2023, 61. [CrossRef]
- Dancer, S.J. Reducing the risk of COVID-19 transmission in hospitals: focus on additional infection control strategies. Surgery 2021, 39, 752-758. [CrossRef]
- Patton, M.J.; Orihuela, C.J.; Harrod, K.S.; Bhuiyan, M.A.N.; Dominic, P.; Kevil, C.G.; Fort, D.; Liu, V.X.; Farhat, M.; Koff, J.L.; et al. COVID-19 bacteremic co-infection is a major risk factor for mortality, ICU admission, and mechanical ventilation. Critical care 2023, 27, 34. [CrossRef]
- SeyedAlinaghi, S.; Afzalian, A.; Pashaei, Z.; Varshochi, S.; Karimi, A.; Mojdeganlou, H.; Mojdeganlou, P.; Razi, A.; Ghanadinezhad, F.; Shojaei, A.; et al. Gut microbiota and COVID-19: A systematic review. Health science reports 2023, 6, e1080. [CrossRef]
- Din, A.U.; Mazhar, M.; Waseem, M.; Ahmad, W.; Bibi, A.; Hassan, A.; Ali, N.; Gang, W.; Qian, G.; Ullah, R.; et al. SARS-CoV-2 microbiome dysbiosis linked disorders and possible probiotics role. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie 2021, 133, 110947. [CrossRef]
- Ceparano, M.; Baccolini, V.; Migliara, G.; Isonne, C.; Renzi, E.; Tufi, D.; De Vito, C.; De Giusti, M.; Trancassini, M.; Alessandri, F.; et al. Acinetobacter baumannii Isolates from COVID-19 Patients in a Hospital Intensive Care Unit: Molecular Typing and Risk Factors. Microorganisms 2022, 10. [CrossRef]
- Montrucchio, G.; Corcione, S.; Lupia, T.; Shbaklo, N.; Olivieri, C.; Poggioli, M.; Pagni, A.; Colombo, D.; Roasio, A.; Bosso, S.; et al. The Burden of Carbapenem-Resistant Acinetobacter baumannii in ICU COVID-19 Patients: A Regional Experience. Journal of clinical medicine 2022, 11. [CrossRef]
- Abubakar, U.; Al-Anazi, M.; Alanazi, Z.; Rodriguez-Bano, J. Impact of COVID-19 pandemic on multidrug resistant gram positive and gram negative pathogens: A systematic review. Journal of infection and public health 2023, 16, 320-331. [CrossRef]
- Fanaei, V.; Validi, M.; Zamanzad, B.; Karimi, A. Isolation and identification of specific bacteriophages against methicillin-resistant Staphylococcus aureus, extended-spectrum beta-lactamases-producing Escherichia coli, extended-spectrum beta-lactamases-producing Klebsiella pneumoniae, and multidrug-resistant Acinetobacter baumanniiin vitro. FEMS microbiology letters 2021, 368. [CrossRef]
- Yahya, R.O. Problems Associated with Co-Infection by Multidrug-Resistant Klebsiella pneumoniae in COVID-19 Patients: A Review. Healthcare 2022, 10. [CrossRef]
- Kumar, S.; Anwer, R.; Azzi, A. Molecular typing methods & resistance mechanisms of MDR Klebsiella pneumoniae. AIMS microbiology 2023, 9, 112-130. [CrossRef]
- Rangel, K.; Chagas, T.P.G.; De-Simone, S.G. Acinetobacter baumannii Infections in Times of COVID-19 Pandemic. Pathogens 2021, 10. [CrossRef]
- Russo, A.; Gavaruzzi, F.; Ceccarelli, G.; Borrazzo, C.; Oliva, A.; Alessandri, F.; Magnanimi, E.; Pugliese, F.; Venditti, M. Multidrug-resistant Acinetobacter baumannii infections in COVID-19 patients hospitalized in intensive care unit. Infection 2022, 50, 83-92. [CrossRef]
- Pustijanac, E.; Hrenovic, J.; Vranic-Ladavac, M.; Mocenic, M.; Karcic, N.; Lazaric Stefanovic, L.; Hrstic, I.; Loncaric, J.; Seruga Music, M.; Drcelic, M.; et al. Dissemination of Clinical Acinetobacter baumannii Isolate to Hospital Environment during the COVID-19 Pandemic. Pathogens 2023, 12. [CrossRef]
- Bongiovanni, M.; Barda, B. Pseudomonas aeruginosa Bloodstream Infections in SARS-CoV-2 Infected Patients: A Systematic Review. Journal of clinical medicine 2023, 12. [CrossRef]
- Marua, A.M.; Shethwala, N.D.; Bhatt, P.; Shah, A. Evaluation of Bacterial Co-Infections and Antibiotic Resistance in Positive COVID-19 Patients. Maedica 2022, 17, 350-356. [CrossRef]
- Jeon, K.; Jeong, S.; Lee, N.; Park, M.J.; Song, W.; Kim, H.S.; Kim, H.S.; Kim, J.S. Impact of COVID-19 on Antimicrobial Consumption and Spread of Multidrug-Resistance in Bacterial Infections. Antibiotics 2022, 11. [CrossRef]
- American Thoracic, S.; Infectious Diseases Society of, A. Guidelines for the management of adults with hospital-acquired, ventilator-associated, and healthcare-associated pneumonia. American journal of respiratory and critical care medicine 2005, 171, 388-416. [CrossRef]
- Sreenath, K.; Batra, P.; Vinayaraj, E.V.; Bhatia, R.; SaiKiran, K.; Singh, V.; Singh, S.; Verma, N.; Singh, U.B.; Mohan, A.; et al. Coinfections with Other Respiratory Pathogens among Patients with COVID-19. Microbiology spectrum 2021, 9, e0016321. [CrossRef]
- Mutua, J.M.; Njeru, J.M.; Musyoki, A.M. Multidrug resistant bacterial infections in severely ill COVID-19 patients admitted in a national referral and teaching hospital, Kenya. BMC infectious diseases 2022, 22, 877. [CrossRef]
- Yoon, S.M.; Lee, J.; Lee, S.M.; Lee, H.Y. Incidence and clinical outcomes of bacterial superinfections in critically ill patients with COVID-19. Frontiers in medicine 2023, 10, 1079721. [CrossRef]
- Pascale, R.; Bussini, L.; Gaibani, P.; Bovo, F.; Fornaro, G.; Lombardo, D.; Ambretti, S.; Pensalfine, G.; Appolloni, L.; Bartoletti, M.; et al. Carbapenem-resistant bacteria in an intensive care unit during the coronavirus disease 2019 (COVID-19) pandemic: A multicenter before-and-after cross-sectional study. Infection control and hospital epidemiology 2022, 43, 461-466. [CrossRef]
- Magnasco, L.; Mikulska, M.; Giacobbe, D.R.; Taramasso, L.; Vena, A.; Dentone, C.; Dettori, S.; Tutino, S.; Labate, L.; Di Pilato, V.; et al. Spread of Carbapenem-Resistant Gram-Negatives and Candida auris during the COVID-19 Pandemic in Critically Ill Patients: One Step Back in Antimicrobial Stewardship? Microorganisms 2021, 9. [CrossRef]
- Tiri, B.; Sensi, E.; Marsiliani, V.; Cantarini, M.; Priante, G.; Vernelli, C.; Martella, L.A.; Costantini, M.; Mariottini, A.; Andreani, P.; et al. Antimicrobial Stewardship Program, COVID-19, and Infection Control: Spread of Carbapenem-Resistant Klebsiella Pneumoniae Colonization in ICU COVID-19 Patients. What Did Not Work? Journal of clinical medicine 2020, 9. [CrossRef]
- Mahoney, A.R.; Safaee, M.M.; Wuest, W.M.; Furst, A.L. The silent pandemic: Emergent antibiotic resistances following the global response to SARS-CoV-2. iScience 2021, 24, 102304. [CrossRef]
- Das, R.; Kotra, K.; Singh, P.; Loh, B.; Leptihn, S.; Bajpai, U. Correction to: Alternative Treatment Strategies for Secondary Bacterial and Fungal Infections Associated with COVID-19. Infectious diseases and therapy 2022, 11, 79-80. [CrossRef]
- Khan, A.; Rao, T.S.; Joshi, H.M. Phage therapy in the Covid-19 era: Advantages over antibiotics. Current research in microbial sciences 2022, 3, 100115. [CrossRef]
- Abedon, S.T.; Garcia, P.; Mullany, P.; Aminov, R. Editorial: Phage Therapy: Past, Present and Future. Frontiers in microbiology 2017, 8, 981. [CrossRef]
- Ling, H.; Lou, X.; Luo, Q.; He, Z.; Sun, M.; Sun, J. Recent advances in bacteriophage-based therapeutics: Insight into the post-antibiotic era. Acta pharmaceutica Sinica. B 2022, 12, 4348-4364. [CrossRef]
- Clokie, M.R.J.; Sicheritz-Ponten, T.E. Phage Therapy: Insights from the Past, the Great Need of the Present, and Glimpses into the Future. Phage 2022, 3, 65-66. [CrossRef]
- Li, C.; Shi, T.; Sun, Y.; Zhang, Y. A Novel Method to Create Efficient Phage Cocktails via Use of Phage-Resistant Bacteria. Applied and environmental microbiology 2022, 88, e0232321. [CrossRef]
- Rimon, A.; Gelman, D.; Yerushalmy, O.; Coppenhagen-Glazer, S.; Katvan, E.; Nir-Paz, R.; Hazan, R. Phage Therapy in Israel, Past, Present, and Future. Phage 2022, 3, 85-94. [CrossRef]
- Grabowski, L.; Lepek, K.; Stasilojc, M.; Kosznik-Kwasnicka, K.; Zdrojewska, K.; Maciag-Dorszynska, M.; Wegrzyn, G.; Wegrzyn, A. Bacteriophage-encoded enzymes destroying bacterial cell membranes and walls, and their potential use as antimicrobial agents. Microbiological research 2021, 248, 126746. [CrossRef]
- Walsh, L.; Johnson, C.N.; Hill, C.; Ross, R.P. Efficacy of Phage- and Bacteriocin-Based Therapies in Combatting Nosocomial MRSA Infections. Frontiers in molecular biosciences 2021, 8, 654038. [CrossRef]
- Liu, H.; Hu, Z.; Li, M.; Yang, Y.; Lu, S.; Rao, X. Therapeutic potential of bacteriophage endolysins for infections caused by Gram-positive bacteria. Journal of biomedical science 2023, 30, 29. [CrossRef]
- Simons, A.; Alhanout, K.; Duval, R.E. Bacteriocins, Antimicrobial Peptides from Bacterial Origin: Overview of Their Biology and Their Impact against Multidrug-Resistant Bacteria. Microorganisms 2020, 8. [CrossRef]
- Hassan, M.; Flanagan, T.W.; Kharouf, N.; Bertsch, C.; Mancino, D.; Haikel, Y. Antimicrobial Proteins: Structure, Molecular Action, and Therapeutic Potential. Pharmaceutics 2022, 15. [CrossRef]
- Cong, W.; Stuart, B.; N, A.I.; Liu, B.; Tang, Y.; Wang, H.; Wang, Y.; Manchundiya, A.; Lambert, H. Antibiotic Use and Bacterial Infection in COVID-19 Patients in the Second Phase of the SARS-CoV-2 Pandemic: A Scoping Review. Antibiotics 2022, 11. [CrossRef]
- Bengoechea, J.A.; Bamford, C.G. SARS-CoV-2, bacterial co-infections, and AMR: the deadly trio in COVID-19? EMBO molecular medicine 2020, 12, e12560. [CrossRef]
- Mirzaei, R.; Goodarzi, P.; Asadi, M.; Soltani, A.; Aljanabi, H.A.A.; Jeda, A.S.; Dashtbin, S.; Jalalifar, S.; Mohammadzadeh, R.; Teimoori, A.; et al. Bacterial co-infections with SARS-CoV-2. IUBMB life 2020, 72, 2097-2111. [CrossRef]
- Lavrinenko, A.; Kolesnichenko, S.; Kadyrova, I.; Turmukhambetova, A.; Akhmaltdinova, L.; Klyuyev, D. Bacterial Co-Infections and Antimicrobial Resistance in Patients Hospitalized with Suspected or Confirmed COVID-19 Pneumonia in Kazakhstan. Pathogens 2023, 12. [CrossRef]
- Russell, C.D.; Fairfield, C.J.; Drake, T.M.; Turtle, L.; Seaton, R.A.; Wootton, D.G.; Sigfrid, L.; Harrison, E.M.; Docherty, A.B.; de Silva, T.I.; et al. Co-infections, secondary infections, and antimicrobial use in patients hospitalised with COVID-19 during the first pandemic wave from the ISARIC WHO CCP-UK study: a multicentre, prospective cohort study. The Lancet. Microbe 2021, 2, e354-e365. [CrossRef]
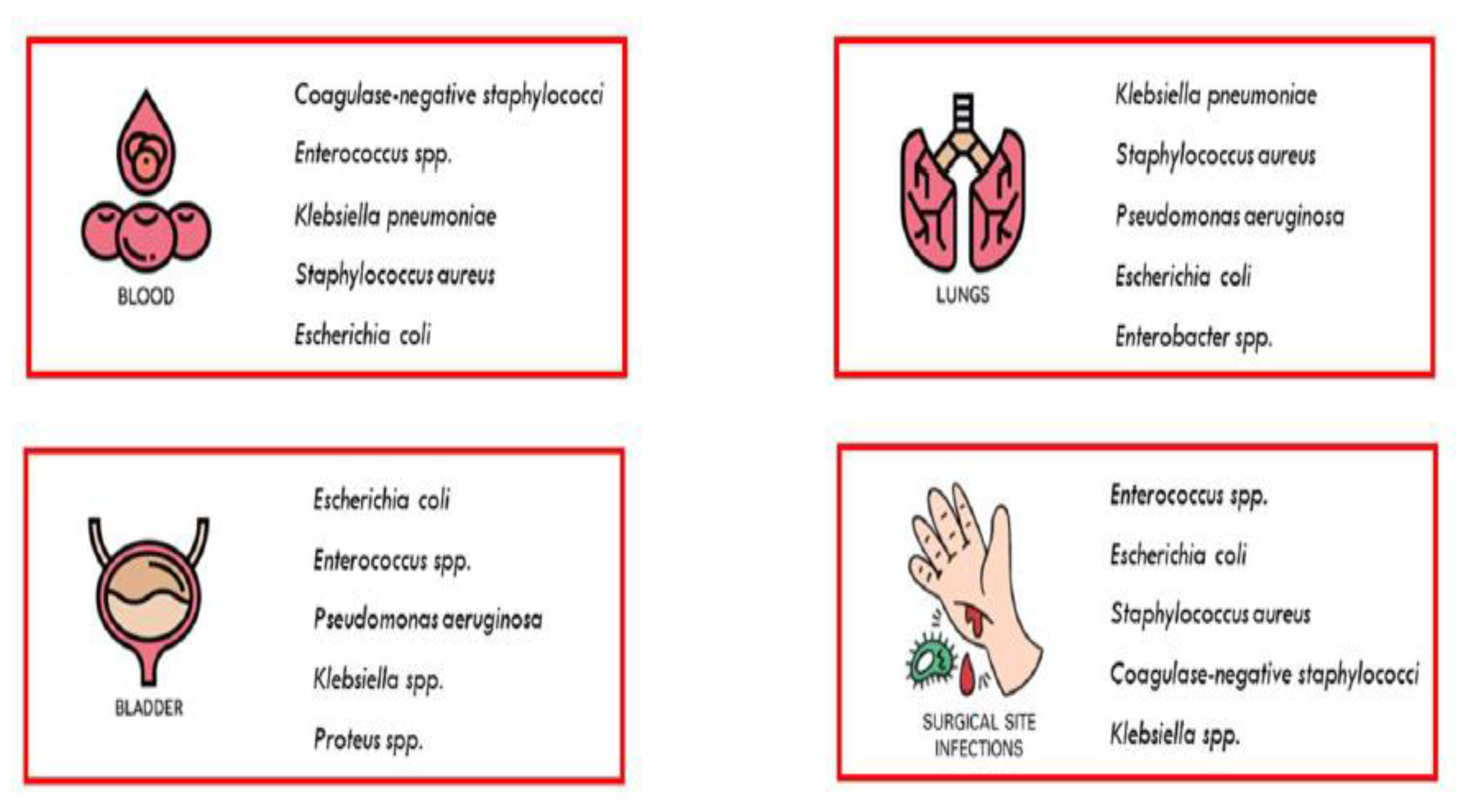
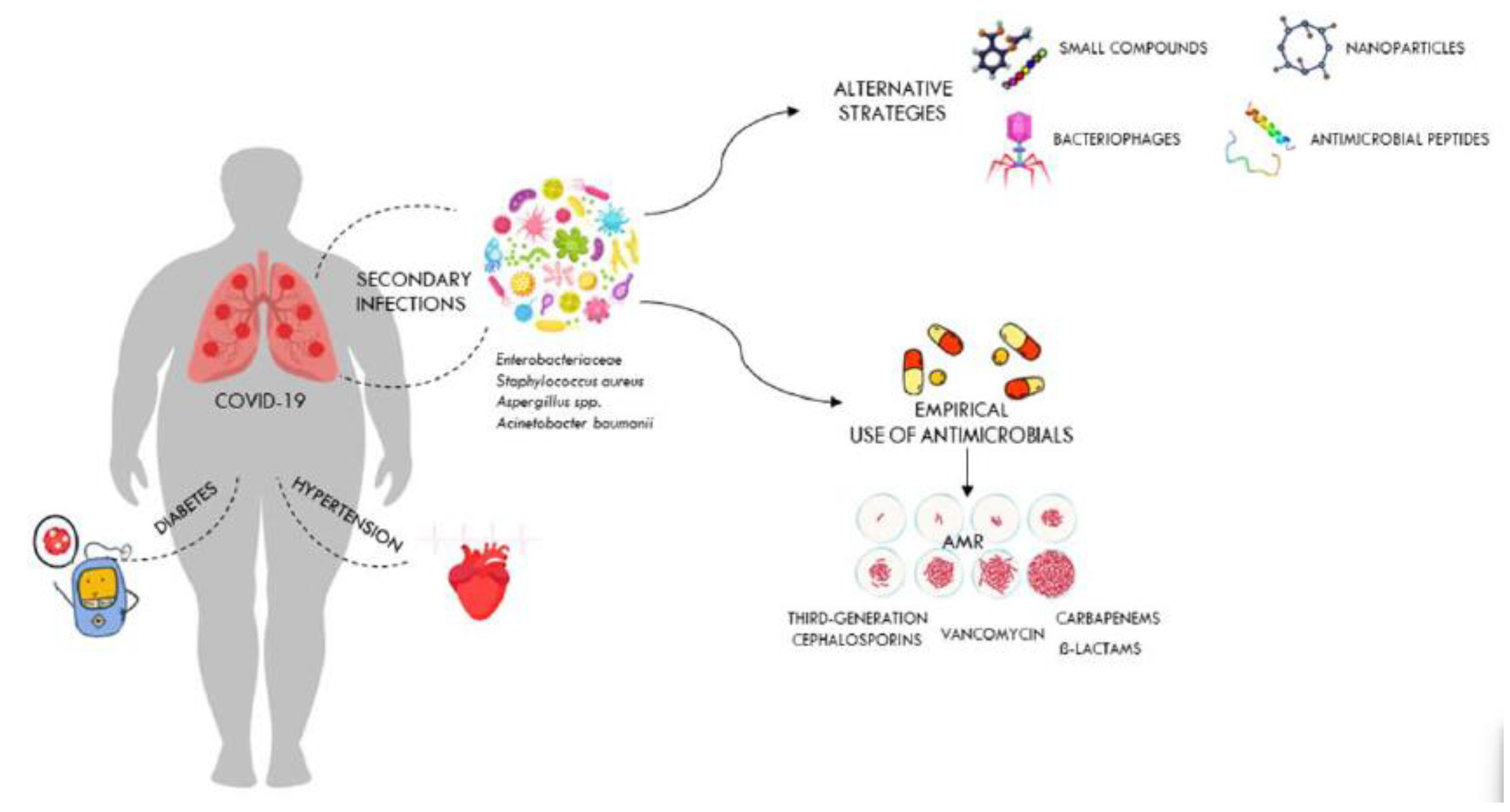
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).



