Submitted:
25 May 2023
Posted:
26 May 2023
You are already at the latest version
Abstract
Keywords:
Introduction
A Brief History of Genome Editing Systems and their Mechanisms
Key Aspects of Fruit Plant Modification through Genome Editing Systems
Delivery of CRISPR/Cas constructs in fruit plants through direct and indirect transformation methods
- Cargo
- Delivery Vehicle
- I.
- DNA plasmid encoding a sgRNA and Cas protein
- II.
- Cas translocation-mediating mRNA and a separate sgRNA
- III.
- Ribonucleoprotein (RNP) complex (sgRNA + Cas protein)
Exploiting the CRISPR/Cas system in fruit plants for biotic and abiotic stress management
Limitations and their possible solutions in engineering climate-smart fruit plants
Conclusions and Future Perspectives
Supplementary Materials
Authors’ Contribution
Data Availability
Acknowledgments
Conflict of interests
References
- Al-Mssallem, Ibrahim S., Songnian Hu, Xiaowei Zhang, Qiang Lin, Wanfei Liu, Jun Tan, Xiaoguang Yu, et al. 2013. “Genome Sequence of the Date Palm Phoenix Dactylifera L.” Nature Communications 2013 4:1 4 (1): 1–9. https://doi.org/10.1038/ncomms3274. [CrossRef]
- Alioto, Tyler, Konstantinos G. Alexiou, Amélie Bardil, Fabio Barteri, Raúl Castanera, Fernando Cruz, Amit Dhingra, et al. 2020. “Transposons Played a Major Role in the Diversification between the Closely Related Almond and Peach Genomes: Results from the Almond Genome Sequence.” The Plant Journal : For Cell and Molecular Biology 101 (2): 455–72. https://doi.org/10.1111/TPJ.14538. [CrossRef]
- Anzalone, Andrew V, Peyton B Randolph, Jessie R Davis, Alexander A Sousa, Luke W Koblan, Jonathan M Levy, Peter J Chen, et al. 2019. “Search-and-Replace Genome Editing without Double-Strand Breaks or Donor DNA.” Nature 576 (7785): 149–57. https://doi.org/10.1038/s41586-019-1711-4. [CrossRef]
- Argout, Xavier, Jerome Salse, Jean Marc Aury, Mark J. Guiltinan, Gaetan Droc, Jerome Gouzy, Mathilde Allegre, et al. 2010. “The Genome of Theobroma Cacao.” Nature Genetics 2010 43:2 43 (2): 101–8. https://doi.org/10.1038/ng.736. [CrossRef]
- Arumuganathan, K., and E. D. Earle. 1991. “Nuclear DNA Content of Some Important Plant Species.” Plant Molecular Biology Reporter 1991 9:3 9 (3): 208–18. https://doi.org/10.1007/BF02672069. [CrossRef]
- Axford, David S., Daniel P. Morris, and Jonathan L McMurry. n.d. “Cell Penetrating Peptide-Mediated Nuclear Delivery of Cas9 to Enhance the Utility of CRISPR/Cas Genome Editing.” The FASEB Journal 31: 909.4-909.4. https://doi.org/10.1096/FASEBJ.31.1_SUPPLEMENT.909.4. [CrossRef]
- Bae, Sangsu, Jeongbin Park, and Jin Soo Kim. 2014. “Cas-OFFinder: A Fast and Versatile Algorithm That Searches for Potential off-Target Sites of Cas9 RNA-Guided Endonucleases.” Bioinformatics (Oxford, England) 30 (10): 1473–75. https://doi.org/10.1093/BIOINFORMATICS/BTU048. [CrossRef]
- Basu, Supratim. 2020. “Toward Development of Climate-Resilient Citrus.” Genomic Designing of Climate-Smart Fruit Crops, 117–34. https://doi.org/10.1007/978-3-319-97946-5_5. [CrossRef]
- Bates, Katie, and Kostas Kostarelos. 2013. “Carbon Nanotubes as Vectors for Gene Therapy: Past Achievements, Present Challenges and Future Goals.” Advanced Drug Delivery Reviews 65 (15): 2023–33. https://doi.org/10.1016/J.ADDR.2013.10.003. [CrossRef]
- Boch, Jens, Heidi Scholze, Sebastian Schornack, Angelika Landgraf, Simone Hahn, Sabine Kay, Thomas Lahaye, Anja Nickstadt, and Ulla Bonas. 2009. “Breaking the Code of DNA Binding Specificity of TAL-Type III Effectors.” Science (New York, N.Y.) 326 (5959): 1509–12. https://doi.org/10.1126/SCIENCE.1178811. [CrossRef]
- Boudichevskaia, Anastassia, Gulshan Kumar, Yogesh Sharma, Ritu Kapoor, and Anil Kumar Singh. 2020. “Challenges and Strategies for Developing Climate-Smart Apple Varieties Through Genomic Approaches.” Genomic Designing of Climate-Smart Fruit Crops, 23–71. https://doi.org/10.1007/978-3-319-97946-5_2. [CrossRef]
- Breitler, Jean Christophe, Eveline Dechamp, Claudine Campa, Leonardo Augusto Zebral Rodrigues, Romain Guyot, Pierre Marraccini, and Hervé Etienne. 2018. “CRISPR/Cas9-Mediated Efficient Targeted Mutagenesis Has the Potential to Accelerate the Domestication of Coffea Canephora.” Plant Cell, Tissue and Organ Culture 134 (3): 383–94. https://doi.org/10.1007/S11240-018-1429-2/FIGURES/3. [CrossRef]
- Bressan, Ray, Hans Bohnert, and Jian Kang Zhu. 2009. “Abiotic Stress Tolerance: From Gene Discovery in Model Organisms to Crop Improvement.” Molecular Plant 2 (1): 1–2. https://doi.org/10.1093/MP/SSN097. [CrossRef]
- Brito, Jose L.R., Faith E. Davies, David Gonzalez, and Gareth J. Morgan. 2008. “Streptolysin-O Reversible Permeabilisation Is an Effective Method to Transfect SiRNAs into Myeloma Cells.” Journal of Immunological Methods 333 (1–2): 147–55. https://doi.org/10.1016/J.JIM.2008.01.009. [CrossRef]
- Brown, Allan, Sebastien C. Carpentier, and Rony Swennen. 2020. “Breeding Climate-Resilient Bananas.” Genomic Designing of Climate-Smart Fruit Crops, 91–115. https://doi.org/10.1007/978-3-319-97946-5_4. [CrossRef]
- Brown, Naomi J, Christine A Newell, Susan Stanley, Jit E Chen, Abigail J Perrin, Kaisa Kajala, and Julian M Hibberd. 2011. “Independent and Parallel Recruitment of Preexisting Mechanisms Underlying C₄ Photosynthesis.” Science (New York, N.Y.) 331 (6023): 1436–39. https://doi.org/10.1126/science.1201248. [CrossRef]
- Brutnell, Thomas P, Lin Wang, Kerry Swartwood, Alexander Goldschmidt, David Jackson, Xin-Guang Zhu, Elizabeth Kellogg, and Joyce Van Eck. 2010. “Setaria Viridis: A Model for C4 Photosynthesis.” The Plant Cell 22 (8): 2537–44. https://doi.org/10.1105/tpc.110.075309. [CrossRef]
- Butiuc-Keul, Anca, Anca Farkas, Rahela Carpa, Cristina Dobrota, and Dumitrana Iordache. 2022. “Development of Smart Fruit Crops by Genome Editing.” Turkish Journal of Agriculture and Forestry 46 (2): 129–40. https://doi.org/10.3906/tar-2012-104. [CrossRef]
- Campoy, Jose A., Jean M. Audergon, D. Ruiz, and Pedro Martínez-Gómez. 2020. “Genomic Designing for New Climate-Resilient Apricot Varieties in a Warming Context.” Genomic Designing of Climate-Smart Fruit Crops, 73–89. https://doi.org/10.1007/978-3-319-97946-5_3. [CrossRef]
- Chagné, David, Ross N. Crowhurst, Massimo Pindo, Amali Thrimawithana, Cecilia Deng, Hilary Ireland, Mark Fiers, et al. 2014. “The Draft Genome Sequence of European Pear (Pyrus Communis L. ’Bartlett’).” PloS One 9 (4). https://doi.org/10.1371/JOURNAL.PONE.0092644. [CrossRef]
- Chakrabarti, Anob M, Tristan Henser-Brownhill, Josep Monserrat, Anna R Poetsch, Nicholas M Luscombe, and Paola Scaffidi. 2019. “Target-Specific Precision of CRISPR-Mediated Genome Editing.” Molecular Cell 73 (4): 699-713.e6. https://doi.org/10.1016/j.molcel.2018.11.031. [CrossRef]
- Charrier, Aurélie, Emilie Vergne, Nicolas Dousset, Andréa Richer, Aurélien Petiteau, and Elisabeth Chevreau. 2019. “Efficient Targeted Mutagenesis in Apple and First Time Edition of Pear Using the CRISPR-Cas9 System.” Frontiers in Plant Science 10 (February): 40. https://doi.org/10.3389/FPLS.2019.00040/BIBTEX. [CrossRef]
- Chen, Fuqiang, Xiao Ding, Yongmei Feng, Timothy Seebeck, Yanfang Jiang, and Gregory D Davis. 2017. “Targeted Activation of Diverse CRISPR-Cas Systems for Mammalian Genome Editing via Proximal CRISPR Targeting.” Nature Communications 8 (1): 14958. https://doi.org/10.1038/ncomms14958. [CrossRef]
- Chen, Janice S., Yavuz S. Dagdas, Benjamin P. Kleinstiver, Moira M. Welch, Alexander A. Sousa, Lucas B. Harrington, Samuel H. Sternberg, J. Keith Joung, Ahmet Yildiz, and Jennifer A. Doudna. 2017. “Enhanced Proofreading Governs CRISPR–Cas9 Targeting Accuracy.” Nature 2017 550:7676 550 (7676): 407–10. https://doi.org/10.1038/nature24268. [CrossRef]
- Cho, Seung Woo, Sojung Kim, Jong Min Kim, and Jin Soo Kim. 2013. “Targeted Genome Engineering in Human Cells with the Cas9 RNA-Guided Endonuclease.” Nature Biotechnology 2013 31:3 31 (3): 230–32. https://doi.org/10.1038/nbt.2507. [CrossRef]
- Conover, Robert A. 1964. “Distortion Ringspot, a Severe Virus Disease of Papaya in Florida.” Proceedings of Florida State Horticultural Society 77: 440–44.
- Crispo, M., A. P. Mulet, L. Tesson, N. Barrera, F. Cuadro, P. C. Dos Santos-Neto, T. H. Nguyen, et al. 2015. “Efficient Generation of Myostatin Knock-Out Sheep Using CRISPR/Cas9 Technology and Microinjection into Zygotes.” PloS One 10 (8). https://doi.org/10.1371/JOURNAL.PONE.0136690. [CrossRef]
- Cui, Hongchang. 2021. “Challenges and Approaches to Crop Improvement Through C3-to-C4 Engineering .” Frontiers in Plant Science . https://www.frontiersin.org/articles/10.3389/fpls.2021.715391. https://doi.org/10.3389/fpls.2021.715391. [CrossRef]
- D’Astolfo, Diego S., Romina J. Pagliero, Anita Pras, Wouter R. Karthaus, Hans Clevers, Vikram Prasad, Robert Jan Lebbink, Holger Rehmann, and Niels Geijsen. 2015. “Efficient Intracellular Delivery of Native Proteins.” Cell 161 (3): 674–90. https://doi.org/10.1016/J.CELL.2015.03.028. [CrossRef]
- Dash, Prasanta K., and Rhitu Rai. 2016. “Translating the ‘Banana Genome’ to Delineate Stress Resistance, Dwarfing, Parthenocarpy and Mechanisms of Fruit Ripening.” Frontiers in Plant Science 7 (OCTOBER2016): 1543. https://doi.org/10.3389/FPLS.2016.01543/BIBTEX. [CrossRef]
- Davis, Kevin M., Vikram Pattanayak, David B. Thompson, John A. Zuris, and David R. Liu. 2015. “Small Molecule–Triggered Cas9 Protein with Improved Genome-Editing Specificity.” Nature Chemical Biology 2015 11:5 11 (5): 316–18. https://doi.org/10.1038/nchembio.1793. [CrossRef]
- Delrot, Serge, Jérôme Grimplet, Pablo Carbonell-Bejerano, Anna Schwandner, Pierre-François Bert, Luigi Bavaresco, Lorenza Dalla Costa, et al. 2020a. “Genetic and Genomic Approaches for Adaptation of Grapevine to Climate Change.” Genomic Designing of Climate-Smart Fruit Crops, 157–270. https://doi.org/10.1007/978-3-319-97946-5_7. [CrossRef]
- ———. 2020b. “Genetic and Genomic Approaches for Adaptation of Grapevine to Climate Change BT - Genomic Designing of Climate-Smart Fruit Crops.” Edited by Chittaranjan Kole, 157–270. https://doi.org/10.1007/978-3-319-97946-5_7. [CrossRef]
- Dong, Chunsheng, Liang Qu, Haoyi Wang, Lin Wei, Yuansu Dong, and Sidong Xiong. 2015. “Targeting Hepatitis B Virus CccDNA by CRISPR/Cas9 Nuclease Efficiently Inhibits Viral Replication.” Antiviral Research 118 (June): 110–17. https://doi.org/10.1016/J.ANTIVIRAL.2015.03.015. [CrossRef]
- Ebina, Hirotaka, Naoko Misawa, Yuka Kanemura, and Yoshio Koyanagi. 2013. “Harnessing the CRISPR/Cas9 System to Disrupt Latent HIV-1 Provirus.” Scientific Reports 3. https://doi.org/10.1038/SREP02510. [CrossRef]
- Fang, Jingping, Jingping Fang, Jingping Fang, Jingping Fang, Andrew Michael Wood, Youqiang Chen, Youqiang Chen, Jingjing Yue, Ray Ming, and Ray Ming. 2020. “Genomic Variation between PRSV Resistant Transgenic SunUp and Its Progenitor Cultivar Sunset.” BMC Genomics 21 (1): 1–21. https://doi.org/10.1186/S12864-020-06804-7/TABLES/8. [CrossRef]
- Feng, Zhengyan, Botao Zhang, Wona Ding, Xiaodong Liu, Dong Lei Yang, Pengliang Wei, Fengqiu Cao, et al. 2013. “Efficient Genome Editing in Plants Using a CRISPR/Cas System.” Cell Research 23 (10): 1229–32. https://doi.org/10.1038/CR.2013.114. [CrossRef]
- Ferdosi, Shayesteh R, Radwa Ewaisha, Farzaneh Moghadam, Sri Krishna, Jin G Park, Mo R Ebrahimkhani, Samira Kiani, and Karen S Anderson. 2019. “Multifunctional CRISPR-Cas9 with Engineered Immunosilenced Human T Cell Epitopes.” Nature Communications 10 (1): 1842. https://doi.org/10.1038/s41467-019-09693-x. [CrossRef]
- Fister, Andrew S., Lena Landherr, Siela N. Maximova, and Mark J. Guiltinan. 2018a. “Transient Expression of CRISPR/Cas9 Machinery Targeting TcNPR3 Enhances Defense Response in Theobroma Cacao.” Frontiers in Plant Science 9 (March): 268. https://doi.org/10.3389/FPLS.2018.00268/BIBTEX. [CrossRef]
- ———. 2018b. “Transient Expression of CRISPR/Cas9 Machinery Targeting TcNPR3 Enhances Defense Response in Theobroma Cacao.” Frontiers in Plant Science 9 (March). https://doi.org/10.3389/FPLS.2018.00268. [CrossRef]
- Fitch, Maureen M.M., Richard M. Manshardt, Dennis Gonsalves, Jerry L. Slightom, and John C. Sanford. 1992. “Virus Resistant Papaya Plants Derived from Tissues Bombarded with the Coat Protein Gene of Papaya Ringspot Virus.” Bio/Technology 1992 10:11 10 (11): 1466–72. https://doi.org/10.1038/nbt1192-1466. [CrossRef]
- Furbank, Robert T. 2016. “Walking the C4 Pathway: Past, Present, and Future.” Journal of Experimental Botany 67 (14): 4057–66.
- Gaj, Thomas, Charles A. Gersbach, and Carlos F. Barbas. 2013. “ZFN, TALEN, and CRISPR/Cas-Based Methods for Genome Engineering.” Trends in Biotechnology 31 (7): 397–405. https://doi.org/10.1016/J.TIBTECH.2013.04.004. [CrossRef]
- Gao, Linyi, David B.T. Cox, Winston X. Yan, John C. Manteiga, Martin W. Schneider, Takashi Yamano, Hiroshi Nishimasu, Osamu Nureki, Nicola Crosetto, and Feng Zhang. 2017. “Engineered Cpf1 Variants with Altered PAM Specificities.” Nature Biotechnology 2017 35:8 35 (8): 789–92. https://doi.org/10.1038/nbt.3900. [CrossRef]
- Gaudelli, Nicole M., Alexis C. Komor, Holly A. Rees, Michael S. Packer, Ahmed H. Badran, David I. Bryson, and David R. Liu. 2017. “Programmable Base Editing of A•T to G•C in Genomic DNA without DNA Cleavage.” Nature 2017 551:7681 551 (7681): 464–71. https://doi.org/10.1038/nature24644. [CrossRef]
- Gogorcena, Yolanda, Gerardo Sánchez, Santiago Moreno-Vázquez, Salvador Pérez, and Najla Ksouri. 2020. “Genomic-Based Breeding for Climate-Smart Peach Varieties.” Genomic Designing of Climate-Smart Fruit Crops, 271–331. https://doi.org/10.1007/978-3-319-97946-5_8. [CrossRef]
- Gonzalez Porras, Maria A., Paul N. Durfee, Ashley M. Gregory, Gary C. Sieck, C. Jeffrey Brinker, and Carlos B. Mantilla. 2016. “A Novel Approach for Targeted Delivery to Motoneurons Using Cholera Toxin-B Modified Protocells.” Journal of Neuroscience Methods 273 (November): 160–74. https://doi.org/10.1016/J.JNEUMETH.2016.09.003. [CrossRef]
- Grünewald, Julian, Ronghao Zhou, Caleb A Lareau, Sara P Garcia, Sowmya Iyer, Bret R Miller, Lukas M Langner, Jonathan Y Hsu, Martin J Aryee, and J Keith Joung. 2020. “A Dual-Deaminase CRISPR Base Editor Enables Concurrent Adenine and Cytosine Editing.” Nature Biotechnology 38 (7): 861–64. https://doi.org/10.1038/s41587-020-0535-y. [CrossRef]
- Guan, Yuting, Yanlin Ma, Qi Li, Zhenliang Sun, Lie Ma, Lijuan Wu, Liren Wang, et al. 2016. “CRISPR/Cas9-Mediated Somatic Correction of a Novel Coagulator Factor IX Gene Mutation Ameliorates Hemophilia in Mouse.” EMBO Molecular Medicine 8 (5): 477–88. https://doi.org/10.15252/EMMM.201506039. [CrossRef]
- He, Ningjia, Chi Zhang, Xiwu Qi, Shancen Zhao, Yong Tao, Guojun Yang, Tae Ho Lee, et al. 2013. “Draft Genome Sequence of the Mulberry Tree Morus Notabilis.” Nature Communications 2013 4:1 4 (1): 1–9. https://doi.org/10.1038/ncomms3445. [CrossRef]
- Heigwer, Florian, Grainne Kerr, and Michael Boutros. 2014. “E-CRISP: Fast CRISPR Target Site Identification.” Nature Methods 2014 11:2 11 (2): 122–23. https://doi.org/10.1038/nmeth.2812. [CrossRef]
- Horii, Takuro, Yuji Arai, Miho Yamazaki, Sumiyo Morita, Mika Kimura, Masahiro Itoh, Yumiko Abe, and Izuho Hatada. 2014. “Validation of Microinjection Methods for Generating Knockout Mice by CRISPR/Cas-Mediated Genome Engineering.” Scientific Reports 4 (March). https://doi.org/10.1038/SREP04513. [CrossRef]
- Hsu, Patrick D., David A. Scott, Joshua A. Weinstein, F. Ann Ran, Silvana Konermann, Vineeta Agarwala, Yinqing Li, et al. 2013. “DNA Targeting Specificity of RNA-Guided Cas9 Nucleases.” Nature Biotechnology 2013 31:9 31 (9): 827–32. https://doi.org/10.1038/nbt.2647. [CrossRef]
- Hu, Johnny H., Shannon M. Miller, Maarten H. Geurts, Weixin Tang, Liwei Chen, Ning Sun, Christina M. Zeina, et al. 2018. “Evolved Cas9 Variants with Broad PAM Compatibility and High DNA Specificity.” Nature 2018 556:7699 556 (7699): 57–63. https://doi.org/10.1038/nature26155. [CrossRef]
- Huang, Shengxiong, Jian Ding, Dejing Deng, Wei Tang, Honghe Sun, Dongyuan Liu, Lei Zhang, et al. 2013. “Draft Genome of the Kiwifruit Actinidia Chinensis.” Nature Communications 4. https://doi.org/10.1038/NCOMMS3640. [CrossRef]
- Ioannidi, Eleonora I., Matthew T. N. Yarnall, Cian Schmitt-Ulms, Rohan N. Krajeski, Justin Lim, Lukas Villiger, Wenyuan Zhou, et al. 2021. “Drag-and-Drop Genome Insertion without DNA Cleavage with CRISPR-Directed Integrases.” BioRxiv, November, 2021.11.01.466786. https://doi.org/10.1101/2021.11.01.466786. [CrossRef]
- J, Ross. 1995. “MRNA Stability in Mammalian Cells.” Microbiological Reviews 59 (3): 423–50. https://doi.org/10.1128/MR.59.3.423-450.1995. [CrossRef]
- Jaganathan, Deepa, Karthikeyan Ramasamy, Gothandapani Sellamuthu, Shilpha Jayabalan, and Gayatri Venkataraman. 2018. “CRISPR for Crop Improvement: An Update Review.” Frontiers in Plant Science 9 (July): 985. https://doi.org/10.3389/FPLS.2018.00985/BIBTEX. [CrossRef]
- Jaillon, Olivier, Jean Marc Aury, Benjamin Noel, Alberto Policriti, Christian Clepet, Alberto Casagrande, Nathalie Choisne, et al. 2007. “The Grapevine Genome Sequence Suggests Ancestral Hexaploidization in Major Angiosperm Phyla.” Nature 2007 449:7161 449 (7161): 463–67. https://doi.org/10.1038/nature06148. [CrossRef]
- Jia, Hongge, and Wang Nian. 2014. “Targeted Genome Editing of Sweet Orange Using Cas9/SgRNA.” PloS One 9 (4). https://doi.org/10.1371/JOURNAL.PONE.0093806. [CrossRef]
- Jia, Hongge, Vladimir Orbovic, Jeffrey B. Jones, and Nian Wang. 2016. “Modification of the PthA4 Effector Binding Elements in Type I CsLOB1 Promoter Using Cas9/SgRNA to Produce Transgenic Duncan Grapefruit Alleviating XccΔpthA4:DCsLOB1.3 Infection.” Plant Biotechnology Journal 14 (5): 1291–1301. https://doi.org/10.1111/PBI.12495. [CrossRef]
- Jia, Hongge, Yunzeng Zhang, Vladimir Orbović, Jin Xu, Frank F. White, Jeffrey B. Jones, and Nian Wang. 2017a. “Genome Editing of the Disease Susceptibility Gene CsLOB1 in Citrus Confers Resistance to Citrus Canker.” Plant Biotechnology Journal 15 (7): 817–23. https://doi.org/10.1111/PBI.12677. [CrossRef]
- ———. 2017b. “Genome Editing of the Disease Susceptibility Gene CsLOB1 in Citrus Confers Resistance to Citrus Canker.” Plant Biotechnology Journal 15 (7): 817–23. https://doi.org/10.1111/PBI.12677. [CrossRef]
- Jiang, Fengchao, Junhuan Zhang, Sen Wang, Li Yang, Yingfeng Luo, Shenghan Gao, Meiling Zhang, et al. 2019. “The Apricot (Prunus Armeniaca L.) Genome Elucidates Rosaceae Evolution and Beta-Carotenoid Synthesis.” Horticulture Research 2019 6:1 6 (1): 1–12. https://doi.org/10.1038/s41438-019-0215-6. [CrossRef]
- Jiang, Wenzhi, Huanbin Zhou, Honghao Bi, Michael Fromm, Bing Yang, and Donald P. Weeks. 2013. “Demonstration of CRISPR/Cas9/SgRNA-Mediated Targeted Gene Modification in Arabidopsis, Tobacco, Sorghum and Rice.” Nucleic Acids Research 41 (20). https://doi.org/10.1093/NAR/GKT780. [CrossRef]
- Kaur, Navneet, Anshu Alok, Shivani, Navjot Kaur, Pankaj Pandey, Praveen Awasthi, and Siddharth Tiwari. 2018. “CRISPR/Cas9-Mediated Efficient Editing in Phytoene Desaturase (PDS) Demonstrates Precise Manipulation in Banana Cv. Rasthali Genome.” Functional & Integrative Genomics 18 (1): 89–99. https://doi.org/10.1007/S10142-017-0577-5. [CrossRef]
- Kaur, Navneet, Praveen Awasthi, and Siddharth Tiwari. 2020. Fruit Crops Improvement Using CRISPR/Cas9 System. Genome Engineering via CRISPR-Cas9 System. Elsevier Inc. https://doi.org/10.1016/b978-0-12-818140-9.00012-x. [CrossRef]
- Kennedy, Edward M., Anand V. R. Kornepati, Michael Goldstein, Hal P. Bogerd, Brigid C. Poling, Adam W. Whisnant, Michael B. Kastan, and Bryan R. Cullen. 2014. “Inactivation of the Human Papillomavirus E6 or E7 Gene in Cervical Carcinoma Cells by Using a Bacterial CRISPR/Cas RNA-Guided Endonuclease.” Journal of Virology 88 (20): 11965–72. https://doi.org/10.1128/JVI.01879-14. [CrossRef]
- Khalil, Ahmad M. 2020. “The Genome Editing Revolution: Review.” Journal, Genetic Engineering & Biotechnology 18 (1): 68. https://doi.org/10.1186/s43141-020-00078-y. [CrossRef]
- Kim, Hui Kwon, Seonwoo Min, Myungjae Song, Soobin Jung, Jae Woo Choi, Younggwang Kim, Sangeun Lee, Sungroh Yoon, and Hyongbum Kim. 2018. “Deep Learning Improves Prediction of CRISPR–Cpf1 Guide RNA Activity.” Nature Biotechnology 2018 36:3 36 (3): 239–41. https://doi.org/10.1038/nbt.4061. [CrossRef]
- Kim, Yang Gyun, Jooyeun Cha, and Srinivasan Chandrasegaran. 1996. “Hybrid Restriction Enzymes: Zinc Finger Fusions to Fok I Cleavage Domain.” Proceedings of the National Academy of Sciences of the United States of America 93 (3): 1156. https://doi.org/10.1073/PNAS.93.3.1156. [CrossRef]
- Kleinstiver, Benjamin P., Michelle S. Prew, Shengdar Q. Tsai, Ved V. Topkar, Nhu T. Nguyen, Zongli Zheng, Andrew P.W. Gonzales, et al. 2015. “Engineered CRISPR-Cas9 Nucleases with Altered PAM Specificities.” Nature 2015 523:7561 523 (7561): 481–85. https://doi.org/10.1038/nature14592. [CrossRef]
- Koike-Yusa, Hiroko, Yilong Li, E. Pien Tan, Martin Del Castillo Velasco-Herrera, and Kosuke Yusa. 2013. “Genome-Wide Recessive Genetic Screening in Mammalian Cells with a Lentiviral CRISPR-Guide RNA Library.” Nature Biotechnology 2013 32:3 32 (3): 267–73. https://doi.org/10.1038/nbt.2800. [CrossRef]
- Kole, Chittaranjan. 2020. “Genomic Designing of Climate-Smart Fruit Crops.” Genomic Designing of Climate-Smart Fruit Crops, January, 1–404. https://doi.org/10.1007/978-3-319-97946-5/COVER. [CrossRef]
- Komor, Alexis C., Yongjoo B. Kim, Michael S. Packer, John A. Zuris, and David R. Liu. 2016. “Programmable Editing of a Target Base in Genomic DNA without Double-Stranded DNA Cleavage.” Nature 2015 533:7603 533 (7603): 420–24. https://doi.org/10.1038/nature17946. [CrossRef]
- Kumar, Prashant, Yashpal Singh Malik, Balasubramanian Ganesh, Somnath Rahangdale, Sharad Saurabh, Senthilkumar Natesan, Ashish Srivastava, et al. 2020. “CRISPR-Cas System: An Approach With Potentials for COVID-19 Diagnosis and Therapeutics.” Frontiers in Cellular and Infection Microbiology 10 (November): 639. https://doi.org/10.3389/FCIMB.2020.576875/BIBTEX. [CrossRef]
- Kumar, Vinay, and Mukesh Jain. 2015. “The CRISPR–Cas System for Plant Genome Editing: Advances and Opportunities.” Journal of Experimental Botany 66 (1): 47–57. https://doi.org/10.1093/JXB/ERU429. [CrossRef]
- Lantican, Darlon V., Susan R. Strickler, Alma O. Canama, Roanne R. Gardoce, Lukas A. Mueller, and Hayde F. Galvez. 2019. “De Novo Genome Sequence Assembly of Dwarf Coconut (Cocos Nucifera L. ‘Catigan Green Dwarf’) Provides Insights into Genomic Variation Between Coconut Types and Related Palm Species.” G3 Genes|Genomes|Genetics 9 (8): 2377–93. https://doi.org/10.1534/G3.119.400215. [CrossRef]
- Lee, Jae Hoon, Hyo Jun Won, Eun-Seok Oh, Man-Ho Oh, and Je Hyeong Jung. 2020. “Golden Gate Cloning-Compatible DNA Replicon/2A-Mediated Polycistronic Vectors for Plants.” Frontiers in Plant Science 11: 559365. https://doi.org/10.3389/fpls.2020.559365. [CrossRef]
- Lee, Joonsun, Min Hee Jung, Euihwan Jeong, and Jungjoon K. Lee. 2019. “Using Sniper-Cas9 to Minimize Off-Target Effects of CRISPR-Cas9 Without the Loss of On-Target Activity Via Directed Evolution.” JoVE (Journal of Visualized Experiments) 2019 (144): e59202. https://doi.org/10.3791/59202. [CrossRef]
- Lee, Jungjoon K., Euihwan Jeong, Joonsun Lee, Minhee Jung, Eunji Shin, Young hoon Kim, Kangin Lee, et al. 2018. “Directed Evolution of CRISPR-Cas9 to Increase Its Specificity.” Nature Communications 2018 9:1 9 (1): 1–10. https://doi.org/10.1038/s41467-018-05477-x. [CrossRef]
- Li, Hongyi, Yang Yang, Weiqi Hong, Mengyuan Huang, Min Wu, and Xia Zhao. 2020. “Applications of Genome Editing Technology in the Targeted Therapy of Human Diseases: Mechanisms, Advances and Prospects.” Signal Transduction and Targeted Therapy 2020 5:1 5 (1): 1–23. https://doi.org/10.1038/s41392-019-0089-y. [CrossRef]
- Li, Jian Feng, Julie E. Norville, John Aach, Matthew McCormack, Dandan Zhang, Jenifer Bush, George M. Church, and Jen Sheen. 2013a. “Multiplex and Homologous Recombination-Mediated Genome Editing in Arabidopsis and Nicotiana Benthamiana Using Guide RNA and Cas9.” Nature Biotechnology 31 (8): 688–91. https://doi.org/10.1038/NBT.2654. [CrossRef]
- ———. 2013b. “Multiplex and Homologous Recombination–Mediated Genome Editing in Arabidopsis and Nicotiana Benthamiana Using Guide RNA and Cas9.” Nature Biotechnology 2013 31:8 31 (8): 688–91. https://doi.org/10.1038/nbt.2654. [CrossRef]
- Li, Meng Yuan, Yun Tong Jiao, Yu Ting Wang, Na Zhang, Bian Bian Wang, Rui Qi Liu, Xiao Yin, Yan Xu, and Guo Tian Liu. 2020. “CRISPR/Cas9-Mediated VvPR4b Editing Decreases Downy Mildew Resistance in Grapevine (Vitis Vinifera L.).” Horticulture Research 2020 7:1 7 (1): 1–11. https://doi.org/10.1038/s41438-020-00371-4. [CrossRef]
- Lin, Qiupeng, Yuan Zong, Chenxiao Xue, Shengxing Wang, Shuai Jin, Zixu Zhu, Yanpeng Wang, et al. 2020. “Prime Genome Editing in Rice and Wheat.” Nature Biotechnology 38 (5): 582–85. https://doi.org/10.1038/s41587-020-0455-x. [CrossRef]
- Lino, Christopher A., Jason C. Harper, James P. Carney, and Jerilyn A. Timlin. 2018. “Delivering Crispr: A Review of the Challenges and Approaches.” Drug Delivery 25 (1): 1234–57. https://doi.org/10.1080/10717544.2018.1474964. [CrossRef]
- Liu, Chaoyang, Chao Feng, Weizhuo Peng, Jingjing Hao, Juntao Wang, Jianjun Pan, and Yehua He. 2020. “Chromosome-Level Draft Genome of a Diploid Plum (Prunus Salicina).” GigaScience 9 (12). https://doi.org/10.1093/GIGASCIENCE/GIAA130. [CrossRef]
- Lundgren, Marjorie R, Pascal-Antoine Christin, Emmanuel Gonzalez Escobar, Brad S Ripley, Guillaume Besnard, Christine M Long, Paul W Hattersley, Roger P Ellis, Richard C Leegood, and Colin P Osborne. 2016. “Evolutionary Implications of C3–C4 Intermediates in the Grass Alloteropsis Semialata.” Plant, Cell & Environment 39 (9): 1874–85. https://doi.org/10.1111/pce.12665. [CrossRef]
- Luo, Xiang, Haoxian Li, Zhikun Wu, Wen Yao, Peng Zhao, Da Cao, Haiyan Yu, et al. 2020. “The Pomegranate (Punica Granatum L.) Draft Genome Dissects Genetic Divergence between Soft- and Hard-seeded Cultivars.” Plant Biotechnology Journal 18 (4): 955. https://doi.org/10.1111/PBI.13260. [CrossRef]
- Maddalo, Danilo, Eusebio Manchado, Carla P. Concepcion, Ciro Bonetti, Joana A. Vidigal, Yoon Chi Han, Paul Ogrodowski, et al. 2014. “In Vivo Engineering of Oncogenic Chromosomal Rearrangements with the CRISPR/Cas9 System.” Nature 516 (7531): 423–28. https://doi.org/10.1038/NATURE13902. [CrossRef]
- Maggio, Ignazio, Luca Stefanucci, Josephine M. Janssen, Jin Liu, Xiaoyu Chen, Vincent Mouly, and Manuel A.F.V. Gonçalves. 2016. “Selection-Free Gene Repair after Adenoviral Vector Transduction of Designer Nucleases: Rescue of Dystrophin Synthesis in DMD Muscle Cell Populations.” Nucleic Acids Research 44 (3): 1449–70. https://doi.org/10.1093/NAR/GKV1540. [CrossRef]
- Malnoy, Mickael, Roberto Viola, Min Hee Jung, Ok Jae Koo, Seokjoong Kim, Jin Soo Kim, Riccardo Velasco, and Chidananda Nagamangala Kanchiswamy. 2016. “DNA-Free Genetically Edited Grapevine and Apple Protoplast Using CRISPR/Cas9 Ribonucleoproteins.” Frontiers in Plant Science 7 (DECEMBER2016): 1904. https://doi.org/10.3389/FPLS.2016.01904/BIBTEX. [CrossRef]
- Mao, Yanfei, Hui Zhang, Nanfei Xu, Botao Zhang, Feng Gou, and Jian Kang Zhu. 2013. “Application of the CRISPR–Cas System for Efficient Genome Engineering in Plants.” Molecular Plant 6 (6): 2008–11. https://doi.org/10.1093/MP/SST121. [CrossRef]
- Martínez-García, Pedro J., Marc W. Crepeau, Daniela Puiu, Daniel Gonzalez-Ibeas, Jeanne Whalen, Kristian A. Stevens, Robin Paul, et al. 2016. “The Walnut (Juglans Regia) Genome Sequence Reveals Diversity in Genes Coding for the Biosynthesis of Non-Structural Polyphenols.” The Plant Journal : For Cell and Molecular Biology 87 (5): 507–32. https://doi.org/10.1111/TPJ.13207. [CrossRef]
- Matano, Mami, Shoichi Date, Mariko Shimokawa, Ai Takano, Masayuki Fujii, Yuki Ohta, Toshiaki Watanabe, Takanori Kanai, and Toshiro Sato. 2015. “Modeling Colorectal Cancer Using CRISPR-Cas9-Mediated Engineering of Human Intestinal Organoids.” Nature Medicine 21 (3): 256–62. https://doi.org/10.1038/NM.3802. [CrossRef]
- “Method of the Year 2011.” 2011. Nature Methods 2012 9:1 9 (1): 1–1. https://doi.org/10.1038/nmeth.1852. [CrossRef]
- Miao, Jin, Dongshu Guo, Jinzhe Zhang, Qingpei Huang, Genji Qin, Xin Zhang, Jianmin Wan, Hongya Gu, and Li Jia Qu. 2013. “Targeted Mutagenesis in Rice Using CRISPR-Cas System.” Cell Research 2013 23:10 23 (10): 1233–36. https://doi.org/10.1038/cr.2013.123. [CrossRef]
- Ming, Ray, Shaobin Hou, Yun Feng, Qingyi Yu, Alexandre Dionne-Laporte, Jimmy H. Saw, Pavel Senin, et al. 2008. “The Draft Genome of the Transgenic Tropical Fruit Tree Papaya (Carica Papaya Linnaeus).” Nature 452 (7190): 991–96. https://doi.org/10.1038/NATURE06856. [CrossRef]
- Ming, Ray, Robert VanBuren, Ching Man Wai, Haibao Tang, Michael C. Schatz, John E. Bowers, Eric Lyons, et al. 2015. “The Pineapple Genome and the Evolution of CAM Photosynthesis.” Nature Genetics 47 (12): 1435–42. https://doi.org/10.1038/NG.3435. [CrossRef]
- Mittal, Amandeep, Inderjit Singh Yadav, Naresh Kumar Arora, Rajbir Singh Boora, Meenakshi Mittal, Parwinder Kaur, William Erskine, Parveen Chhuneja, Manav Indra Singh Gill, and Kuldeep Singh. 2020. “RNA-Sequencing Based Gene Expression Landscape of Guava Cv. Allahabad Safeda and Comparative Analysis to Colored Cultivars.” BMC Genomics 21 (1): 1–19. https://doi.org/10.1186/S12864-020-06883-6/FIGURES/9. [CrossRef]
- Montague, Tessa G., José M. Cruz, James A. Gagnon, George M. Church, and Eivind Valen. 2014. “CHOPCHOP: A CRISPR/Cas9 and TALEN Web Tool for Genome Editing.” Nucleic Acids Research 42 (W1): W401–7. https://doi.org/10.1093/NAR/GKU410. [CrossRef]
- Moreno-Mateos, Miguel A., Charles E. Vejnar, Jean Denis Beaudoin, Juan P. Fernandez, Emily K. Mis, Mustafa K. Khokha, and Antonio J. Giraldez. 2015. “CRISPRscan: Designing Highly Efficient SgRNAs for CRISPR-Cas9 Targeting in Vivo.” Nature Methods 2015 12:10 12 (10): 982–88. https://doi.org/10.1038/nmeth.3543. [CrossRef]
- Mori, Kazuki, Kenta Shirasawa, Hitoshi Nogata, Chiharu Hirata, Kosuke Tashiro, Tsuyoshi Habu, Sangwan Kim, Shuichi Himeno, Satoru Kuhara, and Hidetoshi Ikegami. 2017. “Identification of RAN1 Orthologue Associated with Sex Determination through Whole Genome Sequencing Analysis in Fig (Ficus Carica L.).” Scientific Reports 2017 7:1 7 (1): 1–15. https://doi.org/10.1038/srep41124. [CrossRef]
- Mout, Rubul, Moumita Ray, Gulen Yesilbag Tonga, Yi Wei Lee, Tristan Tay, Kanae Sasaki, and Vincent M. Rotello. 2017. “Direct Cytosolic Delivery of CRISPR/Cas9-Ribonucleoprotein for Efficient Gene Editing.” ACS Nano 11 (3): 2452–58. https://doi.org/10.1021/ACSNANO.6B07600/SUPPL_FILE/NN6B07600_SI_004.AVI. [CrossRef]
- Naim, Fatima, Benjamin Dugdale, Jennifer Kleidon, Anthony Brinin, Kylie Shand, Peter Waterhouse, and James Dale. 2018. “Gene Editing the Phytoene Desaturase Alleles of Cavendish Banana Using CRISPR/Cas9.” Transgenic Research 27 (5): 451–60. https://doi.org/10.1007/S11248-018-0083-0. [CrossRef]
- Naito, Yuki, Kimihiro Hino, Hidemasa Bono, and Kumiko Ui-Tei. 2015. “CRISPRdirect: Software for Designing CRISPR/Cas Guide RNA with Reduced off-Target Sites.” Bioinformatics 31 (7): 1120–23. https://doi.org/10.1093/BIOINFORMATICS/BTU743. [CrossRef]
- Nakajima, Ikuko, Yusuke Ban, Akifumi Azuma, Noriyuki Onoue, Takaya Moriguchi, Toshiya Yamamoto, Seiichi Toki, and Masaki Endo. 2017. “CRISPR/Cas9-Mediated Targeted Mutagenesis in Grape.” PloS One 12 (5). https://doi.org/10.1371/JOURNAL.PONE.0177966. [CrossRef]
- Nakamura, T., H. Akita, Y. Yamada, H. Hatakeyama, and H. Harashima. 2012. “A Multifunctional Envelope-Type Nanodevice for Use in Nanomedicine: Concept and Applications.” Accounts of Chemical Research 45 (7): 1113–21. https://doi.org/10.1021/AR200254S/ASSET/IMAGES/MEDIUM/AR-2011-00254S_0006.GIF. [CrossRef]
- Narayanan, Narayanan, Getu Beyene, Raj Deepika Chauhan, Eliana Gaitán-Solís, Jackson Gehan, Paula Butts, Dimuth Siritunga, et al. 2019. “Biofortification of Field-Grown Cassava by Engineering Expression of an Iron Transporter and Ferritin.” Nature Biotechnology 2019 37:2 37 (2): 144–51. https://doi.org/10.1038/s41587-018-0002-1. [CrossRef]
- Nekrasov, Vladimir, Brian Staskawicz, Detlef Weigel, Jonathan D.G. Jones, and Sophien Kamoun. 2013. “Targeted Mutagenesis in the Model Plant Nicotiana Benthamiana Using Cas9 RNA-Guided Endonuclease.” Nature Biotechnology 31 (8): 691–93. https://doi.org/10.1038/NBT.2655. [CrossRef]
- Newell, Christine A, Naomi J Brown, Zheng Liu, Alexander Pflug, Udo Gowik, Peter Westhoff, and Julian M Hibberd. 2010. “Agrobacterium Tumefaciens-Mediated Transformation of Cleome Gynandra L., a C4 Dicotyledon That Is Closely Related to Arabidopsis Thaliana.” Journal of Experimental Botany 61 (5): 1311–19. https://doi.org/10.1093/jxb/erq009. [CrossRef]
- Nishitani, Chikako, Narumi Hirai, Sadao Komori, Masato Wada, Kazuma Okada, Keishi Osakabe, Toshiya Yamamoto, and Yuriko Osakabe. 2016a. “Efficient Genome Editing in Apple Using a CRISPR/Cas9 System.” Scientific Reports 2016 6:1 6 (1): 1–8. https://doi.org/10.1038/srep31481. [CrossRef]
- ———. 2016b. “Efficient Genome Editing in Apple Using a CRISPR/Cas9 System.” Scientific Reports 6 (August). https://doi.org/10.1038/SREP31481. [CrossRef]
- Noman, Ali, Muhammad Aqeel, and Shuilin He. 2016. “CRISPR-Cas9: Tool for Qualitative and Quantitative Plant Genome Editing.” Frontiers in Plant Science 7 (NOVEMBER2016): 1740. https://doi.org/10.3389/FPLS.2016.01740/BIBTEX. [CrossRef]
- Nunez, Sarahi, Eric Arets, Rob Alkemade, Caspar Verwer, and Rik Leemans. 2019. “Assessing the Impacts of Climate Change on Biodiversity: Is below 2° C Enough?” Climatic Change 154: 351–65.
- Osakabe, Yuriko, Zhenchang Liang, Chong Ren, Chikako Nishitani, Keishi Osakabe, Masato Wada, Sadao Komori, et al. 2018. “CRISPR–Cas9-Mediated Genome Editing in Apple and Grapevine.” Nature Protocols 2018 13:12 13 (12): 2844–63. https://doi.org/10.1038/s41596-018-0067-9. [CrossRef]
- Ousterout, David G., Ami M. Kabadi, Pratiksha I. Thakore, William H. Majoros, Timothy E. Reddy, and Charles A. Gersbach. 2015. “Multiplex CRISPR/Cas9-Based Genome Editing for Correction of Dystrophin Mutations That Cause Duchenne Muscular Dystrophy.” Nature Communications 2015 6:1 6 (1): 1–13. https://doi.org/10.1038/ncomms7244. [CrossRef]
- Peer, Reut, Gil Rivlin, Sara Golobovitch, Moshe Lapidot, Amit Gal-On, Alexander Vainstein, Tzvi Tzfira, and Moshe A. Flaishman. 2015. “Targeted Mutagenesis Using Zinc-Finger Nucleases in Perennial Fruit Trees.” Planta 241 (4): 941–51. https://doi.org/10.1007/S00425-014-2224-X. [CrossRef]
- Peng, Aihong, Shanchun Chen, Tiangang Lei, Lanzhen Xu, Yongrui He, Liu Wu, Lixiao Yao, and Xiuping Zou. 2017a. “Engineering Canker-Resistant Plants through CRISPR/Cas9-Targeted Editing of the Susceptibility Gene CsLOB1 Promoter in Citrus.” Plant Biotechnology Journal 15 (12): 1509–19. https://doi.org/10.1111/PBI.12733. [CrossRef]
- ———. 2017b. “Engineering Canker-Resistant Plants through CRISPR/Cas9-Targeted Editing of the Susceptibility Gene CsLOB1 Promoter in Citrus.” Plant Biotechnology Journal 15 (12): 1509–19. https://doi.org/10.1111/PBI.12733. [CrossRef]
- Platt, Randall J., Sidi Chen, Yang Zhou, Michael J. Yim, Lukasz Swiech, Hannah R. Kempton, James E. Dahlman, et al. 2014. “CRISPR-Cas9 Knockin Mice for Genome Editing and Cancer Modeling.” Cell 159 (2): 440–55. https://doi.org/10.1016/J.CELL.2014.09.014. [CrossRef]
- Prykhozhij, Sergey V., Vinothkumar Rajan, Daniel Gaston, and Jason N. Berman. 2015. “CRISPR MultiTargeter: A Web Tool to Find Common and Unique CRISPR Single Guide RNA Targets in a Set of Similar Sequences.” PLOS ONE 10 (3): e0119372. https://doi.org/10.1371/JOURNAL.PONE.0119372. [CrossRef]
- Ramesh, S. V., V. Arunachalam, and M. K. Rajesh. 2020. “Genomic Designing of Climate-Smart Coconut.” Genomic Designing of Climate-Smart Fruit Crops, 135–56. https://doi.org/10.1007/978-3-319-97946-5_6. [CrossRef]
- Raveux, Aurélien, Sandrine Vandormael-Pournin, and Michel Cohen-Tannoudji. 2017. “Optimization of the Production of Knock-in Alleles by CRISPR/Cas9 Microinjection into the Mouse Zygote.” Scientific Reports 7 (February). https://doi.org/10.1038/SREP42661. [CrossRef]
- Rees, Holly A., and David R. Liu. 2018. “Base Editing: Precision Chemistry on the Genome and Transcriptome of Living Cells.” Nature Reviews Genetics 2018 19:12 19 (12): 770–88. https://doi.org/10.1038/s41576-018-0059-1. [CrossRef]
- “Regulation of Citrus <em>DMR6</Em> via RNA Interference and CRISPR/Cas9-Mediated Gene Editing to Improve Huanglongbing Tolerance.” n.d.
- Rodríguez-Leal, Daniel, Zachary H Lemmon, Jarrett Man, Madelaine E Bartlett, and Zachary B Lippman. 2017. “Engineering Quantitative Trait Variation for Crop Improvement by Genome Editing.” Cell 171 (2): 470-480.e8. https://doi.org/10.1016/j.cell.2017.08.030. [CrossRef]
- Roehm, Pamela C., Masoud Shekarabi, Hassen S. Wollebo, Anna Bellizzi, Lifan He, Julian Salkind, and Kamel Khalili. 2016. “Inhibition of HSV-1 Replication by Gene Editing Strategy.” Scientific Reports 2016 6:1 6 (1): 1–11. https://doi.org/10.1038/srep23146. [CrossRef]
- Rugienius, Rytis, Birutė Frercks, Ingrida Mažeikienė, Neringa Rasiukevičiūtė, Danas Baniulis, and Vidmantas Stanys. 2020. “Development of Climate-Resilient Varieties in Rosaceous Berries.” Genomic Designing of Climate-Smart Fruit Crops, 333–84. https://doi.org/10.1007/978-3-319-97946-5_9. [CrossRef]
- Salonia, Fabrizio, Angelo Ciacciulli, Lara Poles, Helena Domenica Pappalardo, Stefano La Malfa, and Concetta Licciardello. 2020. “New Plant Breeding Techniques in Citrus for the Improvement of Important Agronomic Traits. A Review.” Frontiers in Plant Science 11. https://doi.org/10.3389/fpls.2020.01234. [CrossRef]
- Sattar, Muhammad N., Zafar Iqbal, Muhammad N. Tahir, Muhammad S. Shahid, Muhammad Khurshid, Abdullatif A. Al-Khateeb, and Suliman A. Al-Khateeb. 2017. “CRISPR/Cas9: A Practical Approach in Date Palm Genome Editing.” Frontiers in Plant Science 8 (August). https://doi.org/10.3389/FPLS.2017.01469/FULL. [CrossRef]
- Sauer, Noel J., Jerry Mozoruk, Ryan B. Miller, Zachary J. Warburg, Keith A. Walker, Peter R. Beetham, Christian R. Schöpke, and Greg F.W. Gocal. 2016. “Oligonucleotide-Directed Mutagenesis for Precision Gene Editing.” Plant Biotechnology Journal 14 (2): 496–502. https://doi.org/10.1111/PBI.12496. [CrossRef]
- Scalabrin, Simone, Lucile Toniutti, Gabriele Di Gaspero, Davide Scaglione, Gabriele Magris, Michele Vidotto, Sara Pinosio, et al. 2020. “A Single Polyploidization Event at the Origin of the Tetraploid Genome of Coffea Arabica Is Responsible for the Extremely Low Genetic Variation in Wild and Cultivated Germplasm.” Scientific Reports 2020 10:1 10 (1): 1–13. https://doi.org/10.1038/s41598-020-61216-7. [CrossRef]
- Schwank, Gerald, Bon Kyoung Koo, Valentina Sasselli, Johanna F. Dekkers, Inha Heo, Turan Demircan, Nobuo Sasaki, et al. 2013. “Functional Repair of CFTR by CRISPR/Cas9 in Intestinal Stem Cell Organoids of Cystic Fibrosis Patients.” Cell Stem Cell 13 (6): 653–58. https://doi.org/10.1016/J.STEM.2013.11.002. [CrossRef]
- Sedelnikova, Olga V, Thomas E Hughes, and Jane A Langdale. 2018. “Understanding the Genetic Basis of C4 Kranz Anatomy with a View to Engineering C3 Crops.” Annual Review of Genetics 52: 249–70. https://doi.org/10.1146/annurev-genet-120417-031217. [CrossRef]
- Shan, Qiwei, Yanpeng Wang, Jun Li, Yi Zhang, Kunling Chen, Zhen Liang, Kang Zhang, et al. 2013. “Targeted Genome Modification of Crop Plants Using a CRISPR-Cas System.” Nature Biotechnology 31 (8): 686–88. https://doi.org/10.1038/NBT.2650. [CrossRef]
- Shao, Xiuhong, Shaoping Wu, Tongxin Dou, Haocheng Zhu, Chunhua Hu, Heqiang Huo, Weidi He, et al. 2020. “Using CRISPR/Cas9 Genome Editing System to Create MaGA20ox2 Gene-Modified Semi-Dwarf Banana.” Plant Biotechnology Journal 18 (1): 17–19. https://doi.org/10.1111/pbi.13216. [CrossRef]
- South, Paul F., Amanda P. Cavanagh, Helen W. Liu, and Donald R. Ort. 2019. “Synthetic Glycolate Metabolism Pathways Stimulate Crop Growth and Productivity in the Field.” Science 363 (6422). https://doi.org/10.1126/SCIENCE.AAT9077/SUPPL_FILE/DATASETS_S1-S11_S16-S20.PDF. [CrossRef]
- Stemmer, Manuel, Thomas Thumberger, Maria Del Sol Keyer, Joachim Wittbrodt, and Juan L. Mateo. 2015. “CCTop: An Intuitive, Flexible and Reliable CRISPR/Cas9 Target Prediction Tool.” PLOS ONE 10 (4): e0124633. https://doi.org/10.1371/JOURNAL.PONE.0124633. [CrossRef]
- Su, Jinghan, Huiping Sun, Qingshuo Meng, Pengcheng Zhang, Qi Yin, and Yaping Li. 2017. “Enhanced Blood Suspensibility and Laser-Activated Tumor-Specific Drug Release of Theranostic Mesoporous Silica Nanoparticles by Functionalizing with Erythrocyte Membranes.” Theranostics 7 (3): 523–37. https://doi.org/10.7150/THNO.17259. [CrossRef]
- Suleiman, Ahmed Abdul Jabbar, Walaa Yahya Saedi, and Mohammed Jobair Muhaidi. 2021. “Widely Used Gene Editing Strategies in Cancer Treatment a Systematic Review.” Gene Reports 22 (March): 100983. https://doi.org/10.1016/J.GENREP.2020.100983. [CrossRef]
- Sun, Wujin, Wenyan Ji, Jordan M. Hall, Quanyin Hu, Chao Wang, Chase L. Beisel, and Zhen Gu. 2015. “Self-Assembled DNA Nanoclews for the Efficient Delivery of CRISPR-Cas9 for Genome Editing.” Angewandte Chemie (International Ed. in English) 54 (41): 12029–33. https://doi.org/10.1002/ANIE.201506030. [CrossRef]
- Tabassum, Javaria, Shakeel Ahmad, Babar Hussain, Amos Musyoki Mawia, Aqib Zeb, and Luo Ju. 2021. “Applications and Potential of Genome-Editing Systems in Rice Improvement: Current and Future Perspectives.” Agronomy 2021, Vol. 11, Page 1359 11 (7): 1359. https://doi.org/10.3390/AGRONOMY11071359. [CrossRef]
- Tabebordbar, Mohammadsharif, Kexian Zhu, Jason K.W. Cheng, Wei Leong Chew, Jeffrey J. Widrick, Winston X. Yan, Claire Maesner, et al. 2016. “In Vivo Gene Editing in Dystrophic Mouse Muscle and Muscle Stem Cells.” Science (New York, N.Y.) 351 (6271): 407–11. https://doi.org/10.1126/SCIENCE.AAD5177. [CrossRef]
- Taylor, Rachel A., Sadie J. Ryan, Catherine A. Lippi, David G. Hall, Hossein A. Narouei-Khandan, Jason R. Rohr, and Leah R. Johnson. 2019. “Predicting the Fundamental Thermal Niche of Crop Pests and Diseases in a Changing World: A Case Study on Citrus Greening.” Journal of Applied Ecology 56 (8): 2057–68. https://doi.org/10.1111/1365-2664.13455. [CrossRef]
- Teng, Kai Wen, Yuji Ishitsuka, Pin Ren, Yeoan Youn, Xiang Deng, Pinghua Ge, Andrew S. Belmont, and Paul R. Selvin. 2016. “Labeling Proteins inside Living Cells Using External Fluorophores for Microscopy.” ELife 5 (DECEMBER2016). https://doi.org/10.7554/ELIFE.20378. [CrossRef]
- Tripathi, Jaindra N., Valentine O. Ntui, Mily Ron, Samwel K. Muiruri, Anne Britt, and Leena Tripathi. 2019. “CRISPR/Cas9 Editing of Endogenous Banana Streak Virus in the B Genome of Musa Spp. Overcomes a Major Challenge in Banana Breeding.” Communications Biology 2019 2:1 2 (1): 1–11. https://doi.org/10.1038/s42003-019-0288-7. [CrossRef]
- Truong, Dong Jiunn Jeffery, Karin Kühner, Ralf Kühn, Stanislas Werfel, Stefan Engelhardt, Wolfgang Wurst, and Oskar Ortiz. 2015. “Development of an Intein-Mediated Split-Cas9 System for Gene Therapy.” Nucleic Acids Research 43 (13): 6450–58. https://doi.org/10.1093/NAR/GKV601. [CrossRef]
- Velasco, Riccardo, Andrey Zharkikh, Jason Affourtit, Amit Dhingra, Alessandro Cestaro, Ananth Kalyanaraman, Paolo Fontana, et al. 2010. “The Genome of the Domesticated Apple (Malus × Domestica Borkh.).” Nature Genetics 42 (10): 833–39. https://doi.org/10.1038/NG.654. [CrossRef]
- Verde, Ignazio, Albert G. Abbott, Simone Scalabrin, Sook Jung, Shengqiang Shu, Fabio Marroni, Tatyana Zhebentyayeva, et al. 2013. “The High-Quality Draft Genome of Peach (Prunus Persica) Identifies Unique Patterns of Genetic Diversity, Domestication and Genome Evolution.” Nature Genetics 2013 45:5 45 (5): 487–94. https://doi.org/10.1038/ng.2586. [CrossRef]
- Wan, Dong-Yan, Ye Guo, Yuan Cheng, Yang Hu, Shunyuan Xiao, Yuejin Wang, and Ying-Qiang Wen. 2020. “CRISPR/Cas9-Mediated Mutagenesis of VvMLO3 Results in Enhanced Resistance to Powdery Mildew in Grapevine (Vitis Vinifera).” Horticulture Research 7 (January): 116. https://doi.org/10.1038/s41438-020-0339-8. [CrossRef]
- Wang, Jiawei, Weizhen Liu, Dongzi Zhu, Po Hong, Shizhong Zhang, Shijun Xiao, Yue Tan, et al. 2020. “Chromosome-Scale Genome Assembly of Sweet Cherry (Prunus Avium L.) Cv. Tieton Obtained Using Long-Read and Hi-C Sequencing.” Horticulture Research 2020 7:1 7 (1): 1–11. https://doi.org/10.1038/s41438-020-00343-8. [CrossRef]
- Wang, Xianhang, Mingxing Tu, Dejun Wang, Jianwei Liu, Yajuan Li, Zhi Li, Yuejin Wang, and Xiping Wang. 2018. “CRISPR/Cas9-Mediated Efficient Targeted Mutagenesis in Grape in the First Generation.” Plant Biotechnology Journal 16 (4): 844–55. https://doi.org/10.1111/PBI.12832. [CrossRef]
- Wang, Yan, Tao Sun, Tingting Li, Meng Wang, Guangxiao Yang, and Guangyuan He. 2016. “A CBL-Interacting Protein Kinase TaCIPK2 Confers Drought Tolerance in Transgenic Tobacco Plants through Regulating the Stomatal Movement.” PLOS ONE 11 (12): e0167962. https://doi.org/10.1371/JOURNAL.PONE.0167962. [CrossRef]
- Wang, Zupeng, Shuaibin Wang, Dawei Li, Qiong Zhang, Li Li, Caihong Zhong, Yifei Liu, and Hongwen Huang. 2018a. “Optimized Paired-SgRNA/Cas9 Cloning and Expression Cassette Triggers High-Efficiency Multiplex Genome Editing in Kiwifruit.” Plant Biotechnology Journal 16 (8): 1424–33. https://doi.org/10.1111/PBI.12884. [CrossRef]
- ———. 2018b. “Optimized Paired-SgRNA/Cas9 Cloning and Expression Cassette Triggers High-Efficiency Multiplex Genome Editing in Kiwifruit.” Plant Biotechnology Journal 16 (8): 1424–33. https://doi.org/10.1111/PBI.12884. [CrossRef]
- Wen, Xiao Peng, Xiao Ming Pang, Narumi Matsuda, Masayuki Kita, Hiromichi Inoue, Yu Jin Hao, Chikako Honda, and Takaya Moriguchi. 2008. “Over-Expression of the Apple Spermidine Synthase Gene in Pear Confers Multiple Abiotic Stress Tolerance by Altering Polyamine Titers.” Transgenic Research 17 (2): 251–63. https://doi.org/10.1007/S11248-007-9098-7. [CrossRef]
- Wong, Nathan, Weijun Liu, and Xiaowei Wang. 2015. “WU-CRISPR: Characteristics of Functional Guide RNAs for the CRISPR/Cas9 System.” Genome Biology 16 (1): 1–8. https://doi.org/10.1186/S13059-015-0784-0/FIGURES/4. [CrossRef]
- Wu, Jun, Zhiwen Wang, Zebin Shi, Shu Zhang, Ray Ming, Shilin Zhu, M. Awais Khan, et al. 2013. “The Genome of the Pear (Pyrus Bretschneideri Rehd.).” Genome Research 23 (2): 396–408. https://doi.org/10.1101/GR.144311.112. [CrossRef]
- Xiao, Yong, Pengwei Xu, Haikuo Fan, Luc Baudouin, Wei Xia, Stéphanie Bocs, Junyang Xu, et al. 2017. “The Genome Draft of Coconut (Cocos Nucifera).” GigaScience 6 (11): 1. https://doi.org/10.1093/GIGASCIENCE/GIX095. [CrossRef]
- Xie, Kabin, and Yinong Yang. 2013. “RNA-Guided Genome Editing in Plants Using a CRISPR-Cas System.” Molecular Plant 6 (6): 1975–83. https://doi.org/10.1093/MP/SST119. [CrossRef]
- Xu, Huimin, Qingyi Yu, Yan Shi, Xiuting Hua, Haibao Tang, Long Yang, Ray Ming, and Jisen Zhang. 2018. “PGD: Pineapple Genomics Database.” Horticulture Research 2018 5:1 5 (1): 1–9. https://doi.org/10.1038/s41438-018-0078-2. [CrossRef]
- Xu, Qiang, Ling Ling Chen, Xiaoan Ruan, Dijun Chen, Andan Zhu, Chunli Chen, Denis Bertrand, et al. 2012. “The Draft Genome of Sweet Orange (Citrus Sinensis).” Nature Genetics 2012 45:1 45 (1): 59–66. https://doi.org/10.1038/ng.2472. [CrossRef]
- Yang, Hung Chih, and Pei Jer Chen. 2018. “The Potential and Challenges of CRISPR-Cas in Eradication of Hepatitis B Virus Covalently Closed Circular DNA.” Virus Research 244 (January): 304–10. https://doi.org/10.1016/J.VIRUSRES.2017.06.010. [CrossRef]
- Yang, Yue, Jin Xu, Shuyu Ge, and Liqin Lai. 2021. “CRISPR/Cas: Advances, Limitations, and Applications for Precision Cancer Research.” Frontiers in Medicine 8 (March): 115. https://doi.org/10.3389/FMED.2021.649896/BIBTEX. [CrossRef]
- Yee, Jiing-Kuan, and -K Yee. 2016. “Off-Target Effects of Engineered Nucleases.” The FEBS Journal 283 (17): 3239–48. https://doi.org/10.1111/FEBS.13760. [CrossRef]
- Zambounis, Antonios, Ioannis Ganopoulos, Filippos Aravanopoulos, Zoe Hilioti, Panagiotis Madesis, Athanassios Molassiotis, Athanasios Tsaftaris, and Aliki Xanthopoulou. 2020. “Genomics Opportunities and Breeding Strategies Towards Improvement of Climate-Smart Traits and Disease Resistance Against Pathogens in Sweet Cherry.” Genomic Designing of Climate-Smart Fruit Crops, 385–404. https://doi.org/10.1007/978-3-319-97946-5_10. [CrossRef]
- Zhang, Fei, Chantal LeBlanc, Vivian F. Irish, and Yannick Jacob. 2017. “Rapid and Efficient CRISPR/Cas9 Gene Editing in Citrus Using the YAO Promoter.” Plant Cell Reports 36 (12): 1883–87. https://doi.org/10.1007/S00299-017-2202-4. [CrossRef]
- Zhang, Yi, Karen Massel, Ian D. Godwin, and Caixia Gao. 2018. “Applications and Potential of Genome Editing in Crop Improvement 06 Biological Sciences 0604 Genetics 06 Biological Sciences 0607 Plant Biology 07 Agricultural and Veterinary Sciences 0703 Crop and Pasture Production.” Genome Biology 19 (1): 1–11. https://doi.org/10.1186/S13059-018-1586-Y/FIGURES/2. [CrossRef]
- Zhang, Yingxiao, Aimee A Malzahn, Simon Sretenovic, and Yiping Qi. 2019. “The Emerging and Uncultivated Potential of CRISPR Technology in Plant Science.” Nature Plants 5 (8): 778–94. https://doi.org/10.1038/s41477-019-0461-5. [CrossRef]
- Zhou, Junhui, Guoming Wang, and Zhongchi Liu. 2018. “Efficient Genome Editing of Wild Strawberry Genes, Vector Development and Validation.” Plant Biotechnology Journal 16 (11): 1868–77. https://doi.org/10.1111/PBI.12922. [CrossRef]
- Zhu, Haocheng, Chao Li, and Caixia Gao. 2020. “Applications of CRISPR–Cas in Agriculture and Plant Biotechnology.” Nature Reviews Molecular Cell Biology 21 (11): 661–77. https://doi.org/10.1038/s41580-020-00288-9. [CrossRef]
- Zhu, Qing gang, Yang Xu, Yong Yang, Chang fei Guan, Qiu yun Zhang, Jing wen Huang, Don Grierson, Kun song Chen, Bang chu Gong, and Xue ren Yin. 2019. “The Persimmon (Diospyros Oleifera Cheng) Genome Provides New Insights into the Inheritance of Astringency and Ancestral Evolution.” Horticulture Research 2019 6:1 6 (1): 1–15. https://doi.org/10.1038/s41438-019-0227-2. [CrossRef]
- Zischewski, Julia, Rainer Fischer, and Luisa Bortesi. 2017. “Detection of On-Target and off-Target Mutations Generated by CRISPR/Cas9 and Other Sequence-Specific Nucleases.” Biotechnology Advances 35 (1): 95–104. https://doi.org/10.1016/J.BIOTECHADV.2016.12.003. [CrossRef]
- Zuris, John A., David B. Thompson, Yilai Shu, John P. Guilinger, Jeffrey L. Bessen, Johnny H. Hu, Morgan L. Maeder, J. Keith Joung, Zheng Yi Chen, and David R. Liu. 2014. “Cationic Lipid-Mediated Delivery of Proteins Enables Efficient Protein-Based Genome Editing in Vitro and in Vivo.” Nature Biotechnology 2014 33:1 33 (1): 73–80. https://doi.org/10.1038/nbt.3081. [CrossRef]
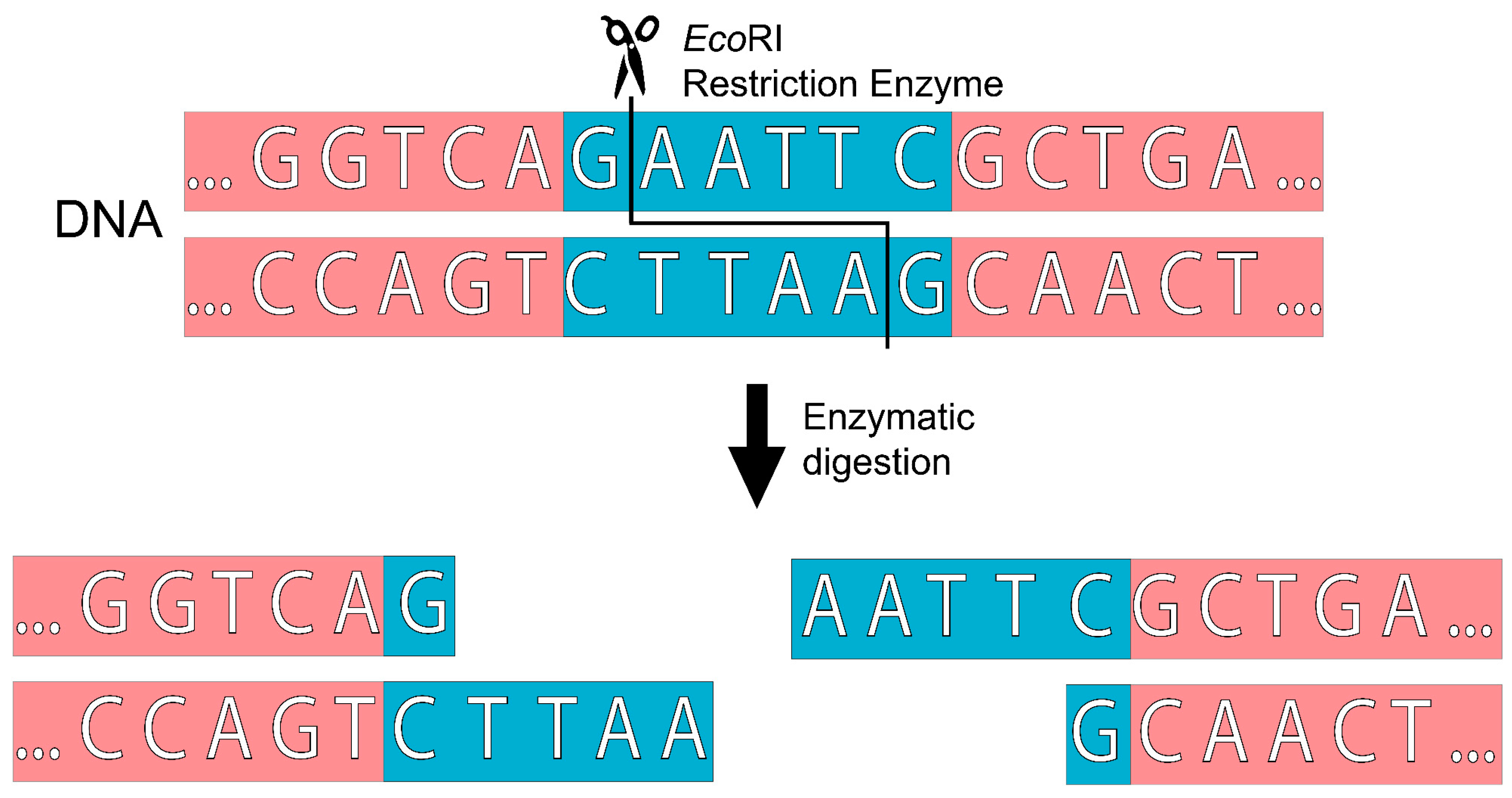
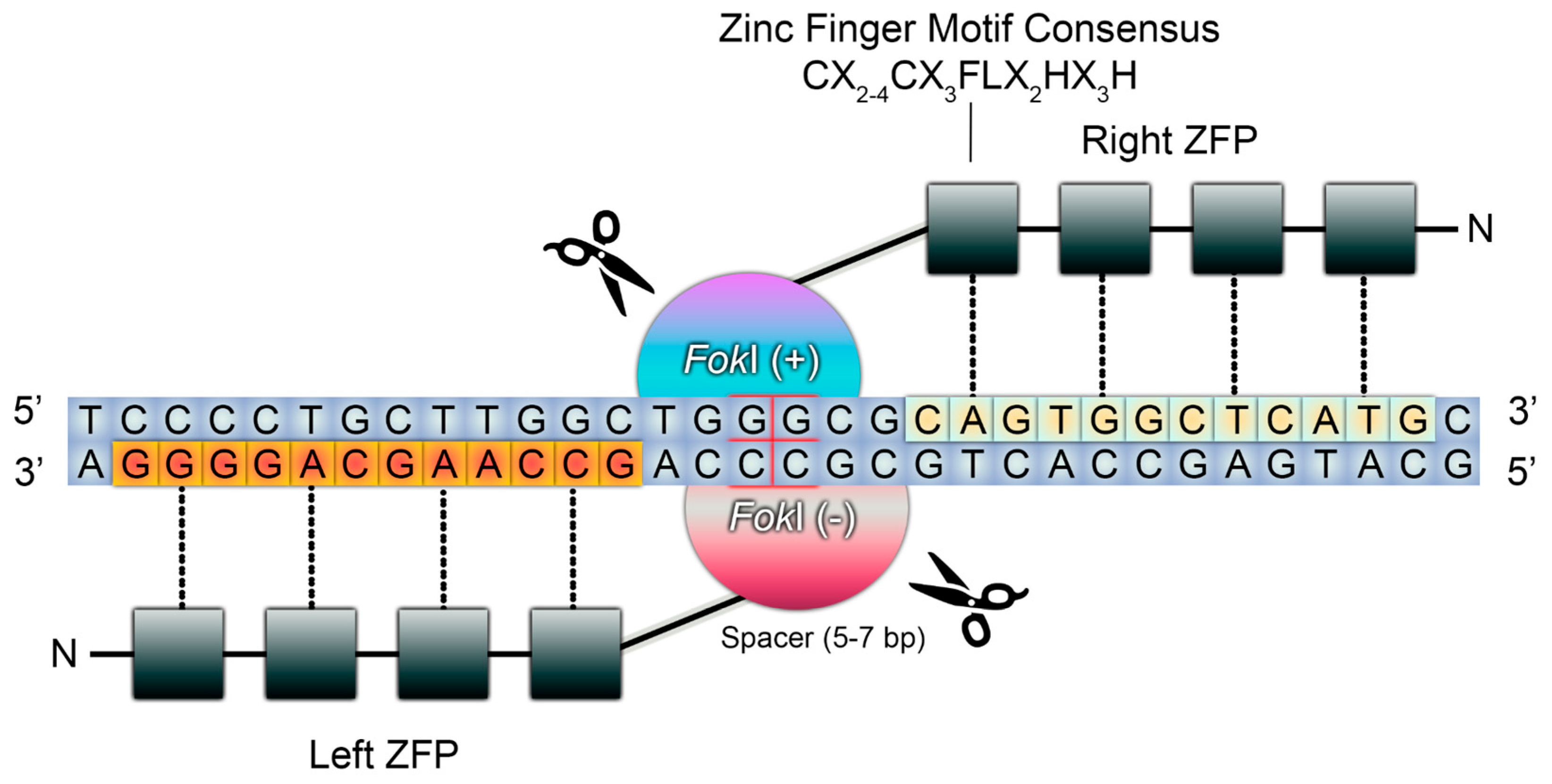
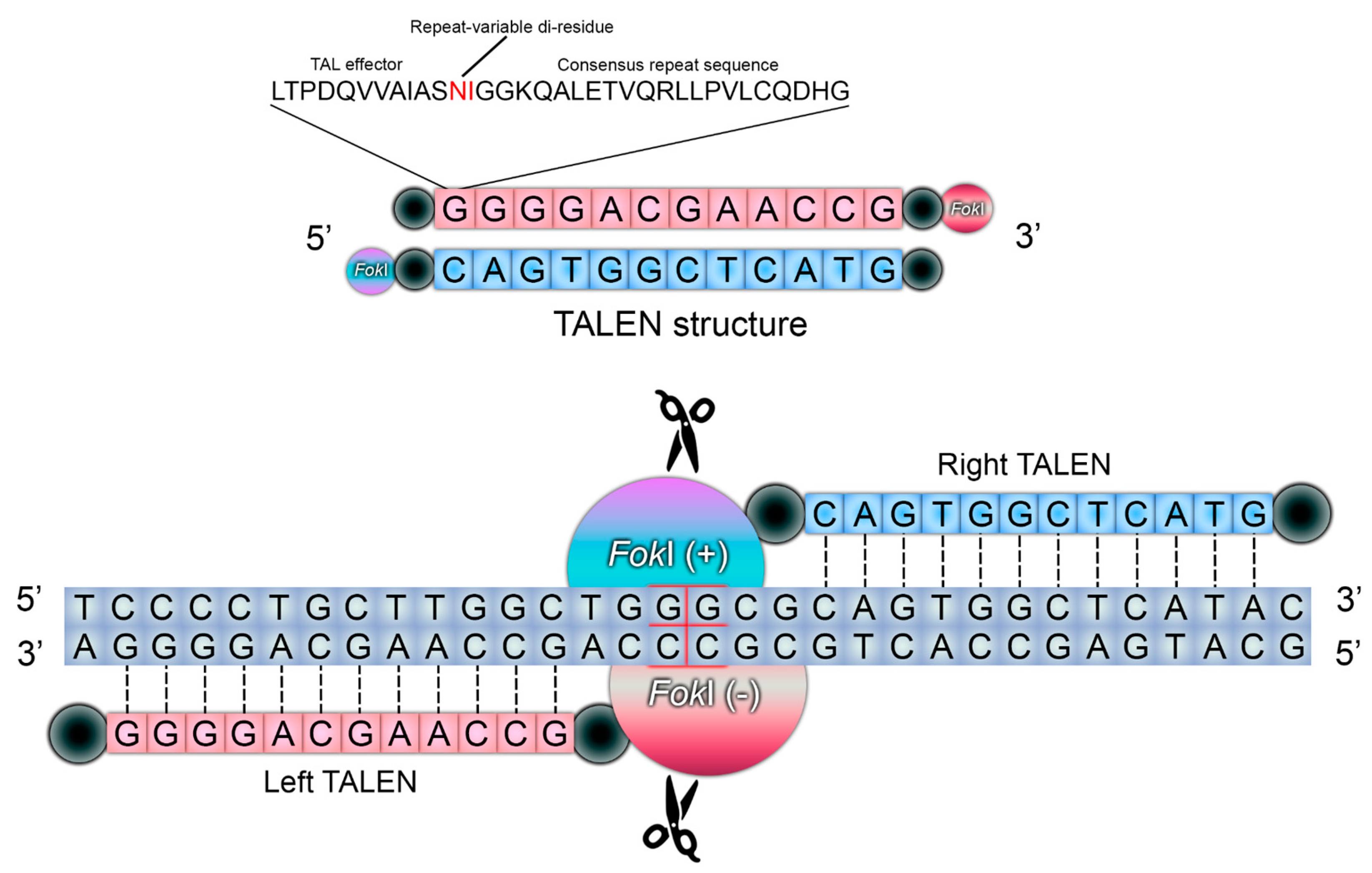
| Properties | ODM | ZFNs | TALENs | CRISPR/Cas9 | Reference(s) |
|---|---|---|---|---|---|
| Multiplexing Capability | Difficult | Difficult | Difficult | Possible | (Mao et al. 2013) (Noman, Aqeel, and He 2016) |
| Mutation Rate | Medium | High | Medium | Low | (Gaj, Gersbach, and Barbas 2013) |
| Mode of Action | Conversion within the target region (sense strand directed) | Breaks in target DNA (Double strand) | Breaks in target DNA (Double strand) | Breaks in target DNA (Double strand) | (Mao et al. 2013) |
| Cloning | Not required | Necessary | Necessary | Not Required (Usually) | (Sauer et al. 2016) |
| Target Sequence Length [bp] | 70-86 | 26-34 | 26-57 | 20-22 | (F. Chen et al. 2017) |
| Targeting Efficiency | Relatively high | High | High | Very high | (Gaj, Gersbach, and Barbas 2013) |
| Components | Chimeraplast | Zn finger FokI nuclease domain (non-specific) | TALE DNA-binding FokI nuclease domain (non-specific) | Cas9 proteins, sgRNA | (V. Kumar and Jain 2015) |
| Structural Protein Composition | Non-protein | Proteins (dimeric) | Proteins (dimeric) | Protein (monomeric) | (J. F. Li et al. 2013b) |
| Catalytic Domains | Catalytic domain absent | Restriction endonuclease FokI | Restriction endonuclease FokI | HNH and RuvC | (Sauer et al. 2016) |
| Large-Scale Libraries | Difficult | Difficult | Very Difficult | Possible | (Hsu et al. 2013) |
| Engineering Steps for Protein | Not required | Required | Required | Required | (Cho et al. 2013) |
| Tool | Web Address | Reference(s) | |
|---|---|---|---|
| CRISPRscan | http://www.crisprscan.org/ | (Moreno-Mateos et al. 2015) | |
| CHOPCHOP | https://chopchop.cbu.uib.no/ | (Montague et al. 2014) | |
| WU-CRISPR | https://crisprdb.org/wu-crispr/ | (Wong, Liu, and Wang 2015) | |
| CRISPRdirect | http://crispr.dbcls.jp/ | (Naito et al. 2015) | |
| CRISPR MultiTargeter | http://www.multicrispr.net/ | (Prykhozhij et al. 2015) | |
| E-CRISP | http://www.e-crisp.org/E-CRISP/ | (Heigwer, Kerr, and Boutros 2014) | |
| CCTop | https://cctop.cos.uni-heidelberg.de/ | (Stemmer et al. 2015) | |
| Cas-OFFinder | http://www.rgenome.net/cas-offinder/ | (Bae, Park, and Kim 2014) |
| Fruits Crop | Loci being Targeted | Type of Modification(s) | Tissue for Modification | Reference(s) |
|---|---|---|---|---|
| Apple | uidA | Activity of β-glucuronidase | Leaf tissue | (Peer et al. 2015) |
| PDS | Biosynthesis of carotenoids | Leaf tissue | (Nishitani et al. 2016a) | |
| IdnDH | Tartaric acid biosynthesis | Leaf tissue | (Osakabe et al. 2018) | |
| DIPM (1, 2, and 4) | Resistance to fire blight | Protoplasm | (Malnoy et al. 2016) | |
| Banana | MaPDS | Biosynthesis of carotenoids | Cell suspension (embryogenic) | (Kaur et al. 2018) |
| eBSV | Viral pathogenesis control | Explant (epicotyl) | (Tripathi et al. 2019) | |
| Kiwifruit | acPDS | Biosynthesis of carotenoids | Leaf tissue | (Z. Wang et al. 2018a) |
| Sweet Orange | CsPDS | Biosynthesis of carotenoids | Leaf tissue | (Jia and Nian 2014) |
| DMR6 | Resistance to Huanglongbing disease | Explant (epicotyl) | (“Regulation of Citrus <em>DMR6</Em> via RNA Interference and CRISPR/Cas9-Mediated Gene Editing to Improve Huanglongbing Tolerance,” n.d.) | |
| Fig | uidA | Activity of β-glucuronidase | Leaf tissue | (Peer et al. 2015) |
| Wanjincheng Orange | CsLOB1 (promoter Sequence) | Resistance against citrus canker | Explant (epicotyl) | (Peng et al. 2017a) |
| Pear | TFL1 | Early flowering | Cell suspension (embryogenic) | (Charrier et al. 2019) |
| Coffee | CcPDS | Biosynthesis of carotenoids | Cell suspension (meristematic) | (Breitler et al. 2018) |
| Cacao | TcNPR3 | Enhanced defense response | Leaf tissue | (Fister et al. 2018a) |
| Grapevine | VvPR4b | Downy mildew resistance | Proembryogenic mass (PEM) cells | (M. Y. Li et al. 2020) |
| VvMLO3 | Powdery mildew | Leaf tissue | (Wan et al. 2020) | |
| Papaya | cp | Resistance against Papaya ringspot virus | immature zygotic embryos | (Fitch et al. 1992) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





