1. Introduction
A comprehensive understanding of the genomic and gene expression relatedness between human and other mammals is necessary to evaluate the common pathways, functional similarities and appropriateness of different animal models to study human diseases. Although mouse is the most commonly used animal model to study human diseases [
1], a smaller size and lifespan, and differences in the latency period for a disease [
2] and drug metabolism [
3], inflammatory response [
4], and other processes [
5,
6,
7,
8] suggest, the need to identify larger animal models to study human diseases. Comparison of genomic sequences among human, mouse and pig indicated that pig sequences were closer to those of the human with more numbers of ultra-conserved regions compared to mouse [
9,
10,
11]. The nonhuman primates are evolutionarily more closely related to human. After the ban on working with Chimpanzees and other great apes, macaques have become the most closely related nonhuman primate model to study human diseases [
12]. Human disease genes and known drug domains have shown high degree of similarity with rhesus macaque genome [
13]. The marmoset, on the other hand, is a useful animal model due to its short gestation and sexual maturation periods while having more sequence similarity with humans than rodents [
12]. It is also a useful model to studying diseases in neurobiology [
14]. A comparative analysis of gene expression and disease-associated genetic variations across commonly used animal models will help understand the similarities leading to appropriate use of specific animal models for disease-specific research.
In this study, we retrieved the protein-coding sequences (CDS) from two rodents (mouse and rat), the pig, two nonhuman primates (rhesus macaque and marmoset) and humans to identify conserved CDS across the six species. Single nucleotide polymorphisms (SNPs) from these CDS were mapped to corresponding disease associations to identify common and unique clinically associated SNPs to better define the relevance of an animal model with various human diseases. Finally, comparative transcriptomic analysis of different tissues was performed for conserved CDS in human, mouse and pig (data from marmoset and rhesus macaque were not available) to identify common tissue-specific expression profiles of the conserved CDS to identify the disease-specific relevance of these animal models.
2. Materials and Methods
2.1. Retrieval of protein-coding sequences
Genomic data for human, rhesus macaque, pig, mouse and rat were retrieved from Ensembl [
15] and for marmoset from National Center for Biotechnology Information (NCBI). Datasets include
Homo sapiens (Human - GCA_000001405.28),
Macaca mulatta (Rhesus macaque - GCA_003339765.3),
Callithrix jacchus (Marmoset – NCBI: GCF_009663435.1),
Sus scrofa (Pig - GCA_000003025.6),
Mus musculus (Mouse - GCA_000001635.9) and
Rattus norvegicus (Rat - GCA_000001895.4). We have extracted a total of 19,962 human, 21,591 rhesus macaque, 22,252 marmoset, 21,280 pig, 21,848 mouse and 22,250 rat coding sequences (CDS) for current analysis.
2.2. Identification of similarity between human CDS and other mammalian sequences
Five pairwise sequence comparisons were performed to identify the conserved human CDS that include human vs. rhesus macaque, human vs. marmoset, human vs. pig, human vs. mouse, and human vs. rat. The Basic Local Alignment Search Tool (BLAST) would align and compare a query DNA sequence with a database of sequences. The BLAST database was constructed for human sequences using the makeblastdb application. CDS from the rhesus macaque, marmoset, pig, mouse, and rat sequences were queried against the human BLAST database using BLASTn (options: -max_hsps 1 -max_target_seqs 1) in BLAST+ (version 2.7.1) [
16]. Based on the pairwise alignment, we identified conserved CDS for other mammals against the human. Some sequences from blast hits showed low coverage. To avoid the false positives, we filtered the sequences with greater than 50% identity and covered at least 50% length of the human CDS. Alignments that fall below these criteria were excluded for further analysis. Based on these similarities, conserved sequences were identified across the five comparisons and plotted using UpSetR [
17]. To understand the synteny block distribution for the human conserved CDS on different chromosomes of the five species, we created circos plots using the R package, shinyCircus [
18] using the human chromosome as the reference.
2.3. Comparison of conserved CDS and identification of SNPs and their associated diseases
Multiple sequence alignment was performed across the six species using 10,316 conserved CDS using ClustalW2 [
19]and SNPs were extracted from the alignment using SNP-sites [
20] and msa2snp (
https://github.com/pinbo/msa2snp). The Ensembl Variant Effect Predictor [
21] was used to identify SNPs with rs ID (RefSeq) Later, Ensembl Post GWAS and SNPnexus (uses Cosmic, ClinVar and GWAS) [
22] were used to predict human SNP-associated diseases. These diseases were further classified under 25 different categories using the DisGeNET [
23]. Further, we have calculated the percentage for each disease category (Number of diseases in each category / Total number of identified diseases) across all the comparisons. We also identified the conserved and species-specific SNPs associated with disease for a particular animal model. Those SNPs were plotted using R packages, such as shinyCircus [
18] and karyoploteR [
24].
2.4. Construction of phylogenetic tree
Multiple sequence alignments of 10,316 conserved CDS from six organisms were concatenated as per their order on the human chromosome (1-22, X and Y) using EMBOSS Union [
25]. The phylogeny was constructed using FastTree (parameter –nt –gtr) version 2.1 [
26] and visualized using Molecular Evolutionary Genetics Analysis (MEGA) version 11 [
27].
2.5. Transcriptomic analysis using tissue-specific data
We retrieved RNA-seq expression data on six tissues (colon, heart, kidney, lung, skeletal muscle and spleen) for human, mouse, rat and pig from the Expression Atlas database [
28] with a default minimum expression value of 0.5 Transcripts Per Million (TPM). Such data for marmoset and rhesus macaque were not available, hence these two species were excluded from the transcriptomic analysis. Conserved genes based on our aforementioned BLAST analysis for each tissue were filtered for the four organisms. Correlation analysis was carried out using the R package, corrplot [
29] to detect tissues with highly similar gene expression profiles between the model organisms and the human. Pearson correlation coefficient was used to measure the linear dependence between two variables.
3. Results
3.1. Identification of conserved CDS with human sequences
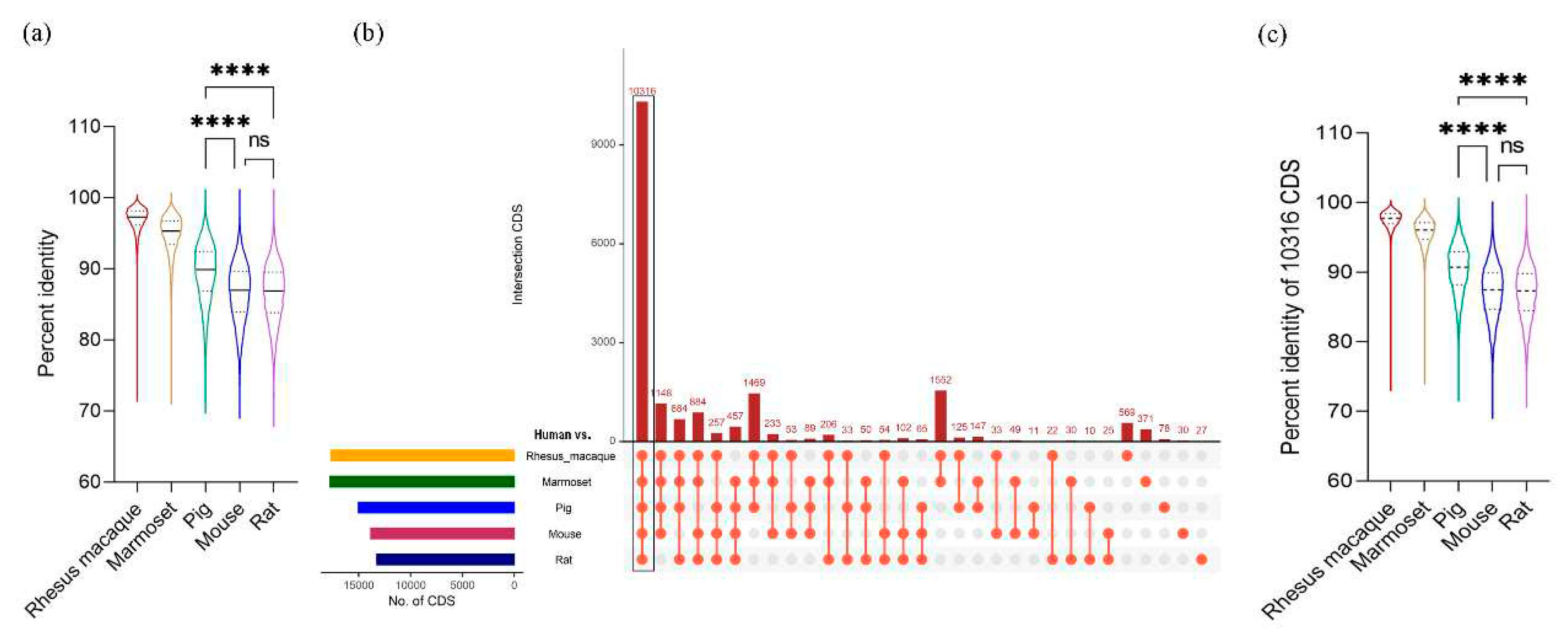
We compared the coding sequences between human and five other species using the BLAST program with cutoffs of at least 50% sequence identity and 50% length match to the human sequences. Results showed that rhesus macaque has the highest average identity (96.82%) followed by marmoset (94.65%), pig (89.37%), mouse (86.65%) and rat (86.53%) (
Table 1). The percent identity ranged from 100 to around 70 that is comparable across all comparison groups (
Table 1). However, the distribution of percent identity of the CDS is not uniform in all comparison groups. In rhesus macaque and marmoset, the identity distribution is skewed towards the median (median for rhesus macaque is: 97.29 and for marmoset is 95.29,
Supplementary Table S1), denoting that the majority of the CDS in these primate species are highly identical to human CDS (
Figure 1a). On the other hand, the identities in pig, mouse and rat are more widely distributed around the median suggesting a varying degree of similarity with certain gene families of human (
Figure 1a and
Supplementary Table S1). Among these three organisms, pigs showed the highest median value (89.89% identity) and significantly higher percent identity with human CDS than those of mouse or rat (
Figure 1a). The complete result of this identity analysis is provided in
Supplementary Table S2.
Based on the pairwise alignment of CDS between human and five other species, 10,316 CDS were found to be conserved across all six species (
Figure 1b and
Supplementary Table S2), which were used for further analyses. In all species, these conserved set of CDS recorded higher percentage identities than those involving all CDS (
Table 1). Among the non-human primates, rhesus macaque showed the highest average identity with the human at 97.53% and pigs showed 90.38% identity, which is significantly higher than those of mice and rats (
Figure 1c and
Supplementary Table S3).
Next, we mapped all conserved CDS from each non-human species to matching positions (by similarity) on human chromosomes to understand their synteny distribution in each species. For this purpose, we used a common color-coding scheme for each chromosome number where the same color represents the same chromosome number in all species. Of note, marmoset has the same number of chromosomes as the human but the other species have fewer. Pig has the least number of with only 18 chromosomes. The circos plots (
Figure 2a) illustrate the mapping of synteny blocks (represented by CDS) from different chromosomes of non-human species using the human chromosomal numbers as a reference. The chromosomes in the non-human species mapped with only color indicate that they contain corresponding human synteny blocks intact and those showing mosaic coloring indicate that the human synteny blocks are distributed on different chromosomes as indicated by different colors. For instance, synteny blocks of conserved CDS from chromosome 1 of macaque (shown in red color) also map to chromosome 1 of the human, but corresponding synteny blocks from other species are mapped to different human chromosomal locations. Similarly, synteny blocks from chromosomes 12 and 13 of the macaque are mapped to chromosome 2 of the human. Of note, the synteny blocks on chromosomes 17, 20 and X are intact in a single chromosome in all the species (as indicated by only one color) while those from other chromosomes are fragmented and distributed in multiple chromosomes (with mosaic color mapping) (
Table 2).
Phylogenetic analysis based on conserved CDS examines the evolutionary distances among the six species. As shown in
Figure 2b, the nonhuman primate (NHP) group has the closest distance to human with the pig positioned in the middle and the rodent group being the farthest from the human. The chromosome specific mapping for all the CDS and conserved CDS were presented in
Supplementary Table S4 and S5, respectively.
3.2. Mapping human disease-relevant SNPs in other species
Mapping of human SNPs to the other species after multiple sequence alignment of 10,316 conserved CDS showed differences in the SNP numbers between the primates and other three organisms. Primates had lower number of predicted SNPs (rhesus macaque: 63,449 and marmoset: 40,181) in the conserved CDS than pig (145,715), mouse (221,383) and rat (220,630). A full list of all the SNPs is provided in the
Supplementary Table S6. The SNPs were then annotated for disease association. Even with a very low number of human SNPs mapped, comparable number of disease associations were identified in rhesus macaque and marmoset as in other three animals (
Table 3). The disease-associated SNPs in each of these six organisms were plotted on each chromosome to easily visualize the distribution and variation of the SNPs across species (
Supplementary Figure S1). The identified diseases with their corresponding rs ID for human versus five animals were listed in
Supplementary Table S7 and
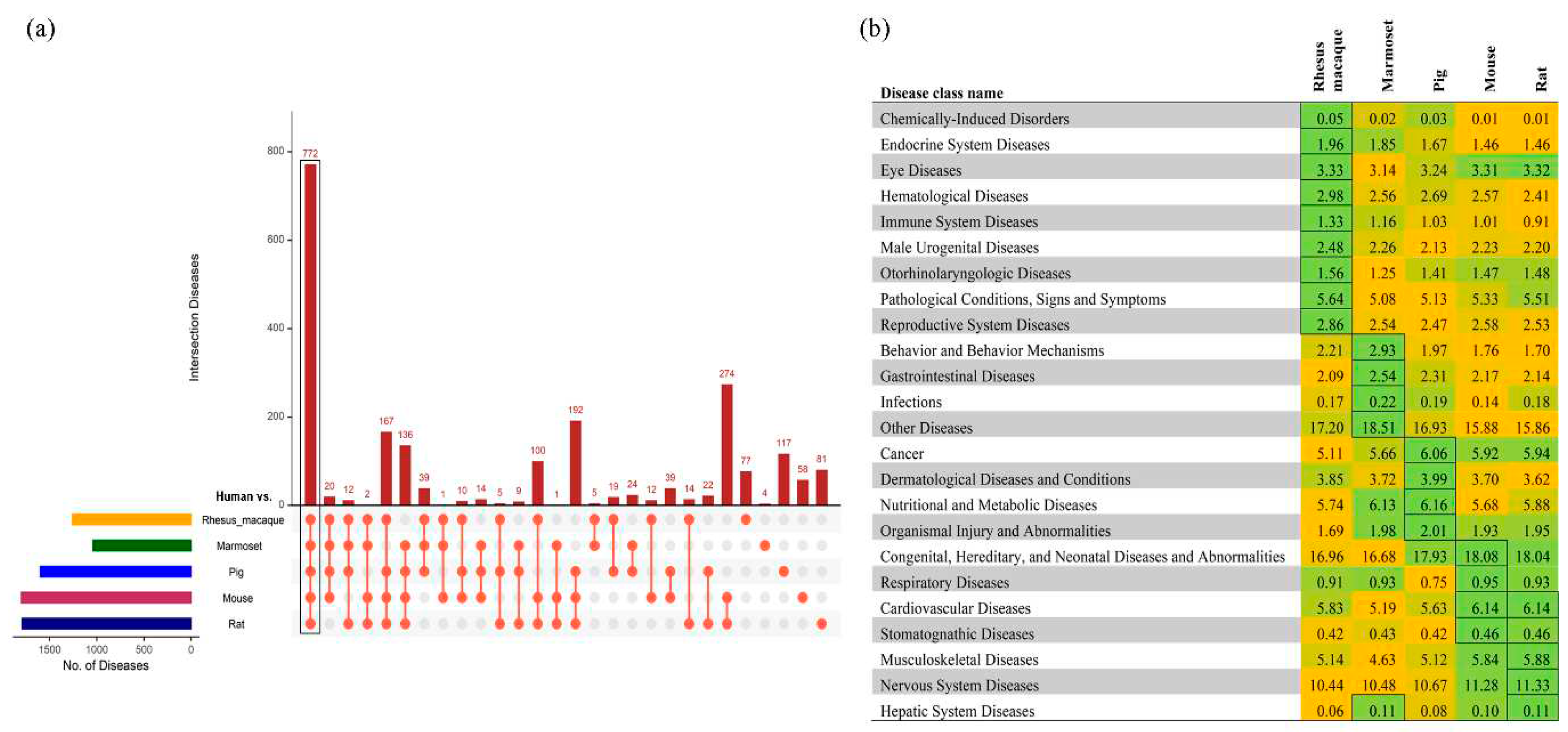
Supplementary Figure S2–S6. Among the predicted diseases based on the human SNPs, 772 were conserved among all six species (
Figure 3a). These 772 diseases were then classified into tissue-specific diseases and compared amongst the five non-human model organisms based on the disease prediction score. The higher the score, the more relevance to the human diseases. Congenital, hereditary and neonatal diseases and abnormalities and nervous system diseases were highly prevalent in all the models (
Figure 3b), while some disease classes were specific to a model organism. SNPs associated with reproductive system diseases were specifically observed in rhesus macaque; behavioral and gastrointestinal diseases in marmoset; cancer and metabolic diseases in pigs; and hepatic and cardiovascular diseases are prevalent in rats and mice (
Figure 3b). These results indicate that human SNPs associated with specific disease classes are prevalent in specific model species, which may provide a basis for selection of appropriate model for a specific disease.
Next, we investigated any model-specific SNP association with human diseases. The pig had the highest number; 117 diseases from 96 SNPs were specific to pigs (
Table 3). The marmoset had the lowest number with only four diseases specifically associated with this species. We have provided a curated list of all model-specific human-associated diseases and identified SNPs with their corresponding genes (
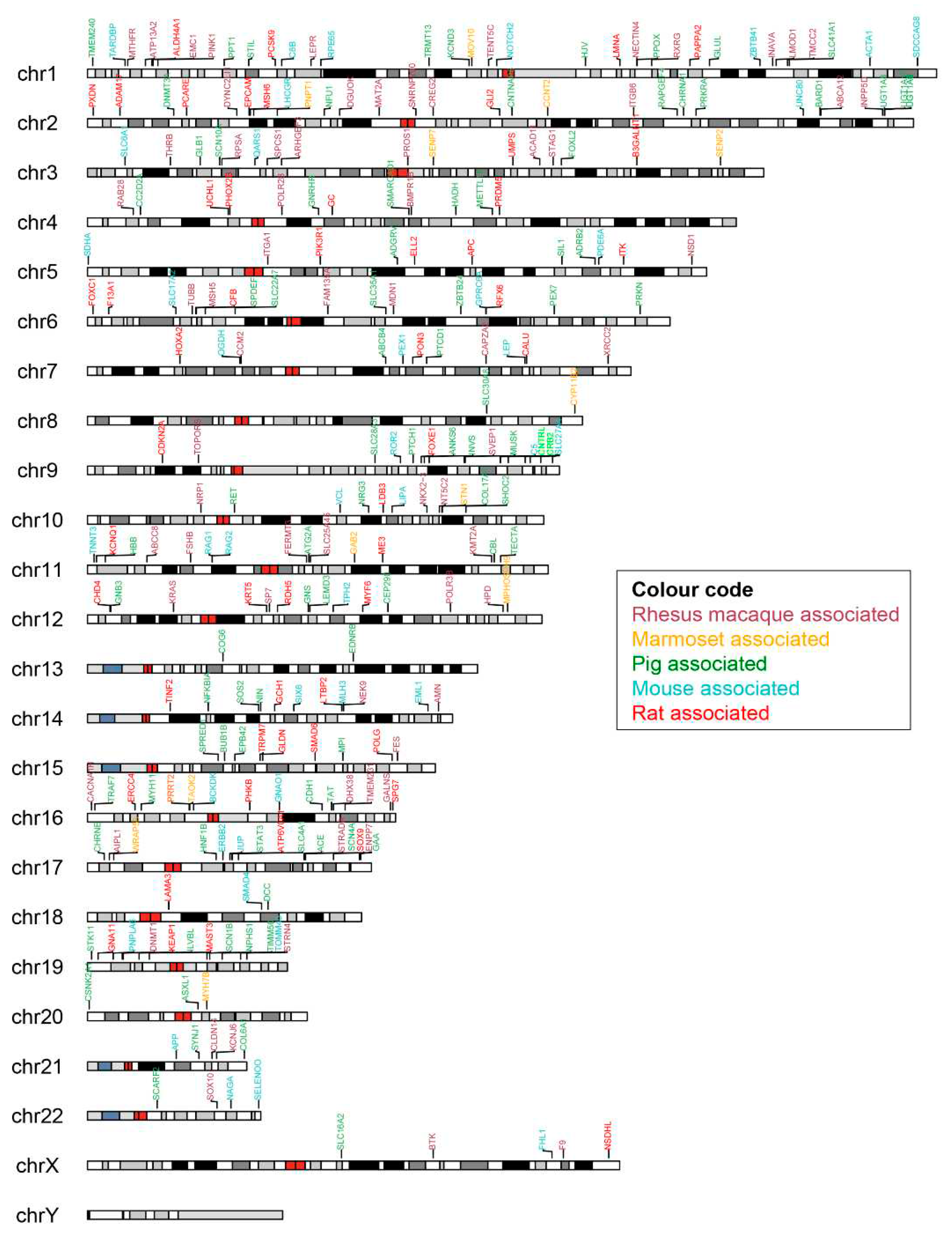
Supplementary Table S8). In our study, none of the disorders were noted in chromosome Y. The pig model had a wide range of distinct diseases spread across all the chromosomes. Exclusively, the two genes detected in chromosome 13, COG6 and EDNRB associated with the pig model cause Shaheen syndrome and Hirschsprung disease (
Figure 4).
Next, we identified the human SNP-bearing genes in the animal models that showed an association with a human disease as shown in the color-coded chromosomal map (
Figure 4). The genes that showed the highest number of disease associations in each animal model corresponding to each human chromosome were listed in
Table 4. The full list of genes per chromosome is provided in
Supplementary Table S7. Six common genes (ATM, POLE, RB1, FBN1, TSC2 and FLNA) were identified across species which were the topmost genes associated with the highest number of diseases in different chromosomes. Two DNA mismatch repair proteins, MSH6 in human chromosome 2 and MLH1 in chromosome 3 were found to have very high disease-association across pig, mouse and rat models. SNPs associated with the Chromosome 7 encoded CFTR (cystic fibrosis transmembrane conductance regulator) gene were high associated in rat and mouse models but not so much in other species. Another gene, PTCH1 (protein patched homolog 1) that is a component of hedgehog pathway is associated with a number of diseases in pigs, especially Holoprosencephaly 7, was identified only in the pig model.
3.3. Transcriptomic analysis of six different tissues
To understand the gene expression similarities between human and the animal models, tissue-specific expression data were accessed from the Expression Atlas database [
28]. Rhesus macaque and marmoset were excluded from the analysis due to non-availability of expression data. Colon, heart, kidney, lung, skeletal muscle and spleen were selected to examine the expression across species. The number of expressed genes in each tissue for the four organisms were identified and listed in
Table 5. Gene expression corresponding to the 10,316 conserved CDS identified from the genomic analysis were taken from each tissue (
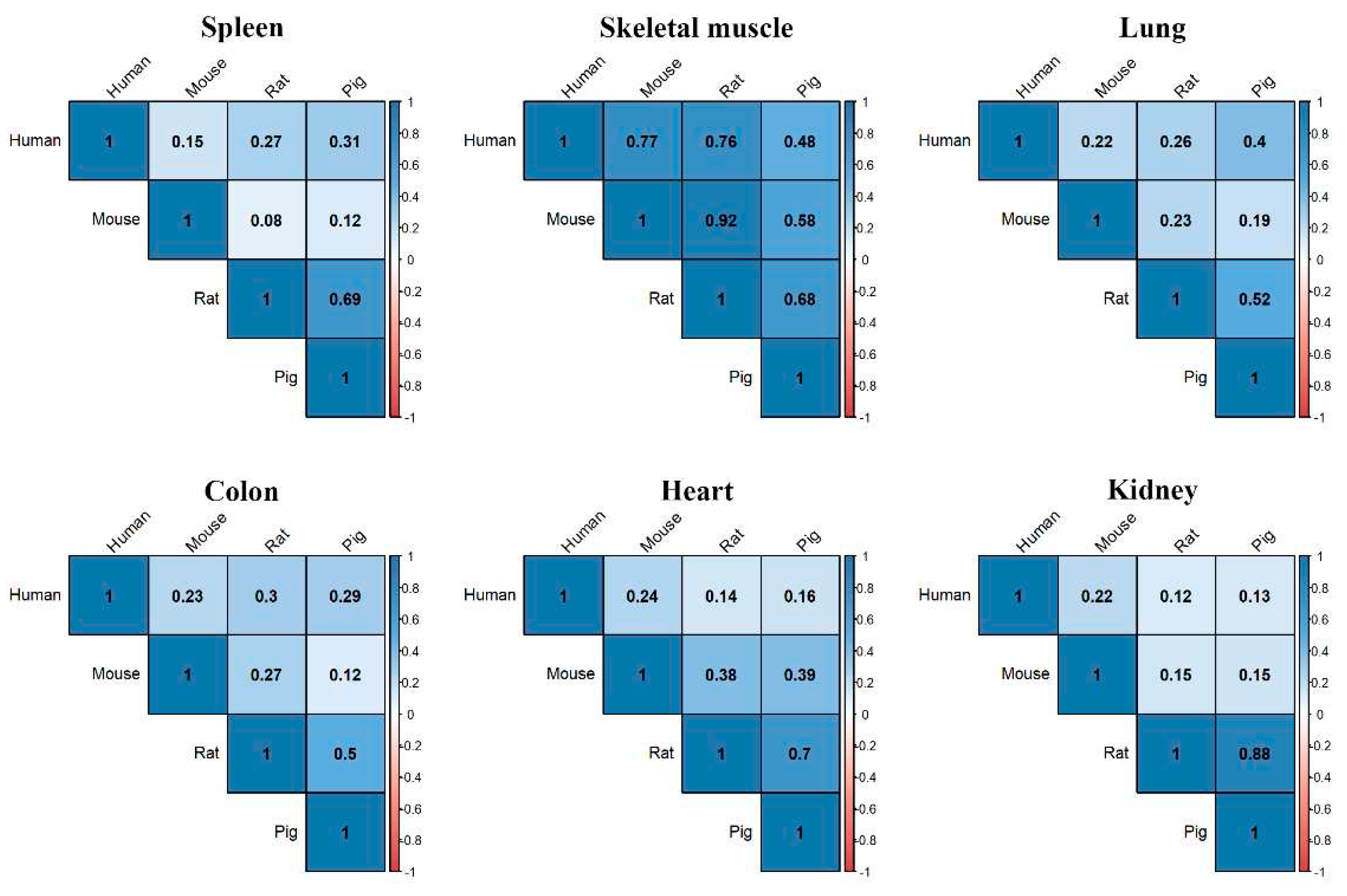
Table 5) and used in subsequent analyses. When we examined the level of gene expression in each tissue and performed correlation analysis with each species, we found that gene expression in spleen of humans was most correlated with the pig followed by rat (
Figure 5). The pig had the highest gene expression correlation with human in the lung and colon, while mouse had the highest in heart, kidney and skeletal muscle (
Figure 5). The gene-level expression for four different organisms in six tissues was plotted in the heatmap and presented in
Supplementary Figure S7–S12.
4. Discussion
The selection of an animal models to study human diseases is based in part on the genetic relatedness to humans. However, species evolution is generally not synchronous with gene evolution, which results in certain human genes having more similarity in certain model organisms, which can influence the selection of different animal models for different research projects. Although primates are known to be evolutionary closer to human and can better mimic the human physiology, use of NHPs is expensive, time-consuming, heavily regulated, and subject to availability [
30,
31]. In this study, we have identified genetic similarity of CDS and mapped disease-associated human SNPs with commonly used animal models, the including the rhesus macaque, marmoset, pig, mouse, and the rat. We also compared human gene expression similarities in multiple tissue types in pig, rat and mouse.
Identification of NHPs as the closest evolutionary neighbors with the human was expected. BLAST-based genome-wide sequence comparison of rhesus macaque and marmoset with the human showed a 95-97% similarity across about 18,000 sequences, and the CDS-level identities also registered 96-98% similarity in these NHP models further supporting the belief that the macaque and marmoset are good animal models to study human diseases (
Table 1). Alternatively, the pig’s genome-level similarity (89.4%) with that of human was higher than that of the rat (86.7%) or mouse (86.5%). This observation remained true at the CDS-level, suggesting that pig is a more suitable model (with respect to genomics) to study human diseases than rodent models (
Table 1).
It should also be noted that pig share the highest number of common genes (96) containing disease-associated SNPs with human than the other examined species (
Table 3), which favors the pig model to study genetically predisposed human diseases. Similarly, these common SNPs between pig and human are also associated with the highest number of human diseases (117), further emphasizing the pig’s strong relatedness to humans. We mapped the SNP- associated diseases on to different disease groups and observed that cancer-associated SNPs had the greatest number in pig; however, the difference in cancer mapping among species was not statistically significant. Nevertheless, examination of multiple levels of relatedness between humans and the pig (genome, CDS, SNPs and cancer disease levels) suggested that pig would be a more accurate genetic model for human cancer research than rodents.
SNP-associated behavioral diseases were mostly observed in the marmoset, which is in concordance with the existing literature [
32,
33]. Our analysis also suggested that the marmoset could also be used to model gastrointestinal diseases, which are found in the marmosets in captivity [
34,
35]. Endocrine and reproductive system diseases were found to be frequent in rhesus macaques, which contributes to the existing evidence supporting the use of the NHP model for adrenal androgen-related and endocrine-based social and reproductive studies [
36,
37]. Nutritional and metabolic disease-associated SNPs were also found to be high in pig. Pigs with high-caloric food intake are prone to developing metabolic syndrome [
38,
39]. In addition, the large body size, omnivorous diet and large gastrointestinal tract in pigs make them a suitable model for nutritional and pharmacological studies [
39]. On the other hand, rodents showed a higher number of SNPs associated with musculoskeletal, cardiovascular and nervous system diseases. Rodents, specifically the rat have been used as hindlimb model to study musculoskeletal parameters [
40]. A rat model for musculoskeletal implant infection was also developed recently [
41]. The mouse has been widely used to study human heart diseases, mainly for myocardial infarction, heart fibrosis, and the cardiomyopathies [
42,
43,
44,
45]. Neurological disorders have also been studied using the rodent models, extensively [
46,
47].
When we specifically analyzed the chromosomes and genes which are associated with human diseases, prevalence of genes in the DNA repair pathway were most commonly found in all of the species. PTCH1 was the topmost SNP-prevalent gene in chromosome 9 for pigs. PTCH1 altered in 2.76% of all cancers was mostly observed to be altered in the colon cancer (TCGA data portal, My cancer genome database [
48]. Similarly in mouse, STK11 was most commonly found in lung cancer appeared as the topmost gene in chromosome 19. Our tissue-based expression analysis also shed more light into the gene expression similarities with human for each species. In pigs, the spleen has the greatest number of genes commonly expressed with human, while the mouse had the lowest among the three species compared. Similarly, the rats had the higher numbers of expressed genes in lung, heart and colon, indicating an advantage of using the rat model for cardiovascular and colonic diseases. With the list of the genes presented in
Table 4, suitable gene-based animal models could be considered to study different human diseases.
5. Conclusions
Using the whole genomic and coding sequence similarities, mapping the human SNPs on to the genomes of five other mammalian species, and tissue-specific expression analysis, this study demonstrated potentially important similarities and differences in genomics and transcriptomics among major model organisms with respect to humans. With respect to this analysis of sequence and expressional data, some species (e.g., NPHs) appeared to be more superior as models of human disease than other species. Overall, it was determined that the pig had greater sequence and expressional homology with humans than rodents had with humans. Based on these data, the pig emerged as a reasonable model to study human diseases, most notably cancer where a related immune system is present. Marmoset models are well positioned to study behavioral and gastrointestinal diseases. Rodents could be a better model for cardiovascular diseases, but have an obvious size discrepancy with humans, and have less overall sequence and expressional homology with humans than the pig has with humans. This study represents the suitability assessment based on the available genomic and expression data only; however, other factors such as cost, feasibility, and individual project goals should be carefully considered in selecting appropriate animal model for each research project.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org, Table S1, title: Distribution analysis of all CDS comparison between human and other species; Table S2, title: The predicted blast hits for human vs. rhesus macaque, marmoset, pig, mouse, and rat and the conserved 10,316 CDS were provided in each sheet; Table S3, title: Distribution analysis of 10,316 commons CDS comparison between human and other species; Table S4, title: Human CDS identified across the rhesus macaque, marmoset, pig, mouse, and rat chromosomes were listed; Table S5, title: Identified 10316 conserved human CDS mapped across the rhesus macaque, marmoset, pig, mouse, and rat chromosomes were listed; Table S6, title: List of predicted SNPs with Refseq (RS) ID in 10,316 CDS in human vs. rhesus macaque, marmoset, pig, mouse, and rat using Ensembl Variant Effect Predictor; Table S7, title: Identified diseases from human vs. rhesus macaque, marmoset, pig, mouse, and rat using Ensembl Post GWAS and SNPnexus; Table S8, title: Human-associated specific diseases in rhesus macaque, marmoset, pig, mouse, and rat with their corresponding genes were listed; Figure S1, title: The number of disease-associated SNPs were plotted in a circus plot. The coloured circle differentiated the six different organisms (outer – inner: human, rhesus macaque, marmoset, pig, mouse, and rat), and the red line inside each circle represents the disease-associated SNPs; Figure S2, title: The number of identified diseases (orange) with their corresponding RefSeq (RS) ID (green), 1255 Variant−Disease Network, was provided for human vs. rhesus macaque; Figure S3, title: The number of identified diseases (orange) with their corresponding RefSeq (RS) ID (green), 1039 Variant−Disease Network, was provided for human vs. marmoset; Figure S4, title: The number of identified diseases (orange) with their corresponding RefSeq (RS) ID (green), 1597 Variant−Disease Network, was provided for human vs. pig; Figure S5, title: The number of identified diseases (orange) with their corresponding RefSeq (RS) ID (green), 1798 Variant−Disease Network, was provided for human vs. mouse; Figure S6, title: The number of identified diseases (orange) with their corresponding RefSeq (RS) ID (green), 1787 Variant−Disease Network, was provided for human vs. rat; Figure S7, title: Heatmap represents the number of expressed genes in spleen tissues across human, mouse, pig and rat; Figure S8, title: Heatmap represents the number of expressed genes in skeletal muscle tissues across human, mouse, pig and rat; Figure S9, title: Heatmap represents the number of expressed genes in lung tissues across human, mouse, pig and rat; Figure S10, title: Heatmap represents the number of expressed genes in colon tissues across human, mouse, pig and rat; Figure S11, title: Heatmap represents the number of expressed genes in heart tissues across human, mouse, pig and rat; Figure S12, title: Heatmap represents the number of expressed genes in kidney tissues across human, mouse, pig and rat.
Author Contributions
Conceptualization, C.G. and M.A.C.; methodology, S.J. and P.M.; software, S.J.; validation, P.M.; formal analysis, X.X.; investigation, C.G. and M.A.C.; resources, B.G.; data curation, S.J.; writing—original draft preparation, S.J. and P.M.; writing—review and editing, C.G and M.A.C.; visualization, S.J.; supervision, C.G. and M.A.C.; project administration, C.G.; funding acquisition, C.G. All authors have read and agreed to the published version of the manuscript.
Funding
This project was partly supported by NIH awards, 5R01CA222907 and 5R01AG062198 to MAC, and 5P20GM103427, 5P30CA036727, 2U54GM115458 to CG; and the Nebraska Research Initiative (NRI) to CG.
Acknowledgments
Authors would like to thank the Bioinformatics and Systems Biology Core (BSBC) facility at UNMC for providing the computational infrastructure and support. BSBC is partly supported by Nebraska Research Initiative (NRI) and NIH awards.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
References
- Hickman, D.L.; Johnson, J.; Vemulapalli, T.H.; Crisler, J.R.; Shepherd, R. Commonly Used Animal Models. In Principles of Animal Research for Graduate and Undergraduate Students; 2017. [Google Scholar]
- Vandamme, T. Use of Rodents as Models of Human Diseases. J Pharm Bioallied Sci 2014, 6. [Google Scholar] [CrossRef]
- Nelson, D.R.; Zeldin, D.C.; Hoffman, S.M.G.; Maltais, L.J.; Wain, H.M.; Nebert, D.W. Comparison of Cytochrome P450 (CYP) Genes from the Mouse and Human Genomes, Including Nomenclature Recommendations for Genes, Pseudogenes and Alternative-Splice Variants. Pharmacogenetics 2004, 14. [Google Scholar] [CrossRef]
- Junhee Seok; H. Shaw Warren; Alex, G.C.; Michael, N.M.; Henry, V.B.; Xu, W.; Richards, D.R.; McDonald-Smith, G.P.; Gao, H.; Hennessy, L.; et al. Genomic Responses in Mouse Models Poorly Mimic Human Inflammatory Diseases. Proc Natl Acad Sci U S A 2013, 110. [Google Scholar] [CrossRef]
- Bailey, K.L.; Cartwright, S.B.; Patel, N.S.; Remmers, N.; Lazenby, A.J.; Hollingsworth, M.A.; Carlson, M.A. Porcine Pancreatic Ductal Epithelial Cells Transformed with KRASG12D and SV40T Are Tumorigenic. Sci Rep 2021, 11. [Google Scholar] [CrossRef]
- Bailey, K.L.; Carlson, M.A. Porcine Models of Pancreatic Cancer. Front Oncol 2019, 9. [Google Scholar] [CrossRef]
- Mondal, P.; Bailey, K.L.; Cartwright, S.B.; Band, V.; Carlson, M.A. Large Animal Models of Breast Cancer. Front Oncol 2022, 12. [Google Scholar] [CrossRef]
- Mondal, P.; Patel, N.S.; Bailey, K.; Aravind, S.; Cartwright, S.B.; Hollingsworth, M.A.; Lazenby, A.J.; Carlson, M.A. Induction of Pancreatic Neoplasia in the KRAS/TP53 Oncopig. Dis Model Mech 2023, 16. [Google Scholar] [CrossRef]
- Wernersson, R.; Schierup, M.H.; Jørgensen, F.G.; Gorodkin, J.; Panitz, F.; Stærfeldt, H.H.; Christensen, O.F.; Mailund, T.; Hornshøj, H.; Klein, A.; et al. Pigs in Sequence Space: A 0.66X Coverage Pig Genome Survey Based on Shotgun Sequencing. BMC Genomics 2005, 6. [Google Scholar] [CrossRef] [PubMed]
- Groenen, M.A.M.; Archibald, A.L.; Uenishi, H.; Tuggle, C.K.; Takeuchi, Y.; Rothschild, M.F.; Rogel-Gaillard, C.; Park, C.; Milan, D.; Megens, H.J.; et al. Analyses of Pig Genomes Provide Insight into Porcine Demography and Evolution. Nature 2012, 491. [Google Scholar] [CrossRef] [PubMed]
- Schook, L.B.; Collares, T. V.; Darfour-Oduro, K.A.; De, A.K.; Rund, L.A.; Schachtschneider, K.M.; Seixas, F.K. Unraveling the Swine Genome: Implications for Human Health. Annu Rev Anim Biosci 2015, 3. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, T.; Fujiwara, K.; Saitou, M.; Tsukiyama, T. Non-Human Primates as a Model for Human Development. Stem Cell Reports 2021, 16. [Google Scholar] [CrossRef]
- Yan, G.; Zhang, G.; Fang, X.; Zhang, Y.; Li, C.; Ling, F.; Cooper, D.N.; Li, Q.; Li, Y.; Van Gool, A.J.; et al. Genome Sequencing and Comparison of Two Nonhuman Primate Animal Models, the Cynomolgus and Chinese Rhesus Macaques. Nat Biotechnol 2011, 29. [Google Scholar] [CrossRef] [PubMed]
- Matsuzaki, M.; Ebina, T. Common Marmoset as a Model Primate for Study of the Motor Control System. Curr Opin Neurobiol 2020, 64. [Google Scholar] [CrossRef]
- Howe, K.L.; Achuthan, P.; Allen, J.; Allen, J.; Alvarez-Jarreta, J.; Ridwan Amode, M.; Armean, I.M.; Azov, A.G.; Bennett, R.; Bhai, J.; et al. Ensembl 2021. Nucleic Acids Res 2021, 49. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and Applications. BMC Bioinformatics 2009, 10. [Google Scholar] [CrossRef]
- Conway, J.R.; Lex, A.; Gehlenborg, N. UpSetR: An R Package for the Visualization of Intersecting Sets and Their Properties. Bioinformatics 2017, 33. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Ouyang, Y.; Yao, W. ShinyCircos: An R/Shiny Application for Interactive Creation of Circos Plot. Bioinformatics 2018, 34. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; Mcgettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X Version 2.0. Bioinformatics 2007, 23. [Google Scholar] [CrossRef]
- Page, A.J.; Taylor, B.; Delaney, A.J.; Soares, J.; Seemann, T.; Keane, J.A.; Harris, S.R. SNP-Sites: Rapid Efficient Extraction of SNPs from Multi-FASTA Alignments. Microb Genom 2016, 2. [Google Scholar] [CrossRef]
- McLaren, W.; Gil, L.; Hunt, S.E.; Riat, H.S.; Ritchie, G.R.S.; Thormann, A.; Flicek, P.; Cunningham, F. The Ensembl Variant Effect Predictor. Genome Biol 2016, 17. [Google Scholar] [CrossRef]
- Oscanoa, J.; Sivapalan, L.; Gadaleta, E.; Dayem Ullah, A.Z.; Lemoine, N.R.; Chelala, C. SNPnexus: A Web Server for Functional Annotation of Human Genome Sequence Variation (2020 Update). Nucleic Acids Res 2020, 48. [Google Scholar] [CrossRef] [PubMed]
- Piñero, J.; Ramírez-Anguita, J.M.; Saüch-Pitarch, J.; Ronzano, F.; Centeno, E.; Sanz, F.; Furlong, L.I. The DisGeNET Knowledge Platform for Disease Genomics: 2019 Update. Nucleic Acids Res 2020, 48. [Google Scholar] [CrossRef]
- Gel, B.; Serra, E. KaryoploteR: An R/Bioconductor Package to Plot Customizable Genomes Displaying Arbitrary Data. Bioinformatics 2017, 33. [Google Scholar] [CrossRef] [PubMed]
- Rice, P.; Longden, L.; Bleasby, A. EMBOSS: The European Molecular Biology Open Software Suite. Trends in Genetics 2000, 16. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2 - Approximately Maximum-Likelihood Trees for Large Alignments. PLoS One 2010, 5. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol Biol Evol 2021, 38. [Google Scholar] [CrossRef]
- Papatheodorou, I.; Fonseca, N.A.; Keays, M.; Tang, Y.A.; Barrera, E.; Bazant, W.; Burke, M.; Füllgrabe, A.; Fuentes, A.M.P.; George, N.; et al. Expression Atlas: Gene and Protein Expression across Multiple Studies and Organisms. Nucleic Acids Res 2018, 46. [Google Scholar] [CrossRef] [PubMed]
- Wei, T.; Simko, V. Corrplot: Visualization of a Correlation Matrix. R Package Version 0.84. Https://Github.Com/Taiyun/Corrplot. Statistician 2017, 56. [Google Scholar]
- Harding, J.D. Nonhuman Primates and Translational Research: Progress, Opportunities, and Challenges. ILAR J 2017, 58. [Google Scholar] [CrossRef]
- Feng, G.; Jensen, F.E.; Greely, H.T.; Okano, H.; Treue, S.; Roberts, A.C.; Fox, J.G.; Caddick, S.; Poo, M.M.; Newsome, W.T.; et al. Opportunities and Limitations of Genetically Modified Nonhuman Primate Models for Neuroscience Research. Proc Natl Acad Sci U S A 2020, 117. [Google Scholar] [CrossRef]
- Miller, C.T.; Freiwald, W.A.; Leopold, D.A.; Mitchell, J.F.; Silva, A.C.; Wang, X. Marmosets: A Neuroscientific Model of Human Social Behavior. Neuron 2016, 90. [Google Scholar] [CrossRef] [PubMed]
- Pomberger, T.; Risueno-Segovia, C.; Gultekin, Y.B.; Dohmen, D.; Hage, S.R. Cognitive Control of Complex Motor Behavior in Marmoset Monkeys. Nat Commun 2019, 10. [Google Scholar] [CrossRef]
- Ludlage, E.; Mansfield, K. Clinical Care and Diseases of the Common Marmoset (Callithrix Jacchus). In Proceedings of the Comparative Medicine; 2003; Vol. 53. [Google Scholar]
- David, J.M.; Dick, E.J.; Hubbard, G.B. Spontaneous Pathology of the Common Marmoset (Callithrix Jacchus) and Tamarins (Saguinus Oedipus, Saguinus Mystax). J Med Primatol 2009, 38. [Google Scholar] [CrossRef] [PubMed]
- Conley, A.J.; Moeller, B.C.; Nguyen, A.D.; Stanley, S.D.; Plant, T.M.; Abbott, D.H. Defining Adrenarche in the Rhesus Macaque (Macaca Mulatta), a Non-Human Primate Model for Adrenal Androgen Secretion. Mol Cell Endocrinol 2011, 336. [Google Scholar] [CrossRef]
- Higham, J.P.; Heistermann, M.; Maestripieri, D. The Endocrinology of Male Rhesus Macaque Social and Reproductive Status: A Test of the Challenge and Social Stress Hypotheses. Behav Ecol Sociobiol 2013, 67. [Google Scholar] [CrossRef] [PubMed]
- Litten-Brown, J.C.; Corson, A.M.; Clarke, L. Porcine Models for the Metabolic Syndrome, Digestive and Bone Disorders: A General Overview. Animal 2010, 4. [Google Scholar] [CrossRef] [PubMed]
- Koopmans, S.J.; Schuurman, T. Considerations on Pig Models for Appetite, Metabolic Syndrome and Obese Type 2 Diabetes: From Food Intake to Metabolic Disease. Eur J Pharmacol 2015, 759. [Google Scholar] [CrossRef]
- Morey-Holton, E.R.; Globus, R.K. Hindlimb Unloading Rodent Model: Technical Aspects. J Appl Physiol 2002, 92. [Google Scholar] [CrossRef]
- Witsø, E.; Hoang, L.; Løseth, K.; Bergh, K. Establishment of an in Vivo Rat Model for Chronic Musculoskeletal Implant Infection. J Orthop Surg Res 2020, 15. [Google Scholar] [CrossRef]
- Grisel, P.; Meinhardt, A.; Lehr, H.A.; Kappenberger, L.; Barrandon, Y.; Vassalli, G. The MRL Mouse Repairs Both Cryogenic and Ischemic Myocardial Infarcts with Scar. Cardiovascular Pathology 2008, 17. [Google Scholar] [CrossRef]
- Unsld, B.; Schotola, H.; Jacobshagen, C.; Seidler, T.; Sossalla, S.; Emons, J.; Klede, S.; Knll, R.; Guan, K.; El-Armouche, A.; et al. Age-Dependent Changes in Contractile Function and Passive Elastic Properties of Myocardium from Mice Lacking Muscle LIM Protein (MLP). Eur J Heart Fail 2012, 14. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, S.; Chawla-Sarkar, M.; Young, D.; Nishiyama, K.; Rayborn, M.E.; Hollyfield, J.G.; Sen, S. Myocardial Cell Death and Regeneration during Progression of Cardiac Hypertrophy to Heart Failure. Journal of Biological Chemistry 2004, 279. [Google Scholar] [CrossRef] [PubMed]
- Elliott, J.F.; Liu, J.; Yuan, Z.N.; Bautista-Lopez, N.; Wallbank, S.L.; Suzuki, K.; Rayner, D.; Nation, P.; Robertson, M.A.; Liu, G.; et al. Autoimmune Cardiomyopathy and Heart Block Develop Spontaneously in HLA-DQ8 Transgenic IAβ Knockout NOD Mice. Proc Natl Acad Sci U S A 2003, 100. [Google Scholar] [CrossRef]
- Xu, Y.; Wu, Z.; Liu, L.; Liu, J.; Wang, Y. Rat Model of Cockayne Syndrome Neurological Disease. Cell Rep 2019, 29, 800–809.e5. [Google Scholar] [CrossRef]
- Harper, A. Mouse Models of Neurological Disorders-A Comparison of Heritable and Acquired Traits. Biochim Biophys Acta Mol Basis Dis 2010, 1802. [Google Scholar] [CrossRef]
- Hutter, C.; Zenklusen, J.C. The Cancer Genome Atlas: Creating Lasting Value beyond Its Data. Cell 2018, 173. [Google Scholar] [CrossRef]
Figure 1.
Similarity of coding sequences (CDS) between human and animal models. a) Distribution of the CDS identity percentages with human in five different models. The line inside the violin plot represents median values. b) The total number of mapped CDS in five different species against human. The upset plot shows intersections across the five comparisons. Each bar represents the number of mapped CDS and the orange dot below the bar indicates their conservation status across each species. c) Distribution of percentage identities of 10,316 conserved CDs between human and five animal models. The line inside the violin plot represents median values. **** denotes p-value <0.0001, ns = statistically non-significant as determined by Kruskal-Wallis non-parametric test.
Figure 1.
Similarity of coding sequences (CDS) between human and animal models. a) Distribution of the CDS identity percentages with human in five different models. The line inside the violin plot represents median values. b) The total number of mapped CDS in five different species against human. The upset plot shows intersections across the five comparisons. Each bar represents the number of mapped CDS and the orange dot below the bar indicates their conservation status across each species. c) Distribution of percentage identities of 10,316 conserved CDs between human and five animal models. The line inside the violin plot represents median values. **** denotes p-value <0.0001, ns = statistically non-significant as determined by Kruskal-Wallis non-parametric test.
Figure 2.
Mapping of the synteny blocks of model organism chromosomes on to the human chromosomes and phylogenetic analysis. a) The Circos plot shows the position of 10,316 conserved CDS for five different animal models using a unique color for each chromosome number. The outermost circle represents the color-coded human chromosomes and each inner circle represents mapping of the synteny blocks of chromosomes from each species on to the human chromosomes showing how they are distributed across the human chromosomes. The chromosome numbers vary across each species with rhesus macaque containing Chr1-20, X, Y; marmoset with Chr1-22, X, Y; pig (Chr1-18, X, Y); mouse (Chr1-21, X, Y); and rat (Chr1-20, X, Y). b) A phylogenetic tree was constructed based on the 10,316 conserved CDS, which showed that the evolutionary distance with human was closest for rhesus macaque, followed by marmoset, pig, mouse and rat. The values above and below the line indicate the bootstrap numbers and evolutionary distance between the species.
Figure 2.
Mapping of the synteny blocks of model organism chromosomes on to the human chromosomes and phylogenetic analysis. a) The Circos plot shows the position of 10,316 conserved CDS for five different animal models using a unique color for each chromosome number. The outermost circle represents the color-coded human chromosomes and each inner circle represents mapping of the synteny blocks of chromosomes from each species on to the human chromosomes showing how they are distributed across the human chromosomes. The chromosome numbers vary across each species with rhesus macaque containing Chr1-20, X, Y; marmoset with Chr1-22, X, Y; pig (Chr1-18, X, Y); mouse (Chr1-21, X, Y); and rat (Chr1-20, X, Y). b) A phylogenetic tree was constructed based on the 10,316 conserved CDS, which showed that the evolutionary distance with human was closest for rhesus macaque, followed by marmoset, pig, mouse and rat. The values above and below the line indicate the bootstrap numbers and evolutionary distance between the species.
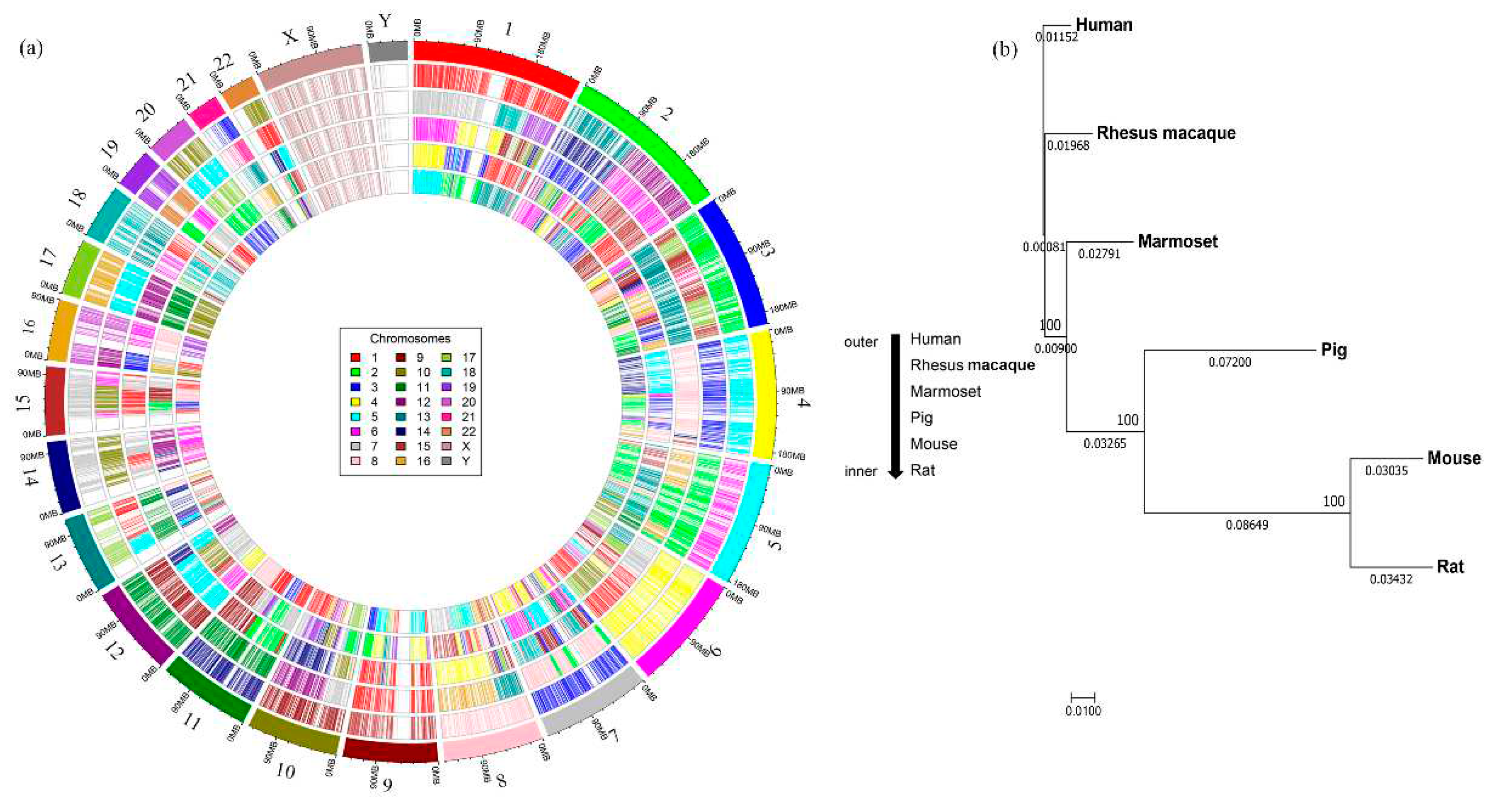
Figure 3.
Comparison of the SNPs associated human diseases across the animal models. a) An upset plot shows the intersections of SNP-associated human diseases across the five species. Each bar represents the number of identified diseases and the orange dot below the bar indicates their conservation across the comparisons. b) The diseases were classified into 25 different categories and the percentages of a specific disease class in each animal model were plotted. Higher to lower percentage numbers within species are colored from green to yellow. The highest value across species for each disease class is indicated by a box.
Figure 3.
Comparison of the SNPs associated human diseases across the animal models. a) An upset plot shows the intersections of SNP-associated human diseases across the five species. Each bar represents the number of identified diseases and the orange dot below the bar indicates their conservation across the comparisons. b) The diseases were classified into 25 different categories and the percentages of a specific disease class in each animal model were plotted. Higher to lower percentage numbers within species are colored from green to yellow. The highest value across species for each disease class is indicated by a box.
Figure 4.
Mapping of human SNP-associated genes in different animal models across the human chromosomes. The identified SNPs associated genes for species-specific diseases were represented with different colors using Karyoplot.
Figure 4.
Mapping of human SNP-associated genes in different animal models across the human chromosomes. The identified SNPs associated genes for species-specific diseases were represented with different colors using Karyoplot.
Figure 5.
Correlation of tissue specific expression across different species. Correlation plot shows the similarity in tissue specific expression between human and other species. The correlation scale ranges from -1 to +1 and all values are in the positive range as indicated by the color.
Figure 5.
Correlation of tissue specific expression across different species. Correlation plot shows the similarity in tissue specific expression between human and other species. The correlation scale ranges from -1 to +1 and all values are in the positive range as indicated by the color.
Table 1.
Comparison of sequence identity among the CDS between human and five other mammalian animal models.
Table 1.
Comparison of sequence identity among the CDS between human and five other mammalian animal models.
| Comparison |
Identified Blast hits* |
Average percentage identity |
Range
of percent
identity |
Average percentage identity for conserved CDS |
| Human vs. Rhesus macaque |
17,638 |
96.82 |
100-71.74 |
97.53 |
| Human vs. Marmoset |
17,787 |
94.65 |
100-71.63 |
95.76 |
| Human vs. Pig |
14,992 |
89.37 |
100-70.81 |
90.38 |
| Human vs. Mouse |
13,806 |
86.65 |
100-70.11 |
87.19 |
| Human vs. Rat |
13,222 |
86.53 |
100-68.93 |
87.04 |
Table 2.
Chromosome-specific mapping of conserved CDS between human and animal model genomes.
Table 2.
Chromosome-specific mapping of conserved CDS between human and animal model genomes.
| Human Chromosomes |
Total CDS |
Conserved
CDS |
Rhesus macaque* |
Marmoset* |
Pig* |
Mouse* |
Rat* |
| Chr1 |
2049 |
1088 |
1 |
7, 18, 19 |
6, 4, 9, 10, 14, 2, 7 |
4, 3, 1, 8 |
5, 2, 13,19, 14, 10, 17, 4 |
| Chr2 |
1244 |
750 |
12, 13 |
6, 14 |
15, 3 |
1, 2, 6, 17, 12, 11 |
9, 6, 3, 4, 14, 13, 20, 18 |
| Chr3 |
1075 |
645 |
2 |
15, 17 |
13 |
9, 16, 3, 6, 14 |
8, 11, 2, 4, 16, 15 |
| Chr4 |
752 |
390 |
5 |
3 |
8, 15, 14 |
5, 3, 8 |
14, 2, 16, 19, 4 |
| Chr5 |
883 |
502 |
6 |
2 |
2, 16 |
13, 18, 11, 15 |
2, 18, 10, 17, 1, 9 |
| Chr6 |
1045 |
574 |
4 |
4 |
7, 1 |
17, 10, 13, 9, 4, 1 |
20, 1, 17, 9, 8, 5 |
| Chr7 |
919 |
470 |
3 |
8,2 |
18, 9, 3 |
5, 6, 12, 11, 13 |
4, 12, 6, 14, 17 |
| Chr8 |
684 |
372 |
8 |
16, 13 |
4, 14,17, 15 |
15, 8, 14, 4, 1, 3 |
7, 5, 16, 15, 2, 11 |
| Chr9 |
779 |
402 |
15 |
1 |
1, 10, 14 3 |
4, 2, 19, 13 |
5, 3, 1, 17 |
| Chr10 |
1309 |
619 |
9 |
12, 7 |
14, 10 |
19, 14, 2, 10, 7, 18, 6, 13 |
1, 17, 20, 16, 15, 4 |
| Chr11 |
727 |
432 |
14 |
11 |
2, 9 |
7, 9, 19, 2 |
1, 8, 3 |
| Chr12 |
1033 |
582 |
11 |
9 |
5, 14 |
10, 5, 6, 15 |
7, 12, 4 |
| Chr13 |
321 |
182 |
17 |
1, 5 |
11 |
14, 8, 5, 3 |
15, 16, 12, 2, 9 |
| Chr14 |
610 |
360 |
7 |
10 |
7, 1 |
12, 14 |
6, 15 |
| Chr15 |
596 |
371 |
7 |
10, 6 |
1, 7 |
9, 2, 7 |
8, 3, 1 |
| Chr16 |
851 |
378 |
20 |
12, 20 |
6, 3 |
8, 7, 16, 17, 11 |
19, 1, 10 |
| Chr17 |
1182 |
637 |
16 |
5 |
12 |
11 |
10 |
| Chr18 |
269 |
157 |
18 |
13 |
1, 6 |
18, 17, 1 |
18, 9, 3 |
| Chr19 |
546 |
282 |
19 |
22 |
6, 2 |
7, 8, 10, 17, 9 |
1, 7, 16, 8, 19, 9, 12 |
| Chr20 |
1469 |
457 |
10 |
5 |
17 |
2 |
3 |
| Chr21 |
234 |
76 |
3 |
21 |
13 |
16, 10, 17 |
11, 20 |
| Chr22 |
444 |
202 |
10 |
1 |
5, 14 |
15, 11, 16, 5, 10 |
7, 14, 11, 12, 20 |
| ChrX |
853 |
381 |
X |
X |
X |
X |
X |
| ChrY |
46 |
7 |
Y |
Y, X |
Y, X |
Y, X |
Y, X |
Table 3.
Number of human SNPs mapped to the conserved CDS across five animal models and their associated disease information.
Table 3.
Number of human SNPs mapped to the conserved CDS across five animal models and their associated disease information.
| Organisms |
Total SNPs in 10,316 CDS with RS number |
SNPs associated with disease |
SNPs identified in genes |
No. of identified diseases |
Species- specific diseases* |
| Human vs. Rhesus macaque |
63,449 |
2198 |
1074 |
1255 |
77 (77) |
| Human vs. Marmoset |
40,181 |
1533 |
867 |
1039 |
4 (12) |
| Human vs. Pig |
145,715 |
4011 |
1428 |
1597 |
117 (96) |
| Human vs. Mouse |
221,383 |
5824 |
1630 |
1798 |
58 (44) |
| Human vs. Rat |
220,631 |
5739 |
1646 |
1787 |
81 (54) |
Table 4.
Genes in each species that are identified with the highest number of human diseases in each human chromosome.
Table 4.
Genes in each species that are identified with the highest number of human diseases in each human chromosome.
| Chromosome |
Rat* |
Diseases |
Mouse* |
Diseases |
Pig* |
Diseases |
Marmoset* |
Diseases |
Rhesus macaque* |
Diseases |
| 1 |
ABCA4 |
41 |
MUTYH |
44 |
SPTA1 |
34 |
NLRP3 |
15 |
SPTA1 |
18 |
| 2 |
MSH6 |
115 |
MSH6 |
97 |
MSH6 |
78 |
APOB |
22 |
APOB |
31 |
| 3 |
MLH1 |
36 |
BAP1 |
34 |
MLH1 |
34 |
ITIH3 |
21 |
ITIH3 |
21 |
| 4 |
WFS1 |
23 |
PDGFRA |
27 |
KIT |
29 |
KIT |
13 |
PDGFRA |
18 |
| 5 |
SDHA |
55 |
SDHA |
75 |
SDHA |
35 |
VCAN |
21 |
SDHA |
14 |
| 6 |
DSP |
40 |
DSP |
52 |
SLC22A7 |
12 |
CFB |
6 |
DSP |
28 |
| 7 |
CFTR |
39 |
CFTR |
38 |
GARS1 |
19 |
GARS1 |
13 |
RELN |
14 |
| 8 |
NBN |
17 |
NBN |
17 |
NBN |
23 |
KCNQ3 |
7 |
FGFR1 |
7 |
| 9 |
NOTCH1 |
95 |
NOTCH1 |
102 |
PTCH1 |
55 |
COL5A1 |
13 |
NOTCH1 |
25 |
| 10 |
RET |
57 |
RET |
55 |
RET |
65 |
RET |
25 |
CUBN |
14 |
| 11 |
ATM |
125 |
ATM |
117 |
ATM |
116 |
ATM |
43 |
ATM |
37 |
| 12 |
POLE |
107 |
POLE |
118 |
POLE |
78 |
POLE |
24 |
POLE |
24 |
| 13 |
RB1 |
15 |
RB1 |
14 |
RB1 |
8 |
RB1 |
5 |
RB1 |
8 |
| 14 |
DYNC1H1 |
43 |
DYNC1H1 |
44 |
DICER1 |
24 |
C14orf39 |
8 |
DICER1 |
10 |
| 15 |
FBN1 |
140 |
FBN1 |
152 |
FBN1 |
120 |
FBN1 |
41 |
FBN1 |
36 |
| 16 |
TSC2 |
209 |
TSC2 |
215 |
TSC2 |
112 |
TSC2 |
38 |
TSC2 |
46 |
| 17 |
SCN4A |
53 |
SCN4A |
58 |
NF1 |
33 |
AC004223.3 |
17 |
AC004223.3 |
21 |
| 18 |
LAMA3 |
13 |
LOXHD1 |
15 |
LAMA3 |
14 |
LAMA3 |
4 |
LOXHD1 |
6 |
| 19 |
LDLR |
72 |
STK11 |
59 |
LDLR |
56 |
LDLR |
22 |
LDLR |
25 |
| 20 |
COL9A3 |
15 |
SLC2A10 |
18 |
JAG1 |
11 |
MYH7B |
9 |
MYH7B |
9 |
| 21 |
COL6A1 |
19 |
COL6A1 |
19 |
CBS |
13 |
CBS |
4 |
CBS |
13 |
| 22 |
NF2 |
18 |
DEPDC5 |
17 |
TMPRSS6 |
29 |
TMPRSS6 |
26 |
TMPRSS6 |
28 |
| X |
FLNA |
110 |
FLNA |
118 |
FLNA |
30 |
FLNA |
14 |
FLNA |
26 |
Table 5.
List of expressed genes in various tissues and their conservation in human, mouse, rat and pig.
Table 5.
List of expressed genes in various tissues and their conservation in human, mouse, rat and pig.
| Tissues |
Expressed genes |
Conserved genes in four organisms based on 10316 CDS |
| Human |
Mouse |
Rat |
Pig |
| Spleen |
10,873 |
9,984 |
16,697 |
17,647 |
4,121 |
| Skeletal muscle |
10,590 |
10,650 |
12,770 |
15,152 |
3,963 |
| Lung |
11,285 |
11,213 |
18,171 |
12,943 |
4,034 |
| Colon |
11,125 |
10,755 |
16,949 |
15,603 |
4,128 |
| Heart |
10,967 |
10,369 |
14,821 |
13,995 |
4,116 |
| Kidney |
11,325 |
10,706 |
16,400 |
16,410 |
4,401 |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).