Submitted:
20 May 2023
Posted:
23 May 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
2.1. Abiotic stress transcriptomes of A. thaliana and P. cheesemanii
2.2. Differentially expressed gene number bias in the three stress responses of A. thaliana and P. cheesemanii
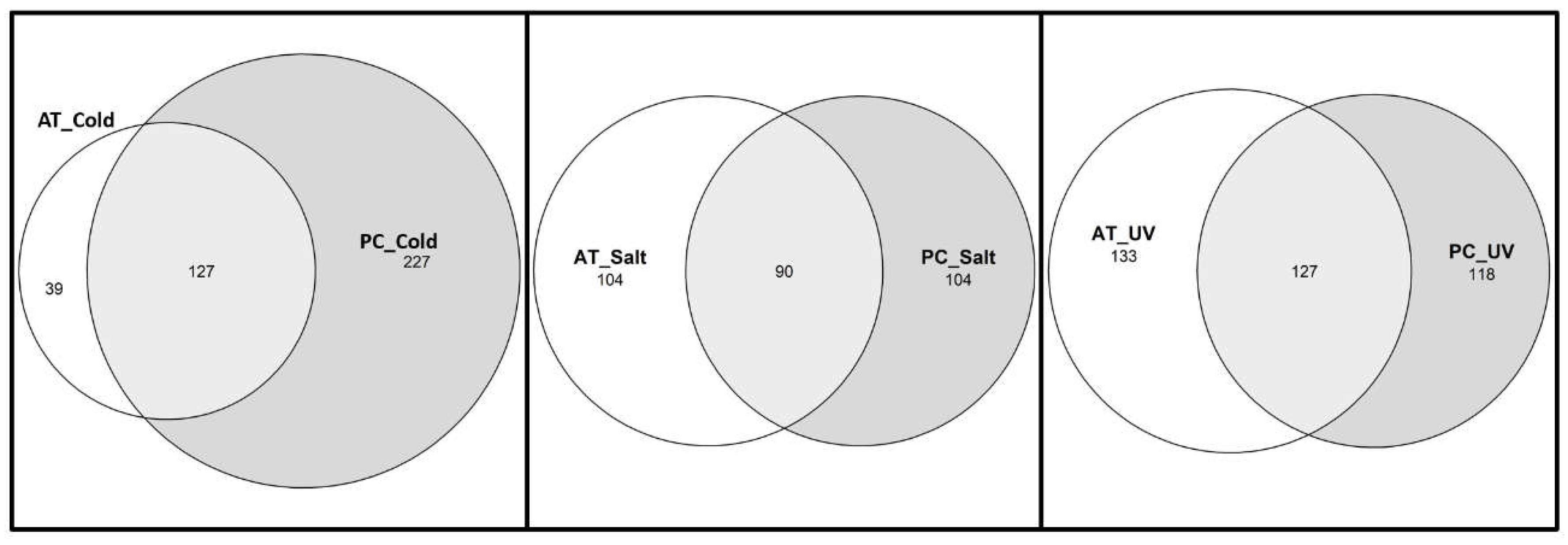
2.3. Comparative analysis of stress-responsive gene sets
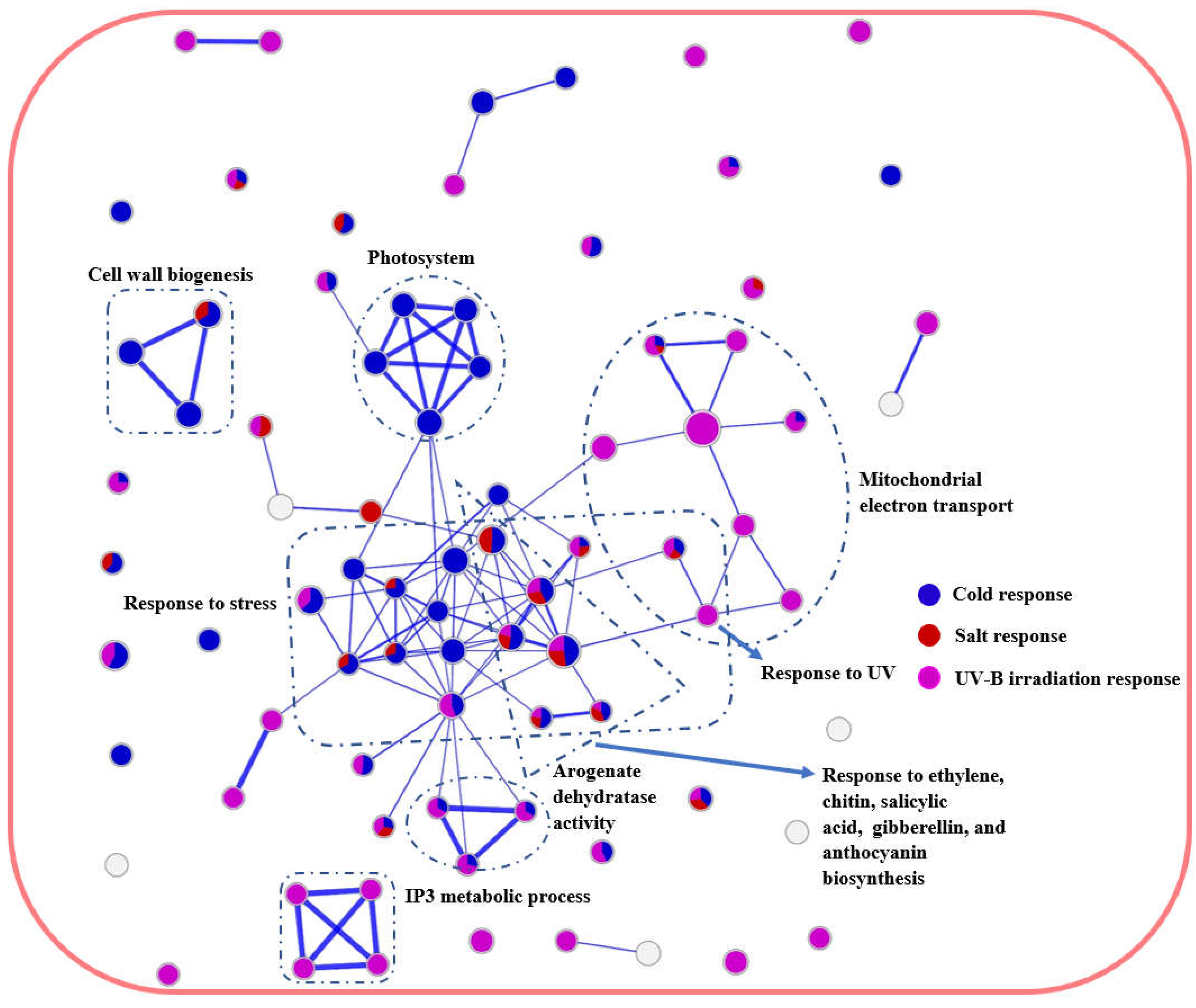
2.4. Network analysis identifies multiple stress-responsive crosstalks in A. thaliana and P. cheesemanii
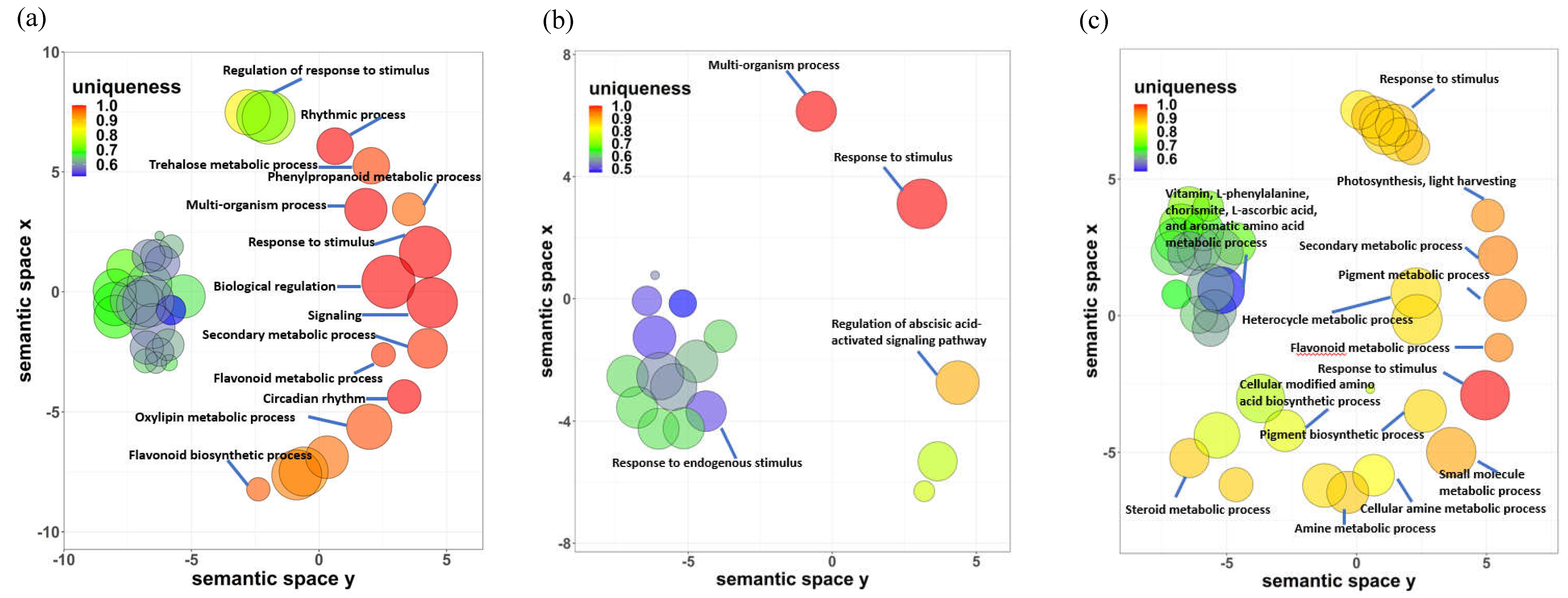
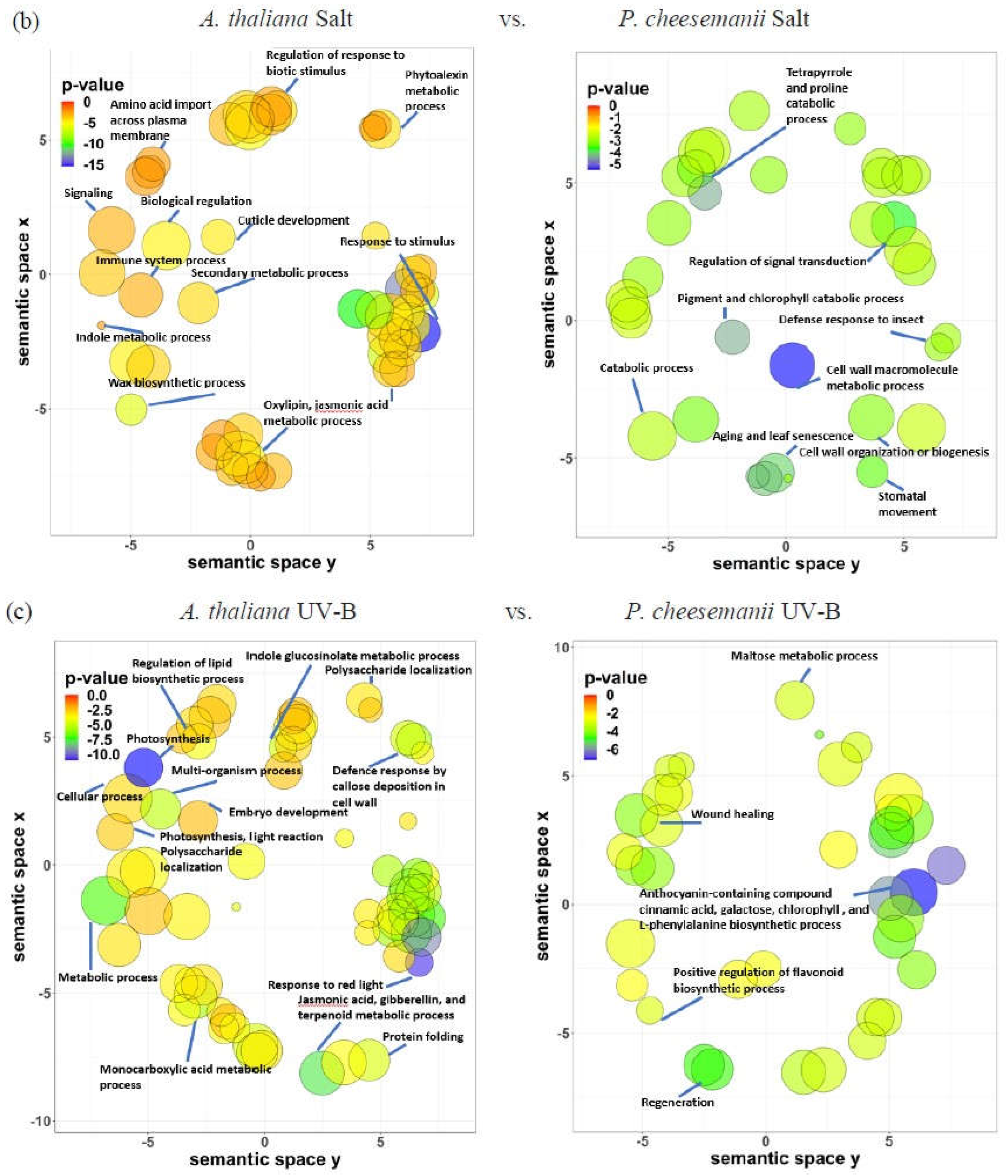
2.5. Identification of biological processes shared between A. thaliana and P. cheesemanii stress responses
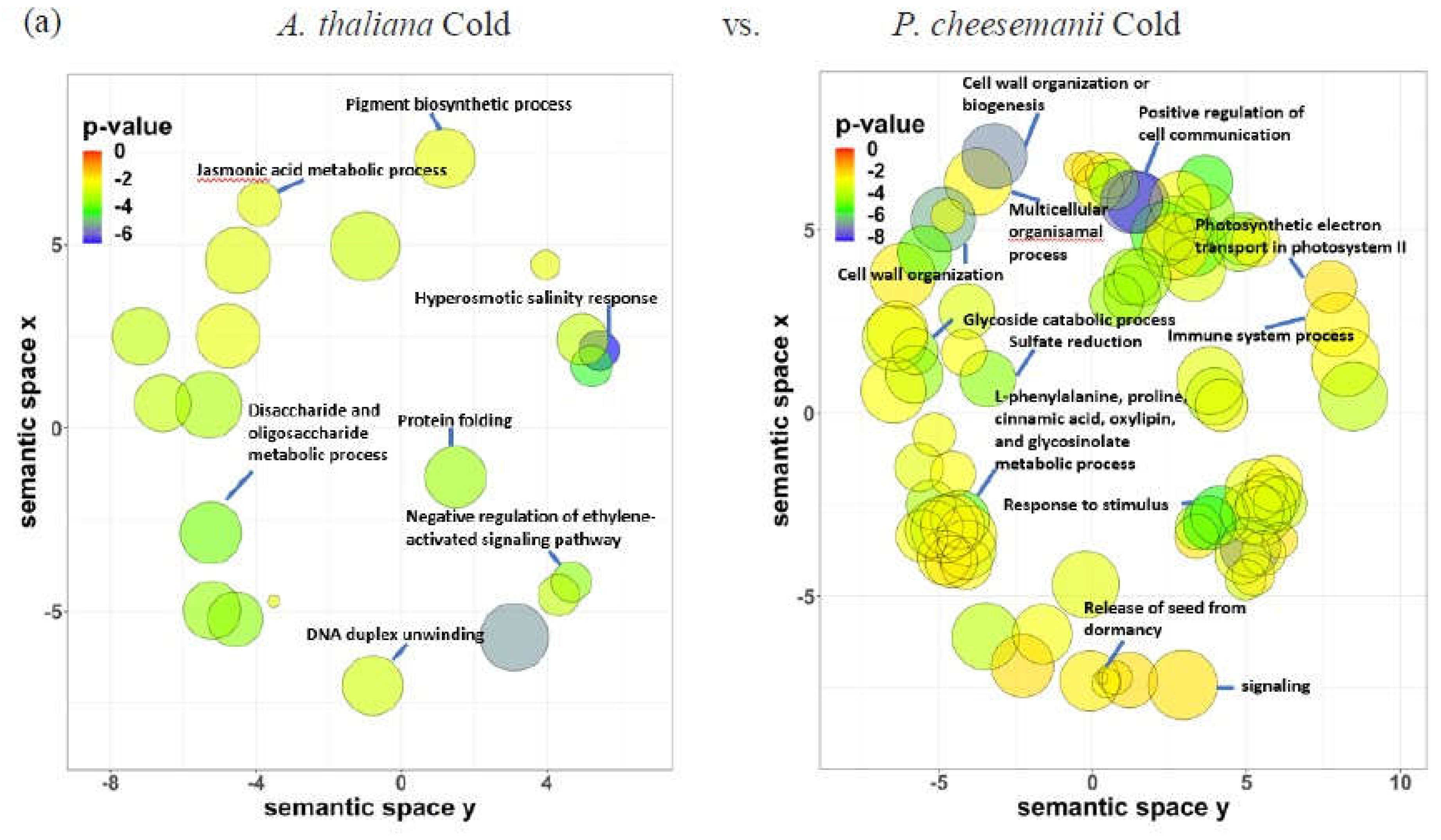
2.6. Identification of unique biological processes in A. thaliana and P. cheesemanii responses to three stresses
3. Discussion
3.1. Classical stress responsive processes are conserved in both A. thaliana and P. cheesemanii
3.2. Unique cold, salt, and UV-B-radiation responses in A. thaliana and P. cheesemanii
4. Materials and methods
4.1. Plant growth and stress treatments
4.2. Library preparation and Illumina transcriptome sequencing
4.3. Pachycladon transcriptome assembly
4.4. Functional annotation of the Pachycladon transcriptome
4.5. Analysis of differential gene expression
4.6. Identification of shared and unique biological processes of species’ stress responses
4.7. Combining weighted correlation network analysis and gene set enrichment analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data availability Statement
Acknowledgments
Conflicts of Interest
References
- Hasanuzzaman, M.; Nahar, K.; Alam, Md.; Roychowdhury, R.; Fujita, M. Physiological, Biochemical, and Molecular Mechanisms of Heat Stress Tolerance in Plants. Int. J. Mol. Sci. 2013, 14(5), 9643–9684. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, P. M.; Bressan, R. A.; Zhu, J.-K.; Bohnert, H. J. Plant cellular and molecular responses to high salinity. Annu. rev. plant physiol. plant mol. biol. 2000, 51(1), 463–499. [Google Scholar] [CrossRef] [PubMed]
- Misra, A. N.; Biswal, A. K.; Misra, M. Physiological, biochemical and molecular aspects of water stress responses in plants, and the bio-technological applications. Proc. Natl. Acad. Sci. India. Sect. B 2002, 72(2), 115–134. [Google Scholar]
- Shanker, A.; Venkateswarlu, B. Abiotic Stress in Plants: Mechanisms and Adaptations. BoD–Books on Demand; IntechOpen: Norderstedt, Germany, 2011; p. 22. [Google Scholar]
- Hasanuzzaman, M.; Nahar, K.; Fujit, M. Extreme Temperature Responses, Oxidative Stress and Antioxidant Defense in Plants. In Abiotic Stress—Plant Responses and Applications in Agriculture; InTechOpen: London, UK, 2013; pp. 169–205. [Google Scholar] [CrossRef]
- Wang, X.; Li, W.; Li, M.; Welti, R. Profiling lipid changes in plant response to low temperatures. Physiol. Plant. 2006, 126(1), 90–96. [Google Scholar] [CrossRef]
- Dong, Y.; Gupta, S.; Sievers, R.; Wargent, J. J.; Wheeler, D.; Putterill, J.; Macknight, R.; Gechev, T.; Mueller-Roeber, B.; Dijkwel, P. P. Genome draft of the Arabidopsis relative Pachycladon cheesemanii reveals novel strategies to tolerate New Zealand’s high ultraviolet B radiation environment. BMC Genom. 2019, 20(1), 838. [Google Scholar] [CrossRef]
- Rabbani, M. A.; Maruyama, K.; Abe, H.; Khan, M. A.; Katsura, K.; Ito, Y.; Yoshiwara, K.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Monitoring Expression Profiles of Rice Genes under Cold, Drought, and High-Salinity Stresses and Abscisic Acid Application Using cDNA Microarray and RNA Gel-Blot Analyses. Plant Physiol. 2003, 133(4), 1755–1767. [Google Scholar] [CrossRef] [PubMed]
- Rensink, W. A.; Iobst, S.; Hart, A.; Stegalkina, S.; Liu, J.; Buell, C. R. Gene expression profiling of potato responses to cold, heat, and salt stress. Funct. Integr. Genom. 2005, 5(4), 201–207. [Google Scholar] [CrossRef]
- Cohen, S. P.; Leach, J. E. Abiotic and biotic stresses induce a core transcriptome response in rice. Sci. Rep. 2019, 9(1), 6273. [Google Scholar] [CrossRef]
- Sharma, R.; Singh, G.; Bhattacharya, S.; Singh, A. Comparative transcriptome meta-analysis of Arabidopsis thaliana under drought and cold stress. PLoS One 2018, 13(9), e0203266. [Google Scholar] [CrossRef]
- Zhu, J.K. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002, 53(1), 247–273. [Google Scholar] [CrossRef]
- Pan, Y.; Wu, L. J.; & Yu, Z.L. Effect of salt and drought stress on antioxidant enzymes activities and SOD isoenzymes of liquorice (Glycyrrhiza uralensis Fisch). Plant Growth Regul. 2006, 49(2), 157–165.
- Sun, C. X.; Li, M. Q.; Gao, X. X.; Liu, L. N.; Wu, X. F.; Zhou, J. H. Metabolic response of maize plants to multi-factorial abiotic stresses. Plant Biol. (Stuttg.) 2016, 18 Suppl 1, 120–129. [Google Scholar] [CrossRef] [PubMed]
- Ni, L.; Wang, Z.; Guo, J.; Pei, X.; Liu, L.; Li, H. ;... Gu, C. Full-Length Transcriptome Sequencing and Comparative Transcriptome Analysis to Evaluate Drought and Salt Stress in Iris lactea var. chinensis. Genes (Basel) 2021, 12(3). [Google Scholar] [CrossRef]
- Xu, Y.; Magwanga, R. O.; Jin, D.; Cai, X.; Hou, Y.; Juyun, Z. ;... Zhou, Z. Comparative transcriptome analysis reveals evolutionary divergence and shared network of cold and salt stress response in diploid D-genome cotton. BMC Plant Biol. 2020, 20(1), 518. [Google Scholar] [CrossRef]
- Dai, Q.; Yan, B.; Huang, S.; Liu, X.; Peng, S.; Miranda, M. L. L.; Chavez, A. Q.; Vergara, B. S.; Olszyk, D. M. Response of oxidative stress defense systems in rice (Oryza sativa) leaves with supplemental UV-B radiation. Physiol. Plant. 1997, 101(2), 301–308. [Google Scholar] [CrossRef]
- Kim, B. C.; Tennessen, D. J.; Last, R. L. UV-B-induced photomorphogenesis in Arabidopsis thaliana. Plant J. 1998, 15(5), 667–674. [Google Scholar] [CrossRef]
- Raghuvanshi, R.; Sharma, R. K. Response of two cultivars of Phaseolus vulgaris L. (French beans) plants exposed to enhanced UV-B radiation under mountain ecosystem. Environ. Sci. Pollut. Res. 2016, 23(1), 831–842. [Google Scholar] [CrossRef]
- Vyšniauskienė, R.; Rančelienė, V. Effect of UV-B radiation on growth and antioxidative enzymes activity in Lithuanian potato (Solanum tuberosum L.) cultivars. Zemdirbyste-Agriculture 2014, 101(1), 51–56. [Google Scholar] [CrossRef]
- Casati, P.; Walbot, V. Rapid transcriptome responses of maize (Zea mays) to UV-B in irradiated and shielded tissues. Genome Biol. 2004, 5(3), R16. [Google Scholar] [CrossRef]
- Gil, M.; Pontin, M.; Berli, F.; Bottini, R.; Piccoli, P. Metabolism of terpenes in the response of grape (Vitis vinifera L.) leaf tissues to UV-B radiation. Phytochem. 2012, 77, 89–98. [Google Scholar] [CrossRef]
- Wang, H.; Li, J.; Tao, W.; Zhang, X.; Gao, X.; Yong, J.; Zhao, J.; Zhang, L.; Li, Y.; Duan, J. Lycium ruthenicum studies: Molecular biology, Phytochemistry and pharmacology. Food Chem. 2018, 240, 759–766. [Google Scholar] [CrossRef]
- Kilian, J.; Whitehead, D.; Horak, J.; Wanke, D.; Weinl, S.; Batistic, O.; D’Angelo, C.; Bornberg-Bauer, E.; Kudla, J.; Harter, K. The AtGenExpress global stress expression data set: Protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J. 2007, 50(2), 347–363. [Google Scholar] [CrossRef] [PubMed]
- Heenan, P.; Mitchell, A. Phylogeny, biogeography and adaptive radiation of Pachycladon (Brassicaceae) in the mountains of South Island, New Zealand. J. Biogeogr. 2003, 30(11), 1737–1749. Available online: https://onlinelibrary.wiley.com/doi/abs/10.1046/j.1365-2699.2003.00941.x. [CrossRef]
- Burghelea, C.; Zaharescu, D.; Dontsova, K.; Maier, R.; Huxman, T.; Chorover, J. Mineral nutrient mobilization by plants from rock: influence of rock type and arbuscular mycorrhiza. Biogeochemistry 2015, 124, 187–203. Available online: https://link.springer.com/article/10.1007/s10533-015-0092-5. [CrossRef]
- Hoffmann, M. H. Biogeography of Arabidopsis thaliana (L.) heynh. (Brassicaceae). J. Biogeogr. 2002, 29(1), 125–134. Available online: https://onlinelibrary.wiley.com/doi/abs/10.1046/j.1365-2699.2002.00647.x. [CrossRef]
- Joly, S.; Heenan, P. B.; Lockhart, P. J. A Pleistocene inter-tribal allopolyploidization event precedes the species radiation of Pachycladon (Brassicaceae) in New Zealand. Mol. Phylogenet. Evol. 2009, 51(2), 365–372. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Witten, D. M.; Johnstone, I. M.; Tibshirani, R. Normalization, testing, and false discovery rate estimation for RNA-sequencing data. Biostatistics 2012, 13(3), 523–538. [Google Scholar] [CrossRef]
- Supek, F.; Bošnjak, M.; Škunca, N.; Šmuc, T. REVIGO Summarizes and Visualizes Long Lists of Gene Ontology Terms. PLoS ONE 2011, 6(7), e21800. [Google Scholar] [CrossRef]
- Abla, M.; Sun, H.; Li, Z.; Wei, C.; Gao, F.; Zhou, Y.; Feng, J. Identification of miRNAs and Their Response to Cold Stress in Astragalus Membranaceus. Biomolecules 2019, 9(5), 182. [Google Scholar] [CrossRef]
- Agurla, S.; Gahir, S.; Munemasa, S.; Murata, Y.; and Raghavendra, A. S. Mechanism of stomatal closure in plants exposed to drought and cold stress. Adv. Exp. Med. Biol. 2018, 1081, 215–232. [Google Scholar] [CrossRef]
- Fei, J.; Wang, Y.; Cheng, H.; Su, Y.; Zhong, Y.; Zheng, L. Cloning and characterization of KoOsmotin from mangrove plant Kandelia obovata under cold stress. BMC Plant Biol. 2021, 21(1), 10. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Z.; Zhang, W.; Marand, A. P.; Zhu, B.; Buell, C. R.; Jiang, J. Cold stress induces enhanced chromatin accessibility and bivalent histone modifications H3K4me3 and H3K27me3 of active genes in potato. Genome Biol. 2019, 20(1), 123. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Li, X.; Zou, J.; Ren, J.; Jin, C.; Zhang, H.; Yu, H.; Jin, H. Comparative transcriptome analysis of genes involved in the drought stress response of two peanut (Arachis hypogaea L.) varieties. BMC Plant Biol. 2021, 21(1), 64. [Google Scholar] [CrossRef] [PubMed]
- Kashyap, S. P.; Prasanna, H. C.; Kumari, N.; Mishra, P.; Singh, B. Understanding salt tolerance mechanism using transcriptome profiling and de novo assembly of wild tomato Solanum chilense. Sci. Rep. 2020, 10(1), 15835. [Google Scholar] [CrossRef]
- Li, X.; Li, M.; Zhou, B.; Yang, Y.; Wei, Q.; Zhang, J. Transcriptome analysis provides insights into the stress response crosstalk in apple (Malus × domestica) subjected to drought, cold and high salinity. Sci. Rep. 2019, 9(1), 9071. [Google Scholar] [CrossRef]
- Inoue, K.; Araki, T.; Endo, M. Circadian clock during plant development. J. Plant Res. 2018, 131(1), 59–66. [Google Scholar] [CrossRef]
- Singh, M.; Mas, P. A Functional Connection between the Circadian Clock and Hormonal Timing in Arabidopsis. Genes 2018, 9(12), 567. [Google Scholar] [CrossRef]
- Xu, X.; Yuan, L.; Yang, X.; Zhang, X.; Wang, L.; Xie, Q. Circadian clock in plants: Linking timing to fitness. J. Integr. Plant Biol. 2022, 64(4), 792–811. [Google Scholar] [CrossRef]
- Andreeva, A. A.; Kudryakova, N. V.; Kuznetsov, Vl. V.; Kusnetsov, V. V. Ontogenetic, Light, and Circadian Regulation of PAP Protein Genes during Seed Germination of Arabidopsis thaliana. Dokl. Biochem. Biophys. 2021, 500(1), 312–316. [Google Scholar] [CrossRef]
- Blair, E. J.; Bonnot, T.; Hummel, M.; Hay, E.; Marzolino, J. M.; Quijada, I. A.; Nagel, D. H. Contribution of time of day and the circadian clock to the heat stress responsive transcriptome in Arabidopsis. Sci. Rep. 2019, 9(1), 4841. [Google Scholar] [CrossRef]
- Cao, L. The Molecular Interactions of Soybean Circadian Clock with Abiotic Stresses and Soybean Cyst Nematode (Heterodera glycines). Ph.D. thesis, Iowa State University, Ames, Iowa, United States. 2022. Available online: https://www.proquest.com/docview/2309795320/abstract/1C0407289E124C0BPQ/1.
- Chang, T.; Zhao, Y.; He, H.; Xi, Q.; Fu, J.; Zhao, Y. Exogenous melatonin improves growth in hulless barley seedlings under cold stress by influencing the expression rhythms of circadian clock genes. PeerJ 2021, 9, e10740. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Zhou, Y.; Fan, F.; Peng, J.; Zhang, J. Coordination of light, circadian clock with temperature: The potential mechanisms regulating chilling tolerance in rice. J. Integr. Plant Biol. 2020, 62(6), 737–760. [Google Scholar] [CrossRef] [PubMed]
- Cortleven, A.; Roeber, V. M.; Frank, M.; Bertels, J.; Lortzing, V.; Beemster, G. T. S.; Schmülling, T. Photoperiod Stress in Arabidopsis thaliana Induces a Transcriptional Response Resembling That of Pathogen Infection. Front. Plant Sci. 2022, 13, 838284. [Google Scholar] [CrossRef]
- Srivastava, D.; Shamim, Md.; Kumar, M.; Mishra, A.; Maurya, R.; Sharma, D.; Pandey, P.; Singh, K. N. Role of circadian rhythm in plant system: An update from development to stress response. Environ. Exp. Bot. 2019, 162, 256–271. [Google Scholar] [CrossRef]
- Legnaioli, T.; Cuevas, J.; Mas, P. TOC1 functions as a molecular switch connecting the circadian clock with plant responses to drought. EMBO Rep. 2009, 28(23), 3745–3757. [Google Scholar] [CrossRef]
- Fowler, S.; Thomashow, M. F. Arabidopsis Transcriptome Profiling Indicates That Multiple Regulatory Pathways Are Activated during Cold Acclimation in Addition to the CBF Cold Response Pathway. Plant Cell 2002, 14(8), 1675–1690. [Google Scholar] [CrossRef]
- Duan, M.; Huang, P.; Yuan, X.; Chen, H.; Huang, J.; Zhang, H. CMYB1 Encoding a MYB Transcriptional Activator Is Involved in Abiotic Stress and Circadian Rhythm in Rice. Sci. World J. 2014, 2014, 1–9. [Google Scholar] [CrossRef]
- Gao, Y.; Yang, X.; Yang, X.; Zhao, T.; An, X.; Chen, Z. Characterization and expression pattern of the trehalose-6-phosphate synthase and trehalose-6-phosphate phosphatase gene families in Populus. Int. J. Biol. Macromol. 2021, 187, 9–23. [Google Scholar] [CrossRef]
- He, Y.; Borrego, E. J.; Gorman, Z.; Huang, P.-C.; Kolomiets, M. V. Relative contribution of LOX10, green leaf volatiles and JA to wound-induced local and systemic oxylipin and hormone signature in Zea mays (maize). Phytochem. 2020, 174, 112334. [Google Scholar] [CrossRef]
- Tyagi, K.; Maoz, I.; Kochanek, B.; Sela, N.; Lerno, L.; Ebeler, S. E.; Lichter, A. Cytokinin but not gibberellin application had major impact on the phenylpropanoid pathway in grape. Hortic. Res. 2021, 8(1), 51. [Google Scholar] [CrossRef]
- Hildreth, S. B.; Littleton, E. S.; Clark, L. C.; Puller, G. C.; Kojima, S.; Winkel, B. S. Crosstalk between flavonoids and the plant circadian clock. BioRxiv 2021, 2021–07. [Google Scholar] [CrossRef]
- Romero, C.; Bellés, J. M.; Vayá, J. L.; Serrano, R.; Culiáñez-Macià, F. A. Expression of the yeast trehalose-6-phosphate synthase gene in transgenic tobacco plants: Pleiotropic phenotypes include drought tolerance. Planta 1997, 201(3), 293–297. [Google Scholar] [CrossRef] [PubMed]
- Lin, Q.; Yang, J.; Wang, Q.; Zhu, H.; Chen, Z.; Dao, Y.; Wang, K. Overexpression of the trehalose-6-phosphate phosphatase family gene AtTPPF improves the drought tolerance of Arabidopsis thaliana. BMC Plant Biol. 2019, 19(1), 381. [Google Scholar] [CrossRef]
- Jang, I.-C.; Oh, S.-J.; Seo, J.-S.; Choi, W.-B.; Song, S. I.; Kim, C. H.; Kim, Y. S.; Seo, H.-S.; Choi, Y. D.; Nahm, B. H.; Kim, J.-K. Expression of a Bifunctional Fusion of the Escherichia coli Genes for Trehalose-6-Phosphate Synthase and Trehalose-6-Phosphate Phosphatase in Transgenic Rice Plants Increases Trehalose Accumulation and Abiotic Stress Tolerance without Stunting Growth. Plant Physiol. 2003, 131(2), 516–524. [Google Scholar] [CrossRef]
- Wang, X.; Du, Y.; Yu, D. Trehalose phosphate synthase 5-dependent trehalose metabolism modulates basal defense responses in Arabidopsis thaliana. J. Integr. Plant Biol. 2019, 61(4), 509–527. [Google Scholar] [CrossRef] [PubMed]
- Ahanger, M. A.; Tomar, N. S.; Tittal, M.; Argal, S.; Agarwal, R. M. Plant growth under water/salt stress: ROS production; antioxidants and significance of added potassium under such conditions. Physiol. Mol. Biol 2017, 23(4), 731–744. [Google Scholar] [CrossRef] [PubMed]
- Harinasut, P.; Poonsopa, D.; Roengmongkol, K.; Charoensataporn, R. Salinity effects on antioxidant enzymes in mulberry cultivar. Sci. 2003, 29(2), 109. [Google Scholar] [CrossRef]
- Parthasarathy, A.; Cross, P. J.; Dobson, R. C. J.; Adams, L. E.; Savka, M. A.; Hudson, A. O. A Three-Ring Circus: Metabolism of the Three Proteogenic Aromatic Amino Acids and Their Role in the Health of Plants and Animals. Front. Mol. Biosci. 2018, 5, 29. [Google Scholar] [CrossRef]
- Kong, J.-Q. Phenylalanine ammonia-lyase, a key component used for phenylpropanoids production by metabolic engineering. RSC Adv. 2015, 5(77), 62587–62603. [Google Scholar] [CrossRef]
- Singh, M.; Bashri, G.; Prasad, S. M.; Singh, V. P. Kinetin Alleviates UV-B-Induced Damage in Solanum lycopersicum: Implications of Phenolics and Antioxidants. J. Plant Growth Regul. 2019, 38(3), 831–841. [Google Scholar] [CrossRef]
- Liu, P.; Li, Q.; Gao, Y.; Wang, H.; Chai, L.; Yu, H.; Jiang, W. A New Perspective on the Effect of UV-B on l -Ascorbic Acid Metabolism in Cucumber Seedlings. J. Agric. Food Chem. 2019, 67(16), 4444–4452. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Fu, X.; Lu, M.; Wei, B. A transcriptomic perspective on the effect of UV irradiation on vitamin C content in pea sprouts. J. Food Sci. Technol. 2022, 42, e09022. [Google Scholar] [CrossRef]
- Megías-Pérez, R.; Hahn, C.; Ruiz-Matute, A. I.; Behrends, B.; Albach, D. C.; Kuhnert, N. Changes in low molecular weight carbohydrates in kale during development and acclimation to cold temperatures determined by chromatographic techniques coupled to mass spectrometry. Food Res. Int. 2020, 127, 108727. [Google Scholar] [CrossRef]
- Selvaraj, M. G.; Ishizaki, T.; Valencia, M.; Ogawa, S.; Dedicova, B.; Ogata, T.; Yoshiwara, K.; Maruyama, K.; Kusano, M.; Saito, K.; Takahashi, F.; Shinozaki, K.; Nakashima, K.; Ishitani, M. Overexpression of an Arabidopsis thaliana galactinol synthase gene improves drought tolerance in transgenic rice and increased grain yield in the field. Plant Biotechnol. J. 2017, 15(11), 1465–1477. [Google Scholar] [CrossRef]
- Morsy, M. R.; Jouve, L.; Hausman, J.-F.; Hoffmann, L.; Stewart, J. McD. Alteration of oxidative and carbohydrate metabolism under abiotic stress in two rice (Oryza sativa L.) genotypes contrasting in chilling tolerance. J. Plant Physiol. 2007, 164(2), 157–167. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Qi, X.; Wang, Z.; Li, P.; Wu, C.; Zhang, H.; Zhao, Y. Overexpression of TsGOLS2, a galactinol synthase, in Arabidopsis thaliana enhances tolerance to high salinity and osmotic stresses. Plant Physiol. Biochem. 2013, 69, 82–89. [Google Scholar] [CrossRef] [PubMed]
- Van den Ende, W.; Peshev, D. Sugars as antioxidants in plants. In Crop Improvement under Adverse Conditions; Springer: Dordrecht, The Netherlands, 2013; pp. 285–307. [Google Scholar] [CrossRef]
- Bolouri-Moghaddam, M. R.; Le Roy, K.; Xiang, L.; Rolland, F.; Van den Ende, W. Sugar signalling and antioxidant network connections in plant cells: Sugar signalling and antioxidant networks in plants. FEBS J. 2010, 277(9), 2022–2037. [Google Scholar] [CrossRef]
- Shen, X.; Wang, Z.; Song, X.; Xu, J.; Jiang, C.; Zhao, Y.; Ma, C.; Zhang, H. Transcriptomic profiling revealed an important role of cell wall remodelling and ethylene signalling pathway during salt acclimation in Arabidopsis. Plant Mol. Biol. 2014, 86(3), 303–317. [Google Scholar] [CrossRef]
- Van den Ende, W.; El-Esawe, S. K. Sucrose signaling pathways leading to fructan and anthocyanin accumulation: A dual function in abiotic and biotic stress responses? Environ. Exp. Bot. 2014, 108, 4–13. [Google Scholar] [CrossRef]
- Doares, S. H.; Syrovets, T.; Weiler, E. W.; Ryan, C. A. Oligogalacturonides and chitosan activate plant defensive genes through the octadecanoid pathway. Proc. Natl. Acad. Sci. U.S.A. 1995, 92(10), 4095–4098. [Google Scholar] [CrossRef]
- Jia, X.; Zeng, H.; Bose, S. K.; Wang, W.; Yin, H. Chitosan oligosaccharide induces resistance to Pst DC3000 in Arabidopsis via a non-canonical N-glycosylation regulation pattern. Carbohydr. Polym. 2020, 250, 116939. [Google Scholar] [CrossRef] [PubMed]
- Linden, J. C.; Phisalaphong, M. Oligosaccharides potentiate methyl jasmonate-induced production of paclitaxel in Taxus canadensis. Plant Sci. 2000, 158(1-2), 41–51. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Li, S.; Zhao, X.; Du, Y.; Ma, X. CDNA microarray analysis of gene expression in Brassica napus treated with oligochitosan elicitor. Plant Physiol. Biochem. 2006, 44(11–12), 910–916. [Google Scholar] [CrossRef] [PubMed]
- Jia, X.; Zeng, H.; Wang, W.; Zhang, F.; Yin, H. Chitosan Oligosaccharide Induces Resistance to Pseudomonas syringae pv. Tomato DC3000 in Arabidopsis thaliana by Activating Both Salicylic Acid– and Jasmonic Acid–Mediated Pathways. Mol. Plant Microbe Interact. 2018, 31(12), 1271–1279. [Google Scholar] [CrossRef] [PubMed]
- Wennberg, M.; Ekvall, J.; Olsson, K.; Nyman, M. Changes in carbohydrate and glucosinolate composition in white cabbage (Brassica oleracea var. Capitata) during blanching and treatment with acetic acid. Food Chem. 2006, 95(2), 226–236. [Google Scholar] [CrossRef]
- Cocetta, G.; Mishra, S.; Raffaelli, A.; Ferrante, A. Effect of heat root stress and high salinity on glucosinolates metabolism in wild rocket. J. Plant Physiol. 2018, 231, 261–270. [Google Scholar] [CrossRef]
- Ljubej, V.; Radojcic Redovnikovic, I.; Salopek-Sondi, B.; Smolko, A.; Roje, S.; Samec, D. Chilling and Freezing Temperature Stress Differently Influence Glucosinolates Content in Brassica oleracea var. acephala. Plants (Basel) 2021, 10(7). [Google Scholar] [CrossRef]
- Salehin, M.; Li, B.; Tang, M.; Katz, E.; Song, L.; Ecker, J. R. ;... Estelle, M. Auxin-sensitive Aux/IAA proteins mediate drought tolerance in Arabidopsis by regulating glucosinolate levels. Nat. Commun. 2019, 10(1), 4021. [Google Scholar] [CrossRef]
- Troufflard, S.; Mullen, W.; Larson, T. R.; Graham, I. A.; Crozier, A.; Amtmann, A.; Armengaud, P. Potassium deficiency induces the biosynthesis of oxylipins and glucosinolates in Arabidopsis thaliana. BMC Plant Biol. 2010, 10, 172. [Google Scholar] [CrossRef]
- Hara, M.; Harazaki, A.; Tabata, K. Administration of isothiocyanates enhances heat tolerance in Arabidopsis thaliana. Plant Growth Regul. 2013, 69(1), 71–77. [Google Scholar] [CrossRef]
- Ludwig-Müller, J.; Krishna, P.; Forreiter, C. A Glucosinolate Mutant of Arabidopsis Is Thermosensitive and Defective in Cytosolic Hsp90 Expression after Heat Stress. Plant Physiol. 2000, 123(3), 949–958. [Google Scholar] [CrossRef] [PubMed]
- More, P.; Agarwal, P.; Joshi, P. S.; Agarwal, P. K. The JcWRKY tobacco transgenics showed improved photosynthetic efficiency and wax accumulation during salinity. Sci. Rep. 2019, 9(1), 19617. [Google Scholar] [CrossRef]
- Shepherd, T.; Wynne Griffiths, D. The effects of stress on plant cuticular waxes. New Phytol. 2006, 171(3), 469–499. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Li, D.; Li, Z.; Hu, Q.; Yang, C.; Zhu, L.; Luo, H. Constitutive Expression of a miR319 Gene Alters Plant Development and Enhances Salt and Drought Tolerance in Transgenic Creeping Bentgrass. Plant Physiol. 2013, 161(3), 1375–1391. [Google Scholar] [CrossRef] [PubMed]
- Djemal, R.; Khoudi, H. TdSHN1, a WIN1/SHN1-type transcription factor, imparts multiple abiotic stress tolerance in transgenic tobacco. Environ. Exp. Bot. 2016, 131, 89–100. [Google Scholar] [CrossRef]
- Zhu, X.; Xiong, L. Putative megaenzyme DWA1 plays essential roles in drought resistance by regulating stress-induced wax deposition in rice. Proc. Natl. Acad. Sci. U.S.A. 2013, 110(44), 17790–17795. [Google Scholar] [CrossRef] [PubMed]
- Kunst, L.; Samuels, L. Plant cuticles shine: Advances in wax biosynthesis and export. Curr. Opin. Plant 2009, 12(6), 721–727. [Google Scholar] [CrossRef]
- Bi, H.; Kovalchuk, N.; Langridge, P.; Tricker, P. J.; Lopato, S.; Borisjuk, N. The impact of drought on wheat leaf cuticle properties. BMC Plant Biol. 2017, 17(1), 85. [Google Scholar] [CrossRef]
- Geng, G.; Li, R.; Stevanato, P.; Lv, C.; Lu, Z.; Yu, L.; Wang, Y. Physiological and Transcriptome Analysis of Sugar Beet Reveals Different Mechanisms of Response to Neutral Salt and Alkaline Salt Stresses. Front. Plant Sci. 2020, 11, 571864. [Google Scholar] [CrossRef]
- Aharoni, A.; Dixit, S.; Jetter, R.; Thoenes, E.; van Arkel, G.; Pereira, A. The SHINE Clade of AP2 Domain Transcription Factors Activates Wax Biosynthesis, Alters Cuticle Properties, and Confers Drought Tolerance when Overexpressed in Arabidopsis. Plant Cell 2004, 16(9), 2463–2480. [Google Scholar] [CrossRef]
- Zhang, J.Y.; Broeckling, C. D.; Blancaflor, E. B.; Sledge, M. K.; Sumner, L. W.; Wang, Z.Y. Overexpression of WXP1, a putative Medicago truncatula AP2 domain-containing transcription factor gene, increases cuticular wax accumulation and enhances drought tolerance in transgenic alfalfa (Medicago sativa). Plant J. 2005, 42(5), 689–707. [Google Scholar] [CrossRef] [PubMed]
- Xiong, J.L.; Wang, H.C.; Tan, X.Y.; Zhang, C.L.; Naeem, M. S. 5-aminolevulinic acid improves salt tolerance mediated by regulation of tetrapyrrole and proline metabolism in Brassica napus L. seedlings under NaCl stress. Plant Physiol. Biochem. 2018, 124, 88–99. [Google Scholar] [CrossRef] [PubMed]
- Korkmaz, A.; Şirikçi, R.; Kocaçınar, F.; Değer, Ö.; Demirkırıan, A. R. Alleviation of salt-induced adverse effects in pepper seedlings by seed application of glycinebetaine. Sci. Hortic. 2012, 148, 197–205. [Google Scholar] [CrossRef]
- Pingle, S. N.; Suryawanshi, S. T.; Pawar, K. R.; Harke, S. N. The Effect of Salt Stress on Proline Content in Maize (Zea mays). LAFOBA2 2022, 64. [Google Scholar] [CrossRef]
- Sivritepe, N.; Sivritepe, H. O.; Eris, A. The effects of NaCl priming on salt tolerance in melon seedlings grown under saline conditions. Sci. Hortic. 2003, 97(3-4), 229–237. [Google Scholar] [CrossRef]
- Sobahan, M. A.; Akter, N.; Ohno, M.; Okuma, E.; Hirai, Y.; Mori, I. C.; Nakamura, Y.; Murata, Y. Effects of Exogenous Proline and Glycinebetaine on the Salt Tolerance of Rice Cultivars. Biosci. Biotechnol. Biochem. 2012, 76(8), 1568–1570. [Google Scholar] [CrossRef]
- Weimberg, R.; Lerner, H. R.; Poljakoff-Mayber, A. A relationship between potassium and proline accumulation in salt-stressed Sorghum bicolor. Physiol. Plant. 1982, 55(1), 5–10. [Google Scholar] [CrossRef]
- Kravets, E. A.; Zelena, L. B.; Zabara, E. P.; Blume, Ya. B. Adaptation strategy of barley plants to UV-B radiation. Emir. J. Food Agric. 2012, 24(6). [Google Scholar] [CrossRef]
- Mintoff, S. J. L.; Rookes, J. E.; Cahill, D. M. Sub-lethal UV-C radiation induces callose, hydrogen peroxide and defence-related gene expression in A rabidopsis thaliana. Plant Biol 2015, 17(3), 703–711. [Google Scholar] [CrossRef]
- Kulich, I.; Vojtíková, Z.; Glanc, M.; Ortmannová, J.; Rasmann, S.; Žárský, V. Cell wall maturation of Arabidopsis trichomes is dependent on exocyst subunit EXO70H4 and involves callose deposition. Plant Physiol. 2015, 168(1), 120–131. [Google Scholar] [CrossRef]
- Clay, N. K.; Adio, A. M.; Denoux, C.; Jander, G.; Ausubel, F. M. Glucosinolate Metabolites Required for an Arabidopsis Innate Immune Response. Science 2009, 323(5910), 95–101. [Google Scholar] [CrossRef] [PubMed]
- Ahuja, I.; Rohloff, J.; Bones, A.M. Defence mechanisms of Brassicaceae: Implications for plant-insect interactions and potential for integrated pest management. A review. Agron. Sustain. Dev. 2010, 30, 311–348. [Google Scholar] [CrossRef]
- De Coninck, B.; Timmermans, P.; Vos, C.; Cammue, B. P. A.; Kazan, K. What lies beneath: Belowground defense strategies in plants. Trends Plant Sci. 2015, 20(2), 91–101. [Google Scholar] [CrossRef] [PubMed]
- Stass, A.; Horst, W. J. Callose in Abiotic Stress. In Chemistry, Biochemistry, and Biology of 1-3 Beta Glucans and Related Polysaccharides, 1st ed.; Antony Bacic, Geoffrey B. Fincher, Bruce A. Stone, Ed.; Elsevier: Oxford, UK, 2009; pp. 499–524. [Google Scholar] [CrossRef]
- Foyer, C. H.; Rasool, B.; Davey, J. W.; Hancock, R. D. Cross-tolerance to biotic and abiotic stresses in plants: A focus on resistance to aphid infestation. J. Exp. Bot. 2016, 67(7), 2025–2037. [Google Scholar] [CrossRef] [PubMed]
- Martin, C.; Gerats, T. Control of Pigment Biosynthesis Genes during Petal Development. Plant Cell 1993, 1253–1264. [Google Scholar] [CrossRef]
- Berli, F. J.; Moreno, D.; Piccoli, P.; HESPANHOL-VIANA, L. E. A. N. D. R. O.; Silva, M. F.; BRESSAN-SMITH, R. I. C. A. R. D. O. . Bottini, R. Abscisic acid is involved in the response of grape (Vitis vinifera L.) cv. Malbec leaf tissues to ultraviolet-B radiation by enhancing ultraviolet-absorbing compounds, antioxidant enzymes and membrane sterols. Plant Cell Environ. 2010, 33(1), 1–10. [Google Scholar] [CrossRef]
- Krueger, F. Trim galore: a wrapper tool around Cutadapt and FastQC to consistently apply quality and adapter trimming to FastQ files. 2015. Available online: http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/.
- Kopylova, E.; Noé, L.; Touzet, H. SortMeRNA: Fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics 2012, 28(24), 3211–3217. [Google Scholar] [CrossRef]
- Grabherr, M. G.; Haas, B. J.; Yassour, M.; Levin, J. Z.; Thompson, D. A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; Chen, Z.; Mauceli, E.; Hacohen, N.; Gnirke, A.; Rhind, N.; di Palma, F.; Birren, B. W.; Nusbaum, C.; Lindblad-Toh, K.; … Regev, A. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29(7), 644–652. [Google Scholar] [CrossRef]
- Zhao, Q.Y.; Wang, Y.; Kong, Y.M.; Luo, D.; Li, X.; Hao, P. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: A comparative study. BMC Bioinform. 2011, 12(S14), S2. [Google Scholar] [CrossRef]
- Schulz, M. H.; Zerbino, D. R.; Vingron, M.; Birney, E. Oases: Robust de novo RNA-seq assembly across the dynamic range of expression levels. Bioinformatics 2012, 28(8), 1086–1092. [Google Scholar] [CrossRef]
- Zerbino, D. R.; Birney, E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008, 18(5), 821–829. [Google Scholar] [CrossRef] [PubMed]
- Robertson, G.; Schein, J.; Chiu, R.; Corbett, R.; Field, M.; Jackman, S. D.; Mungall, K.; Lee, S.; Okada, H. M.; Qian, J. Q.; Griffith, M.; Raymond, A.; Thiessen, N.; Cezard, T.; Butterfield, Y. S.; Newsome, R.; Chan, S. K.; She, R.; Varhol, R.; … Birol, I. De novo assembly and analysis of RNA-seq data. Nat. Methods 2010, 7(11), 909–912. [Google Scholar] [CrossRef] [PubMed]
- Simão, F. A.; Waterhouse, R. M.; Ioannidis, P.; Kriventseva, E. V.; Zdobnov, E. M. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31(19), 3210–3212. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Madan, A. CAP3: A DNA Sequence Assembly Program. Genome Res. 1999, 9(9), 868–877. [Google Scholar] [CrossRef]
- Gilbert, D. EvidentialGene Genome Informatics Software May 2013.
- Altschul, S. F.; Gish, W.; Miller, W.; Myers, E. W.; Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 1990, 215(3), 403–410. [Google Scholar] [CrossRef]
- Yi, L.; Liu, L.; Melsted, P.; Pachter, L. A direct comparison of genome alignment and transcriptome pseudoalignment. BioRxiv 2018, 444620. [Google Scholar] [CrossRef]
- Robinson, M. D.; McCarthy, D. J.; Smyth, G. K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26(1), 139–140. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V. K.; Mukherjee, S.; Ebert, B. L.; Gillette, M. A.; Paulovich, A.; Pomeroy, S. L.; Golub, T. R.; Lander, E. S.; Mesirov, J. P. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 2005, 102(43), 15545–15550. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N. S.; Wang, J. T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13(11), 2498–2504. [Google Scholar] [CrossRef]
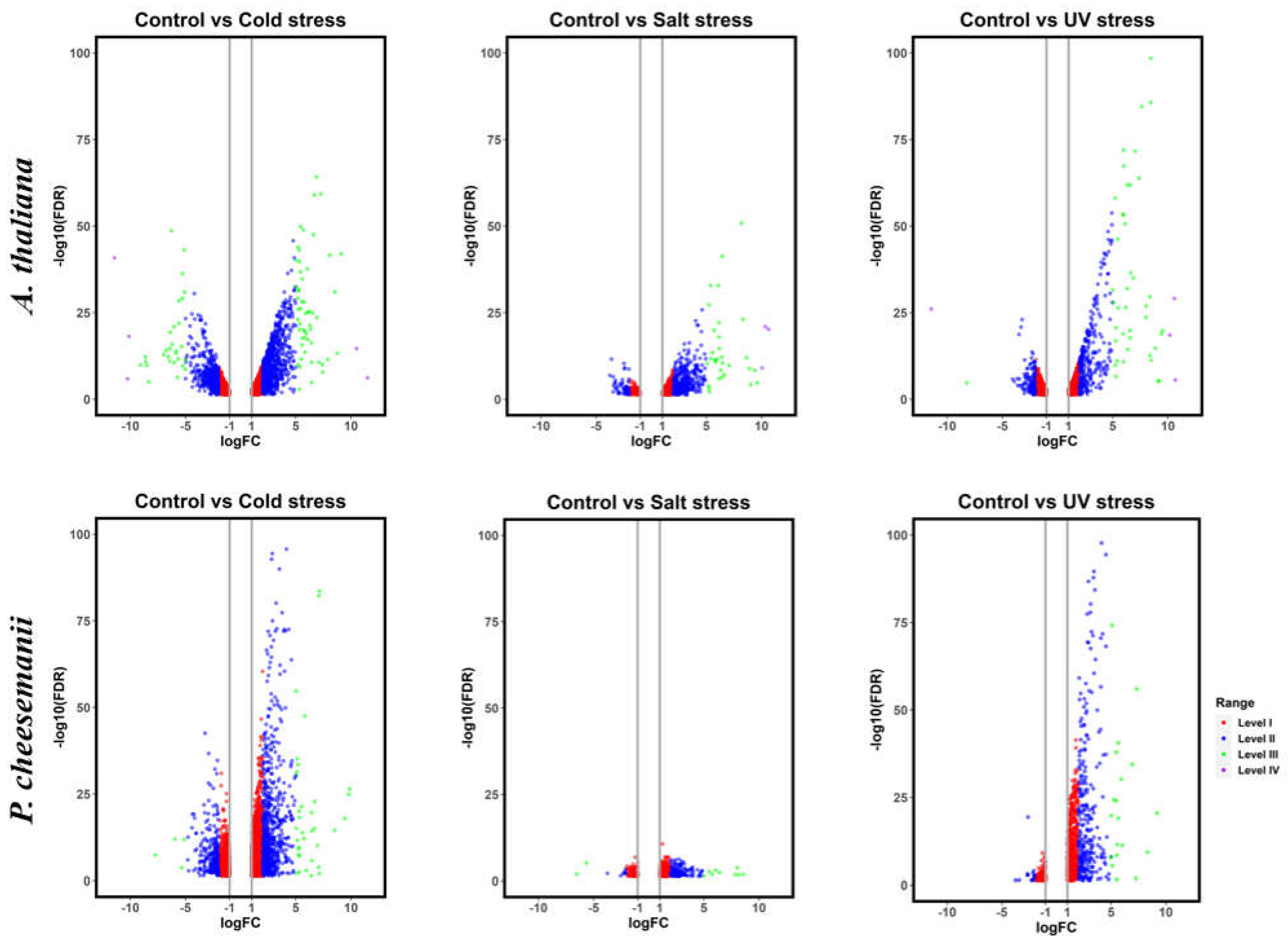
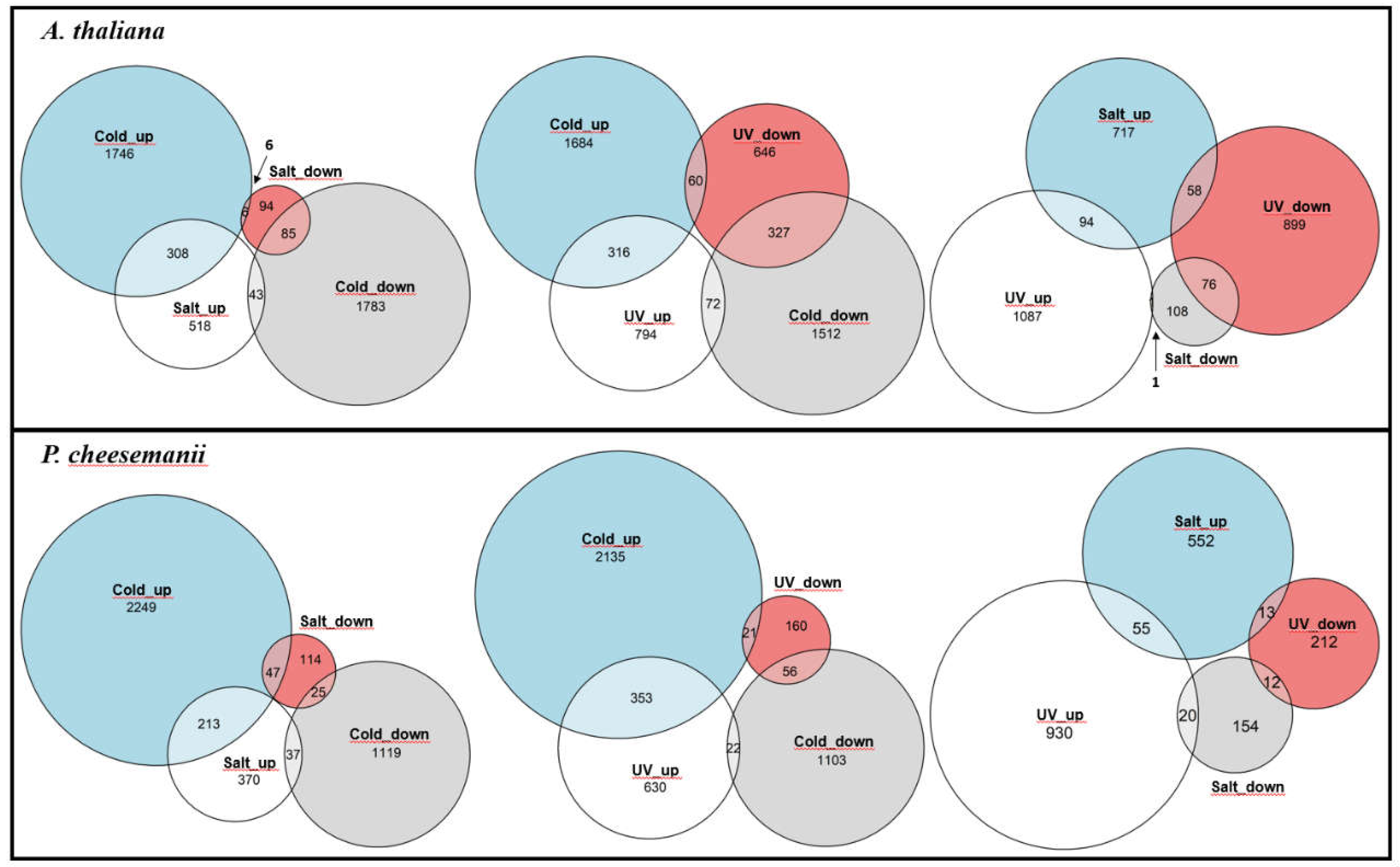
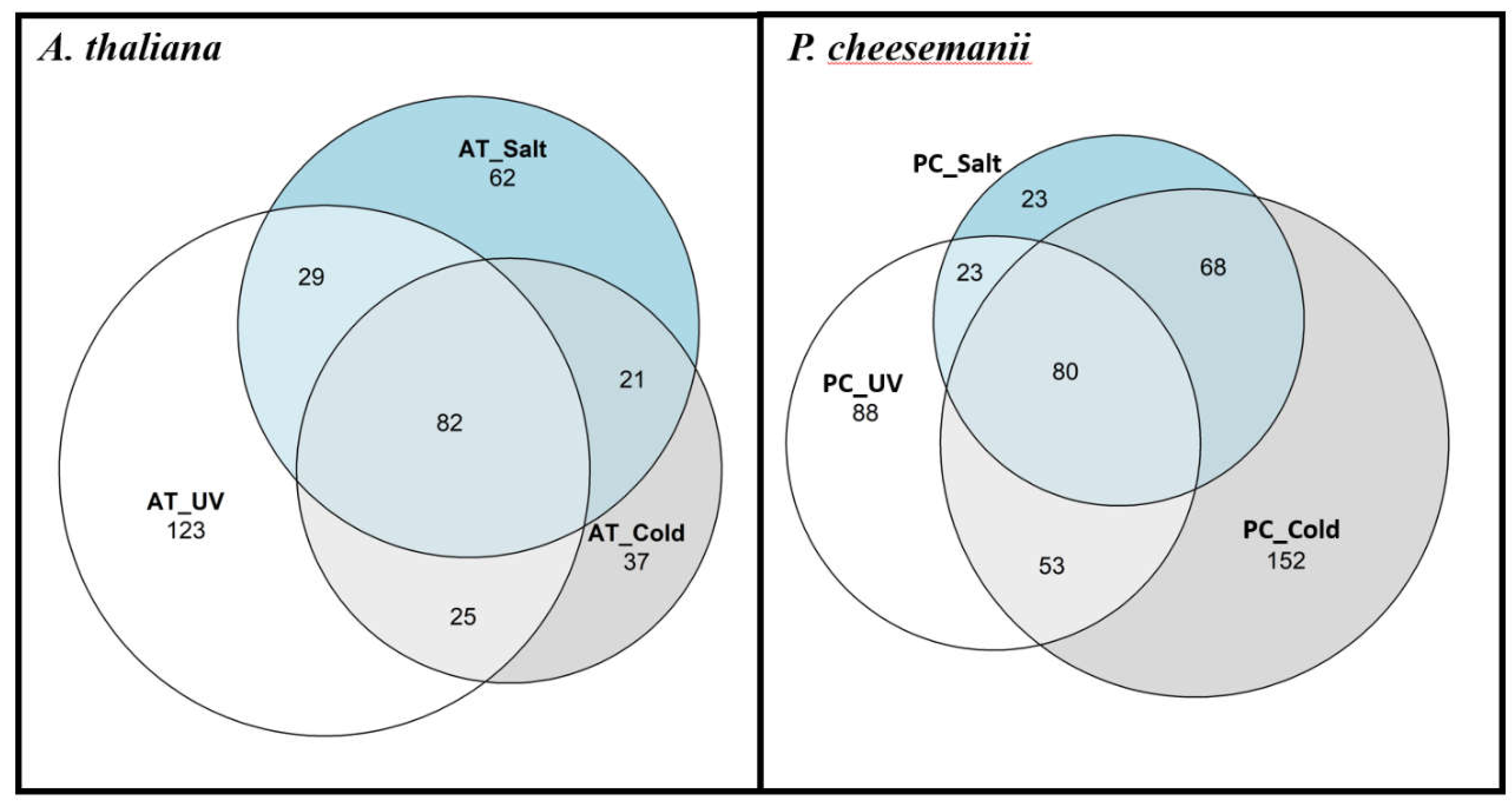
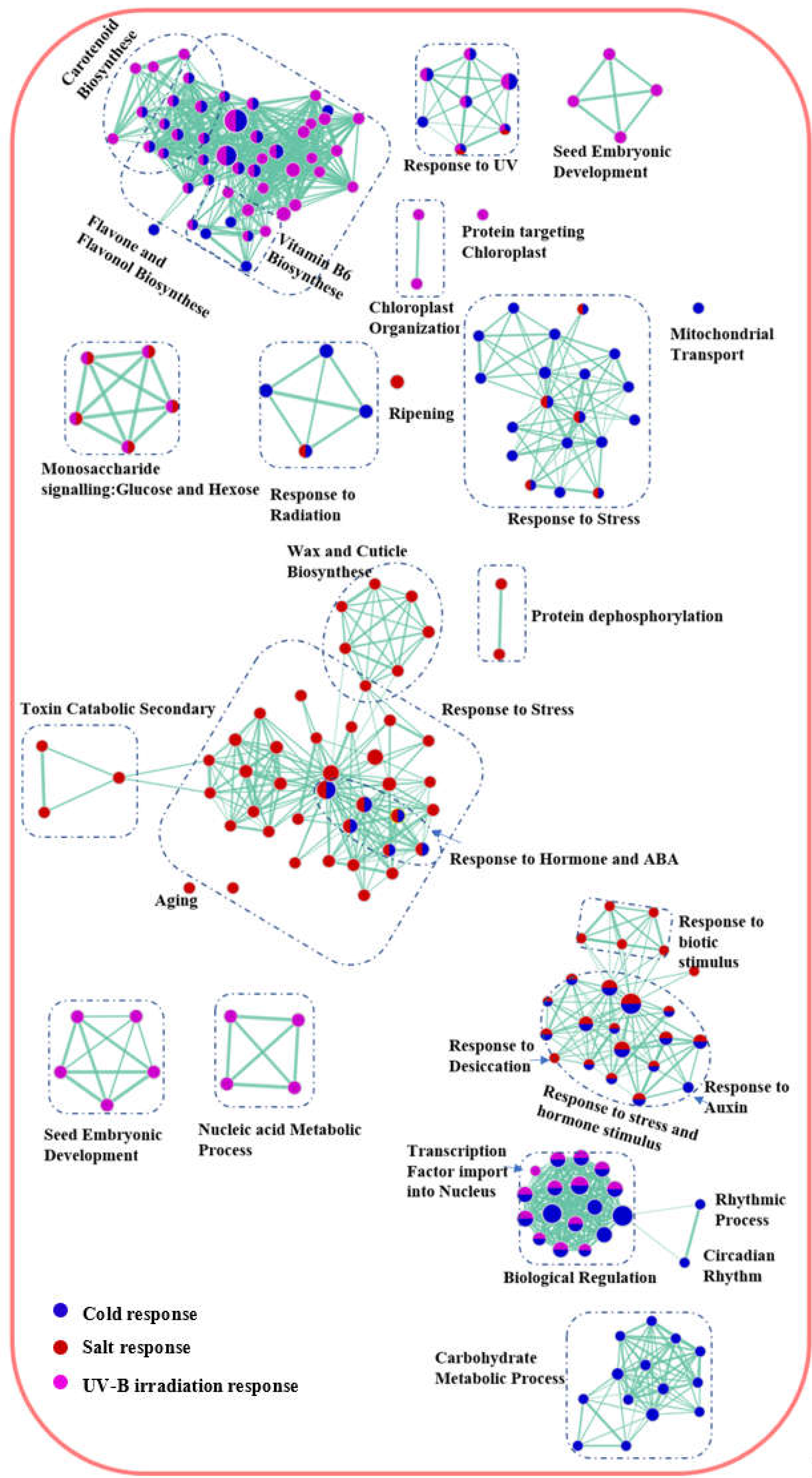
| Species | Stress | Upregulated Genes | Downregulated Genes | Level I | % | Level II | % | Level III | % | Level IV | % | Total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A. thaliana | Cold | 2,060 | 1,911 | 2,641 | 66.5 | 1,242 | 31.3 | 83 | 2.1 | 5 | 0.1 | 3,971 |
| Salt | 869 | 185 | 596 | 56.5 | 423 | 40.1 | 32 | 3.0 | 3 | 0.3 | 1,054 | |
| UV-B | 1,181 | 1,033 | 1,763 | 79.6 | 404 | 18.2 | 43 | 1.9 | 4 | 0.2 | 2,214 | |
| P. cheesemanii | Cold | 2,509 | 1,181 | 2,730 | 74.0 | 918 | 24.9 | 42 | 1.1 | 0 | 0 | 3,690 |
| Salt | 620 | 186 | 635 | 78.8 | 160 | 19.9 | 11 | 1.4 | 0 | 0 | 806 | |
| UV-B | 1,005 | 237 | 937 | 75.5 | 284 | 22.9 | 20 | 1.6 | 0 | 0 | 1,242 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





