1. Introduction
Enterococci are ubiquitous Gram-positive bacteria in nature and commensals in humans, constituting approximately 1% of the fecal microbiota [
1]. The species
Enterococcus faecalis and
Enterococcus faecium are the most common in humans, they are commensals in the oral cavity, intestine, and genitals, but can become opportunistic pathogens causing bacteriemia, endocarditis, meningitis as well as wound, soft tissue, prosthetic and urinary infections.
E. faecalis is also involved in periodontitis, in periimplantitis and endodontic infections [1-3]. Enterococci are naturally resistant to many classes of antibiotics, and resistance is often acquired by selective pressure caused by the inappropriate use of antibiotics. Vancomycin-resistant enterococci (VRE) are a frequent cause of epidemics and have been declared as "high priority" and "serious treat" pathogens by the World Health Organization. About 20 thousand deaths were caused by VRE worldwide in 2019 [
4].
Vancomycin was isolated from
Streptomyces orientalis in 1957. It is a glycopeptide that inhibits wall synthesis of Gram-positive bacteria [
5]. The low toxicity of the drug and its effectiveness in the treatment of methicillin-resistant Staphylococcus aureus (MRSA) epidemics have favored both the adoption and widespread use of vancomycin.
The molecular mechanism of acquiring vancomycin resistance is linked to the presence of operons that can be horizontally transferred by mobile genetic elements. Although there are a variety of operons that confer vancomycin resistance in enterococci, only the
vanA- and
vanB-types are widely spread and thus receive greater public health attention [
6]. The operon
vanA is found on the transposon Tn1546 and takes its name from one of its seven genes, the gene of the ligase
vanA [
7]. The
vanB operon, characterized by
vanB ligase, is often integrated into the host genome [
8]. The prevalence of v
anA and
vanB types varies worldwide;
vanA is more common in North America and Europe, while
vanB is common in Oceania and is increasing in Europe [9-11]. Mobile elements can often simultaneously transfer multiple antibiotic resistances. This complicates the clinical treatment of infections and management of surveillance and containment programs.
Clinically relevant bacterial species that frequently exhibit vancomycin resistance include
E. faecium,
E. faecalis, and
S. aureus. According to the Surveillance Atlas of the Infectious Diseases European Centre for Disease Prevention and Control website (
http://atlas.ecdc.europa.eu/public/index.aspx), the prevalence of VRE is increasing rapidly. In particular, the percentage of vancomycin-resistant
E. faecium infections has more than doubled in the last five years, reaching 28.2% in 2021. Vancomycin resistance in
E. faecalis and
S. aureus was rarer, with values of approximately 1% in Italy.
Acquired vancomycin resistance is most prevalent among enterococci and is still rare in other pathogenic bacteria, such as
S. aureus and
Clostridium difficile [
5,
12]. Although the number of cases of vancomycin-resistant
S. aureus (VRSA) infection is limited, this poses a potential threat to public health, because vancomycin is a key bactericidal drug used for the treatment of invasive MRSA infection. It was observed that most of the VRSA strains acquired vancomycin resistance by the transposon Tn1546 from
E. faecalis [
5]. Successful transfer of the van element from
E. faecalis to an MRSA strain in a mixed infection was confirmed
in vitro and in mice [
13].
Nosocomial infections represent only a part of the field in which the game for containment of antibiotic resistance is played. The zootechnical field is equally relevant, where the wide use of antibiotics generates strong positive selective pressure and could allow the spread of resistant bacteria to the general population through foodstuff [14-16].
The general population may represent a significant reservoir for bacterial species and virulent strains that need to be monitored. An increasing number of reports claim that enterococci routinely inhabit the oral cavity [
17,
18].
E faecalis seems also involved in periodontitis, periimplantitis, and endodontic infections [
2,
3]. Interestingly, it was suggested that oral plaque could be an environment facilitating horizontal gene transfer and the spread of antibiotic resistance genes among biofilm inhabitants [
17]. This provides a rationale for investigating the prevalence of
E. faecalis and
E. faecium in a large sample of healthy Italian subjects and for monitoring the presence of vancomycin resistance genes to understand whether the oral cavity could represent a silent reservoir of virulent enterococci.
2. Results
A sample of 862 participants was investigated to detect sequences of
E. faecium,
E. faecalis and operons that confer vancomycin resistance of the
vanA and
vanB types. Biological specimens obtained from throat swabs were selected and validated in a previous study [
19]. The sample study was randomly selected and not representative of the population; indeed, it was enriched in females (61%) and young adults. As expected,
E. faecalis was found more frequently than
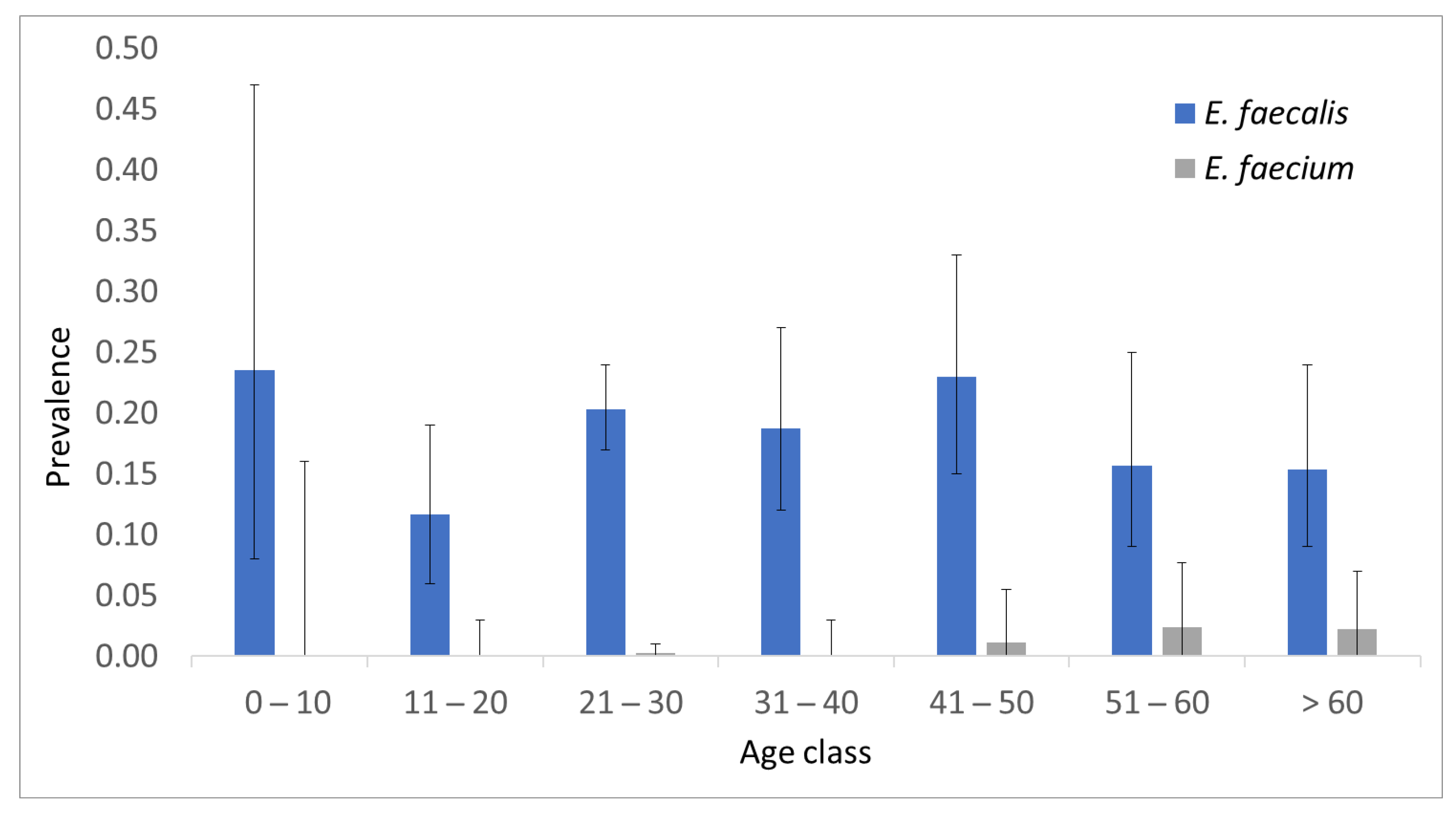
E. faecium. The overall prevalence of these two species was 0.185 (95% C.I. 0.161-0.213) and 0.007 (95% C.I. 0.003-0.014), respectively. The prevalence stratified by age group is shown in
Figure 1.
The age class 11-20 showed the lower prevalence of E. faecalis, but the difference was not significant with respect to the rest of the sample (p value = 0.06). Five out of six E. faecium-positive samples came from people older than 49 years; the prevalence of E. faecium in more senior volunteers (0.0192) was significantly higher than that in younger people (0.0017; p value = 0.005).
The occurrence of
E. faecalis and
E. faecium was not associated with sex, smoking habits, or alcoholic beverage consumption (
Table 1 and
Table 2).
The sequence of the vanA gene was not detected, whereas only one sample was positive in the vanB assay. Interestingly, this sample tested negative for both E. faecalis and E. faecium.
3. Discussion
Enterococci are both common commensals and major opportunistic human pathogens.
E. faecalis and
E. faecium, that are the most common species in humans, frequently cause nosocomial infections. Since the fraction of VRE infections in Italy is rapidly increasing, we evaluated whether healthy population could represent a silent reservoir of these virulent species. A sample of 862 non-hospitalized volunteers was tested to detect
E. faecalis and
E. faecium specific DNA sequences, as well as sequences of
vanA and
vanB genes conferring vancomycin resistance. The overall prevalence of
E. faecalis in oral specimens was 18.5%. This value was similar to that observed in Brazil, where 17% of the samples were positive for enterococci, mostly being
E. faecalis [
20], or in the USA, where 20% of patients with healthy periodontium tested positive for
E. faecalis [
21]; however, it was much lower than that found in periodontal patients (70%) or in insulin treated diabetics [
22]. The prevalence of
E. faecalis in our sample was not influenced by alcohol consumption or smoking. Age was also not influential, in contrast to that observed in the Brazilian cohort, where an increasing degree of carriage in adults and the elderly was observed.
The prevalence of
E. faecium in the present study was 0.7%. Few data regarding oral prevalence of this bacteria in healthy subjects was published. In the above-mentioned Brazilian study,
E. faecium was 50 times rarer than
E. faecalis, being detected only two times out of 240 subjects. Enterococci were never detected among 30 healthy controls from India, but were common in the subgingival biofilm of patients with chronic periodontitis, where
E. faecium was found in 10% of patients and
E. faecalis in 85% of patients [
23].
The screening for vancomycin resistance identified only one sample that was positive for the
vanB type and none for the
vanA type. The
vanA and
vanB genes characterize the mobile genetic elements responsible for acquired vancomycin resistance in clinically relevant enterococci. In several European countries,
vanA enterococci have been isolated from the community and from sewage, feces from farm animals, and raw meat for human consumption, acquired at retail [14,24-26]. Intestinal colonization with VRE in the healthy population has been reported in Belgium, Morocco, and Taiwan at a rate of > 20% [27-29]. On the other hand, no VRE was found in healthy Iranian children [
30]. Unlike these studies, which examined stool samples, our study was based on samples from the oral cavity. We found that the prevalence of
E. faecium infection was relatively low in this anatomical region. Considering that this species is the most relevant among the VRE species, this fact may partially explain our results. Overall, we can conclude that the oral cavity of healthy subjects with good oral hygiene is not a significant reservoir for VRE. With the limitation of the small sample size, a confirmation comes from a study in which
vanA was not found in a small sample of 20 healthy individuals and 20 periodontitis patients [
31].
In the present study, the detection of enterococci and vancomycin resistance was conducted using PCR. Compared to traditional cell culture methods, molecular detection of DNA is more suitable for large-scale screening projects because it is faster and less expensive. When compared, PCR appeared to be more sensitive, possibly because it can detect DNA also from dead cells [
20,
21]. Traditional microbiological methods can provide precise phenotyping of the cultured species. This could be an advantage, particularly for antibiotic resistance. In this study, a single sample carrying the vancomycin resistance
vanB gene was detected; however, this sample was negative for
E. faecium and
E. faecalis. Other species can acquire vancomycin resistance operons, for instance
S. aureus and
C. difficile, but without culture phenotyping, it is impossible to discriminate between different hypotheses.
Overall, this investigation indicates that the oral cavity can be considered a reservoir of clinically relevant enterococci; however, the results suggest that healthy individuals rarely carry vancomycin-resistant strains.
4. Materials and Methods
4.1. Sample Collection
The research sample included 862 oral flora specimens collected from healthy human volunteers. Subjects were randomly selected from the dental clinics of the University of Tor Vergata (Italy) and University of L’Aquila (Italy) between December 2017 and March 2019. All enrolled subjects signed an informed consent form before sample collection. Parents signed an additional consent form for the participating children. The L’Aquila Ethics Committee approved this study (approval number 26/2017).
Patients with systemic disease, facial trauma, those who had undergone radio and/or chemotherapy, or poor oral hygiene were also excluded.
A sample of oral flora swabbing the surface of the tonsils and oropharynx was collected from each subject. The swabs were immediately placed in a test tube containing silica gel capsules to dry the specimens, which were then briefly stored at 4 °C until DNA purification. Briefly, cells were lysed in two steps using lysozyme and proteinase K, then DNA was automatically purified on silica filters using the QIAcube HT instrument and the QIAamp 96 DNA QIAcube HT Kit (Qiagen, Hilden, Germany).
4.2. Molecular Analysis
E. faecalis,
E. faecium,
vanA, and
vanB genes were detected using real-time quantitative polymerase chain reaction (real-time qPCR). The absolute quantification assay for each specific target was performed using hydrolysis probes in the ABI PRISM 7500 thermal cycler (Applied Biosystems, Foster City, CA, USA). Highly specific primers-probe sets were designed with the Primer-BLAST online tool [
32]. The primer quality was further checked with MFEprimer v3.0 software, which also helped to set the multiplex PCR assays [
19]. Oligonucleotides were synthesized by Biomers.net (Ulm, Germany), and their sequences are reported in
Table 3.
Each 20 µL reaction contained 10 µL of 2× qPCRBIO Probe Mix Lo-ROX, 100 ng of template DNA purified from samples, 200 nM of each primer, and 100 nM fluorescent probe. The thermal protocol started with 10′ at 95 °C to activate the polymerase, followed by 40 cycles of two-step amplification: 15′′ at 95 °C and 60′′ at 57 °C. Cloned synthetic DNA target sequences (Eurofin MWG Operon) were used as standards for the quantitative analysis. Serial dilutions of plasmids were used to set PCR reactions with a scalar number of target copies, from 10 to 10,000,000, to check the amplification efficiency and to quantify the sample by comparison with the standard curves, that is, threshold cycle values against the log of the copy number.
4.3. Statistical Analysis
Data analysis and descriptive statistics were performed using SPSS software v.25 (IBM, New York, NY, USA). Associations between variables were analyzed using two-way contingency tables and Pearson’s chi square test; two-sided
p values < 0.05 were considered significant. Level of association between variables were evaluated using the odds ratio (OR). Prevalence confidence limits were calculated with the binomial proportions tool of the OpenEpi web site (
www.openepi.com, accessed on 2 April 2023).
Author Contributions
Conceptualization, Francesco Carinci and Luca Scapoli; Data curation, Claudio Arcuri and Luigi Baggi; Methodology, Annalisa Palmieri and Agnese Pellati; Software, Luca Scapoli; Validation, Marcella Martinelli, Dorina Lauritano and Luca Scapoli; Visualization, Roberto Gatto; Writing – original draft, Luca Scapoli; Writing – review & editing, Annalisa Palmieri and Marcella Martinelli.
Funding
This research received no external funding.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki and approved by the L’Aquila Ethics Committee (approval number 26/2017).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Sghir, A.; Gramet, G.; Suau, A.; Rochet, V.; Pochart, P.; Dore, J. Quantification of bacterial groups within human fecal flora by oligonucleotide probe hybridization. Appl Environ Microbiol 2000, 66, 2263–2266. [Google Scholar] [CrossRef] [PubMed]
- Jepsen, K.; Falk, W.; Brune, F.; Cosgarea, R.; Fimmers, R.; Bekeredjian-Ding, I.; Jepsen, S. Prevalence and Antibiotic Susceptibility Trends of Selected Enterobacteriaceae, Enterococci, and Candida albicans in the Subgingival Microbiota of German Periodontitis Patients: A Retrospective Surveillance Study. Antibiotics (Basel) 2022, 11. [Google Scholar] [CrossRef] [PubMed]
- Kensara, A.; Saito, H.; Mongodin, E.F.; Masri, R. Microbiological profile of peri-implantitis: Analyses of microbiome within dental implants. J Prosthodont 2023. [Google Scholar] [CrossRef]
- Antimicrobial Resistance, C. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- Cong, Y.; Yang, S.; Rao, X. Vancomycin resistant Staphylococcus aureus infections: A review of case updating and clinical features. J Adv Res 2020, 21, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Cetinkaya, Y.; Falk, P.; Mayhall, C.G. Vancomycin-resistant enterococci. Clin Microbiol Rev 2000, 13, 686–707. [Google Scholar] [CrossRef] [PubMed]
- Arthur, M.; Molinas, C.; Depardieu, F.; Courvalin, P. Characterization of Tn1546, a Tn3-related transposon conferring glycopeptide resistance by synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J Bacteriol 1993, 175, 117–127. [Google Scholar] [CrossRef]
- Quintiliani, R., Jr.; Courvalin, P. Conjugal transfer of the vancomycin resistance determinant vanB between enterococci involves the movement of large genetic elements from chromosome to chromosome. FEMS Microbiol Lett 1994, 119, 359–363. [Google Scholar] [CrossRef]
- Lee, T.; Pang, S.; Abraham, S.; Coombs, G.W. Molecular characterization and evolution of the first outbreak of vancomycin-resistant Enterococcus faecium in Western Australia. Int J Antimicrob Agents 2019, 53, 814–819. [Google Scholar] [CrossRef]
- Azzam, A.; Elkafas, H.; Khaled, H.; Ashraf, A.; Yousef, M.; Elkashef, A.A. Prevalence of Vancomycin-resistant enterococci (VRE) in Egypt (2010-2022): a systematic review and meta-analysis. J Egypt Public Health Assoc 2023, 98, 8. [Google Scholar] [CrossRef]
- Werner, G.; Neumann, B.; Weber, R.E.; Kresken, M.; Wendt, C.; Bender, J.K.; group, V.R.E.s. Thirty years of VRE in Germany - "expect the unexpected": The view from the National Reference Centre for Staphylococci and Enterococci. Drug Resist Updat 2020, 53, 100732. [Google Scholar] [CrossRef] [PubMed]
- Ammam, F.; Marvaud, J.C.; Lambert, T. Distribution of the vanG-like gene cluster in Clostridium difficile clinical isolates. Can J Microbiol 2012, 58, 547–551. [Google Scholar] [CrossRef] [PubMed]
- Noble, W.C.; Virani, Z.; Cree, R.G. Co-transfer of vancomycin and other resistance genes from Enterococcus faecalis NCTC 12201 to Staphylococcus aureus. FEMS Microbiol Lett 1992, 72, 195–198. [Google Scholar] [CrossRef] [PubMed]
- Klare, I.; Heier, H.; Claus, H.; Bohme, G.; Marin, S.; Seltmann, G.; Hakenbeck, R.; Antanassova, V.; Witte, W. Enterococcus faecium strains with vanA-mediated high-level glycopeptide resistance isolated from animal foodstuffs and fecal samples of humans in the community. Microb Drug Resist 1995, 1, 265–272. [Google Scholar] [CrossRef]
- Devriese, L.A.; Ieven, M.; Goossens, H.; Vandamme, P.; Pot, B.; Hommez, J.; Haesebrouck, F. Presence of vancomycin-resistant enterococci in farm and pet animals. Antimicrob Agents Chemother 1996, 40, 2285–2287. [Google Scholar] [CrossRef] [PubMed]
- Wegener, H.C.; Aarestrup, F.M.; Jensen, L.B.; Hammerum, A.M.; Bager, F. Use of antimicrobial growth promoters in food animals and Enterococcus faecium resistance to therapeutic antimicrobial drugs in Europe. Emerg Infect Dis 1999, 5, 329–335. [Google Scholar] [CrossRef] [PubMed]
- Sedgley, C.M.; Nagel, A.C.; Shelburne, C.E.; Clewell, D.B.; Appelbe, O.; Molander, A. Quantitative real-time PCR detection of oral Enterococcus faecalis in humans. Arch Oral Biol 2005, 50, 575–583. [Google Scholar] [CrossRef]
- Roberts, A.P.; Mullany, P. Oral biofilms: a reservoir of transferable, bacterial, antimicrobial resistance. Expert Rev Anti Infect Ther 2010, 8, 1441–1450. [Google Scholar] [CrossRef]
- Scapoli, L.; Palmieri, A.; Pellati, A.; Carinci, F.; Lauritano, D.; Arcuri, C.; Baggi, L.; Gatto, R.; Martinelli, M. Prevalence of Staphylococcus aureus and mec-A Cassette in the Throat of Non-Hospitalized Individuals Randomly Selected in Central Italy. Antibiotics (Basel) 2022, 11. [Google Scholar] [CrossRef]
- Komiyama, E.Y.; Lepesqueur, L.S.; Yassuda, C.G.; Samaranayake, L.P.; Parahitiyawa, N.B.; Balducci, I.; Koga-Ito, C.Y. Enterococcus Species in the Oral Cavity: Prevalence, Virulence Factors and Antimicrobial Susceptibility. PLoS One 2016, 11, e0163001. [Google Scholar] [CrossRef]
- Sedgley, C.; Buck, G.; Appelbe, O. Prevalence of Enterococcus faecalis at multiple oral sites in endodontic patients using culture and PCR. J Endod 2006, 32, 104–109. [Google Scholar] [CrossRef] [PubMed]
- Chomicz, L.; Szubinska, D.; Piekarczyk, J.; Wojtowicz, A.; Piekarczyk, B.; Starosciak, B.; Fiedor, P. [Occurrence of oral subclinical infections in insulin treated diabetics]. Wiad Parazytol 2004, 50, 177–180. [Google Scholar] [PubMed]
- Bhardwaj, S.B.; Mehta, M.; Sood, S. Enterococci in the oral cavity of periodontitis patients from different urban socioeconomic groups. Dent Res J (Isfahan) 2020, 17, 147–151. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Migura, L.; Liebana, E.; Jensen, L.B.; Barnes, S.; Pleydell, E. A longitudinal study to assess the persistence of vancomycin-resistant Enterococcus faecium (VREF) on an intensive broiler farm in the United Kingdom. FEMS Microbiol Lett 2007, 275, 319–325. [Google Scholar] [CrossRef]
- Talebi, M.; Sadeghi, J.; Rahimi, F.; Pourshafie, M.R. Isolation and Biochemical Fingerprinting of Vancomycin-Resistant Enterococcus faecium From Meat, Chicken and Cheese. Jundishapur J Microbiol 2015, 8, e15815. [Google Scholar] [CrossRef]
- Nilsson, O.; Alm, E.; Greko, C.; Bengtsson, B. The rise and fall of a vancomycin-resistant clone of Enterococcus faecium among broilers in Sweden. J Glob Antimicrob Resist 2019, 17, 233–235. [Google Scholar] [CrossRef]
- Van der Auwera, P.; Pensart, N.; Korten, V.; Murray, B.E.; Leclercq, R. Influence of oral glycopeptides on the fecal flora of human volunteers: selection of highly glycopeptide-resistant enterococci. J Infect Dis 1996, 173, 1129–1136. [Google Scholar] [CrossRef]
- Wang, J.T.; Chang, S.C.; Wang, H.Y.; Chen, P.C.; Shiau, Y.R.; Lauderdale, T.L.; Hospitals, T. High rates of multidrug resistance in Enterococcus faecalis and E. faecium isolated from inpatients and outpatients in Taiwan. Diagn Microbiol Infect Dis 2013, 75, 406–411. [Google Scholar] [CrossRef]
- Hannaoui, I.; Barguigua, A.; Serray, B.; El Mdaghri, N.; Timinouni, M.; Ait Chaoui, A.; El Azhari, M. Intestinal carriage of vancomycin-resistant enterococci in a community setting in Casablanca, Morocco. J Glob Antimicrob Resist 2016, 6, 84–87. [Google Scholar] [CrossRef]
- Jannati, E.; Amirmozaffari, N.; Saadatmand, S.; Arzanlou, M. Faecal carriage of high-level aminoglycoside-resistant and ampicillin-resistant Enterococcus species in healthy Iranian children. J Glob Antimicrob Resist 2020, 20, 135–144. [Google Scholar] [CrossRef]
- Kim, S.M.; Kim, H.C.; Lee, S.W. Characterization of antibiotic resistance determinants in oral biofilms. J Microbiol 2011, 49, 595–602. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics 2012, 13, 134. [Google Scholar] [CrossRef] [PubMed]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).