Submitted:
29 May 2023
Posted:
30 May 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Microbial Sampling
2.2. Identification of Bacteria
2.3. Genome Sequencing and Analysis
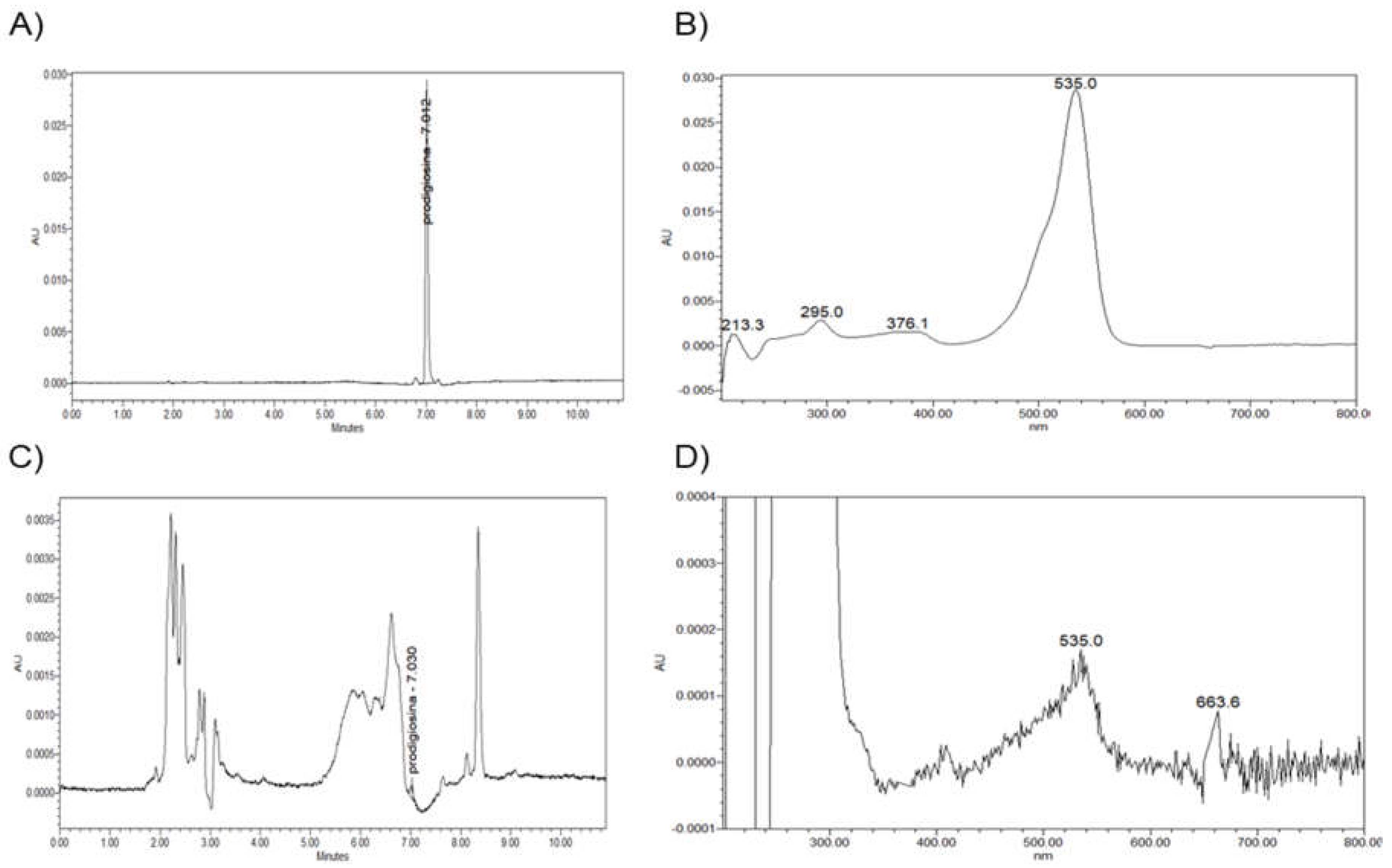
2.4. HPLC Analysis of the Red Pigment
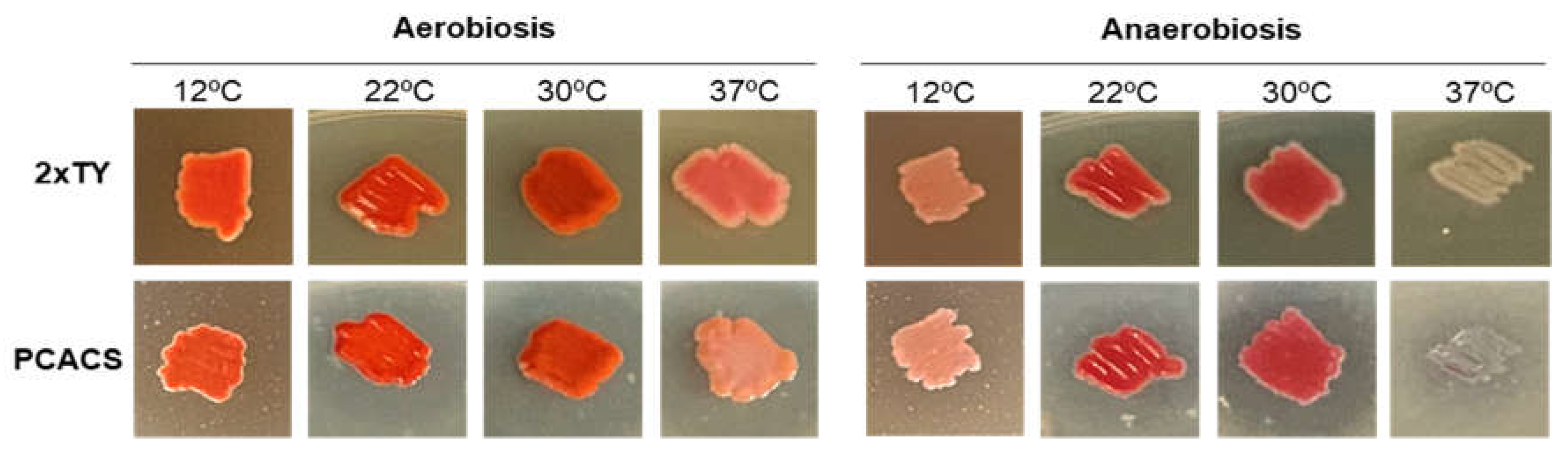
2.5. Optimal Conditions for Prodigiosin Production
2.6. Antimicrobial Activity of Prodigiosin
2.7. Susceptibility to Cheesemaking Conditions
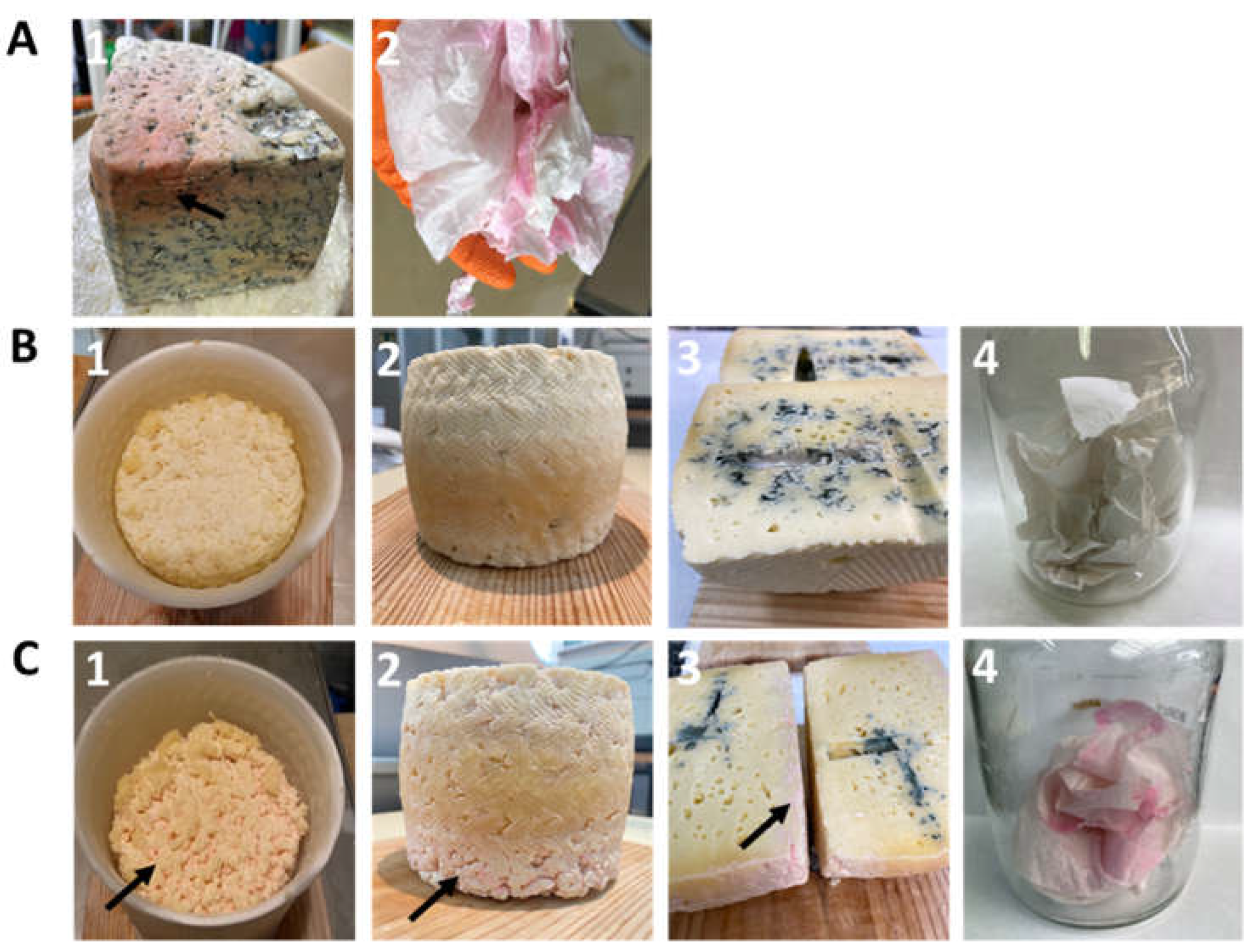
2.8. Experimental Cheese Manufacture and Analysis
3. Results
3.1. Bacterial Association with the Red Colour Defect in Cheese
3.2. Prodigiosin Production by S. marcescens RO1
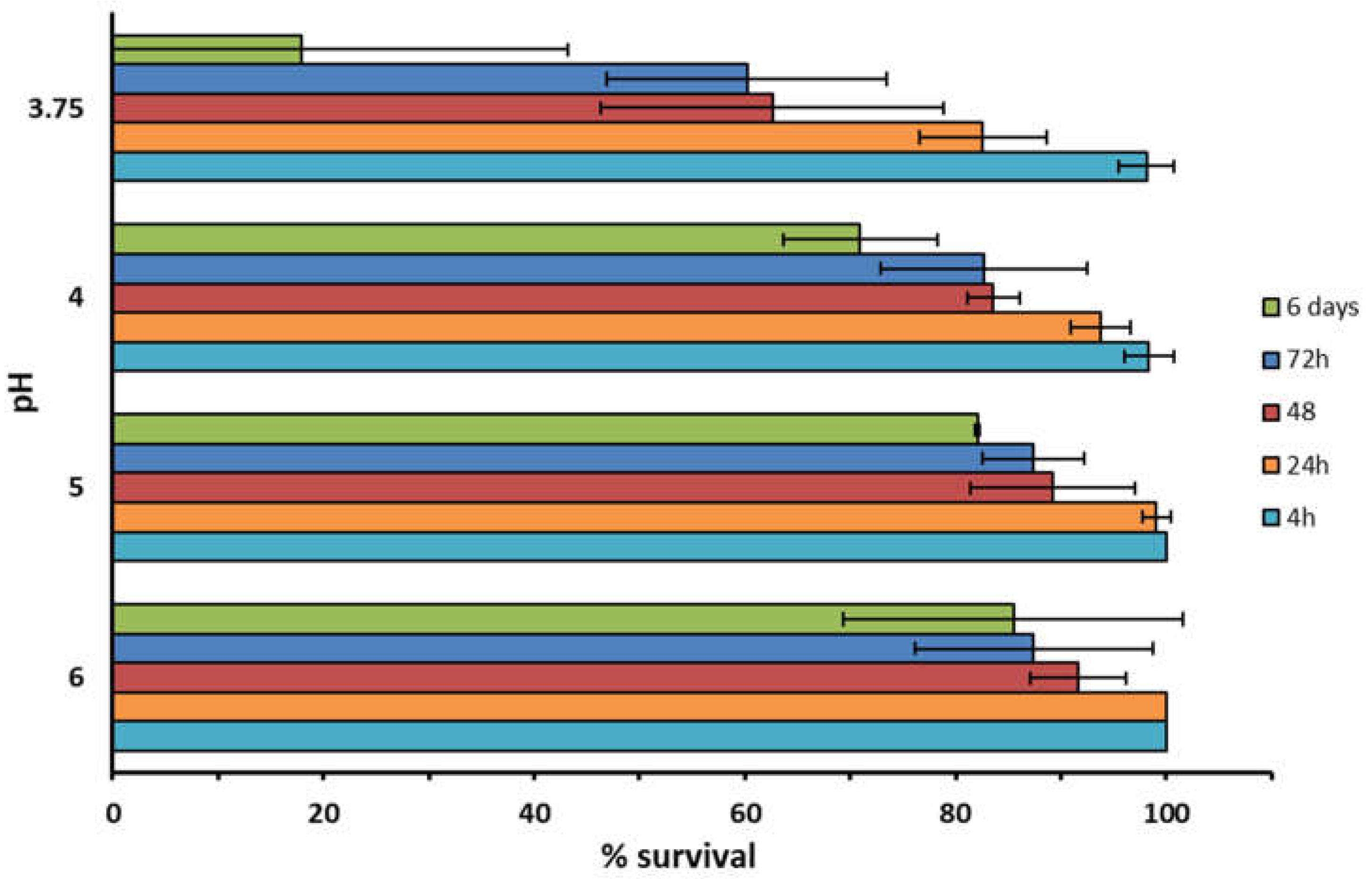
3.3. Impact of pH and NaCl on RO1 Survival
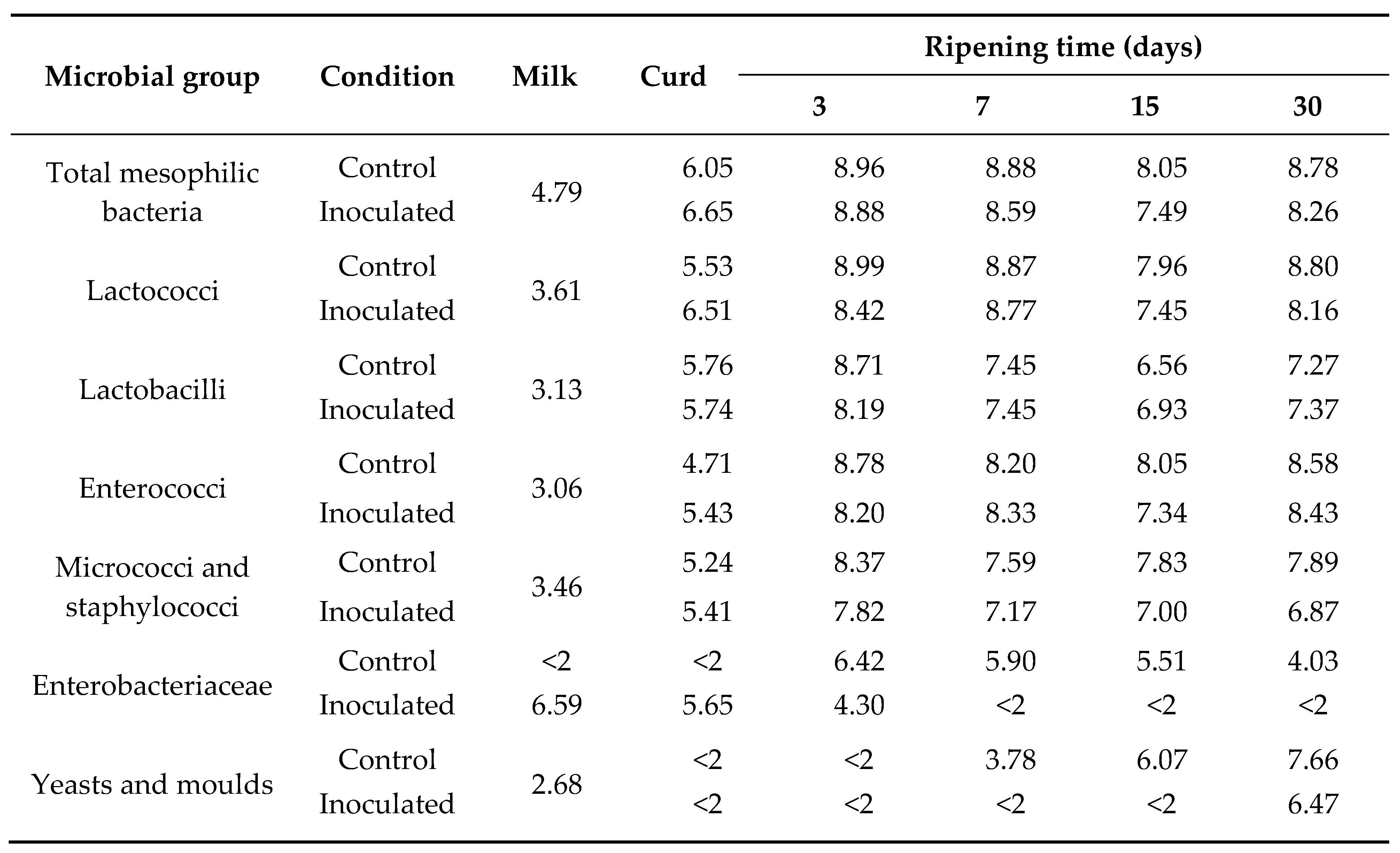
3.4. Recreation of the Red Colour Defect in Experimental Cheeses
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Tilocca, B.; Costanzo, N.; Morittu, V.M.; Spina, A.A.; Soggiu, A.; Britti, D.; Roncada, P.; Piras, C. Milk microbiota: Characterization methods and role in cheese production. J. Proteomics 2020, 210, 103534. [Google Scholar] [CrossRef]
- Smid, E.J.; Kleerebezem, M. Production of aroma compounds in lactic fermentations. Annu. Rev. Food Sci. Technol. 2014, 5, 313–326. [Google Scholar] [CrossRef] [PubMed]
- Ropars, J.; Didiot, E.; Rodríguez de la Vega, R.C.; Bennetot, B.; Coton, M.; Poirier, E.; Coton, E.; Snirc, A.; Le Prieur, S.; Giraud, T. Domestication of the emblematic white cheese-making fungus Penicillium camemberti and its diversification into two varieties. Curr. Biol. 2020, 30, 4441–4453e4. [Google Scholar] [CrossRef] [PubMed]
- Caron, T.; Piver, M.L.; Péron, AC.; Lieben, P.; Lavigne, R.; Brunel, S.; Roueyre, D.; Place, M.; Bonnarme, P.; Giraud, T.; Branca, A.; Landaud, S.; Chassard, C. Strong effect of Penicillium roqueforti populations on volatile and metabolic compounds responsible for aromas, flavor and texture in blue cheeses. Int. J. Food Microbiol. 2021, 354, 109174. [Google Scholar] [CrossRef] [PubMed]
- Giuffrida, D.; Monnet, C.; Laurent, F.; Cacciola, F.; Oteri, M.; Le Piver, M.; Caro, Y.; Donato, P.; Mondello, L.; Roueyre, D.; Dufossé, L. Carotenoids from the ripening bacterium Brevibacterium linens impart color to the rind of the French cheese, Fourme de Montbrison (PDO). Nat. Prod Res. 2020, 34, 10–15. [Google Scholar] [CrossRef]
- Kamelamela, N.; Zalesne, M.; Morimoto, J.; Robbat, A.; Wolfe, BE. Indigo- and indirubin-producing strains of Proteus and Psychrobacter are associated with purple rind defect in a surface-ripened cheese. Food Microbiol. 2018, 76, 543–552. [Google Scholar] [CrossRef]
- Guzzon, R.; Carafa, I.; Tuohy, K.; Cervantes, G.; Vernetti, L.; Barmaz, A.; Larcher, R.; Franciosi, E. Exploring the microbiota of the red-brown defect in smear-ripened cheese by 454-pyrosequencing and its prevention using different cleaning systems. Food Microbiol. 2017, 62, 160–168. [Google Scholar] [CrossRef]
- Quigley, L.; O'Sullivan, D.J.; Daly, D.; O'Sullivan, O.; Burdikova, Z.; Vana, R.; Beresford, T.P.; Ross, R.P.; Fitzgerald, G.F.; McSweeney, P.L.; Giblin, L.; Sheehan, J.J.; Cotter, P.D. Thermus and the pink discoloration defect in cheese. mSystems 2016, 1, e00023–16. [Google Scholar] [CrossRef]
- Cleary, J.L.; Kolachina, S.; Wolfe, B.E.; Sanchez, L.M. Coproporphyrin iii produced by the bacterium Glutamicibacter arilaitensis binds zinc and is upregulated by fungi in cheese rinds. mSystems 2018, 3, e00036–18. [Google Scholar] [CrossRef]
- Flórez, A.B.; Mayo, B. Microbial diversity and succession during the manufacture and ripening of traditional, Spanish, blue-veined Cabrales cheese, as determined by PCR-DGGE. Int. J. Food Microbiol. 2006, 110, 165–171. [Google Scholar] [CrossRef] [PubMed]
- Flórez, A.B.; Ruas-Madiedo, P.; Alonso, L.; Mayo, B. Microbial, chemical and sensorial variables of the Spanish traditional blue-veined Cabrales cheese, as affected by inoculation with commercial Penicillium roqueforti spores. Eur. Food Res. Technol. 2006, 222, 250–257. [Google Scholar] [CrossRef]
- Verraes, C.; Vlaemynck, G.; Van Weyenberg, S.; De Zutter, L.; Daube, G.; Sindic, M.; Uyttendaele, M.; Herman, L. A review of the microbiological hazards of dairy products made from raw milk. Int. Dairy J. 2015, 50, 32–44. [Google Scholar] [CrossRef]
- Rodríguez, J.; González-Guerra, A.; Vázquez, L.; Fernández-López, R.; Flórez, A. B.; de la Cruz, F.; Mayo, B. solation and phenotypic and genomic characterization of Tetragenococcus spp. from two Spanish traditional blue-veined cheeses made of raw milk. Int. J. Food Microbiol. 2022, 371, 109670. [Google Scholar] [CrossRef] [PubMed]
- Harris, A.K.P.; Williamson, N.R.; Slater, H.; Cox, A.; Abbasi, S.; Foulds, I.; Simonsen, H.T.; Leeper, F. J.; Salmond, G.P.C. The Serratia gene cluster encoding biosynthesis of the red antibiotic, prodigiosin, shows species- and strain-dependent genome context variation. Microbiology (Reading, England) 2004, 150 Pt 11, 3547–3560. [Google Scholar] [CrossRef] [PubMed]
- Yip, C.H.; Mahalingam, S.; Wan, K.L.; Nathan, S. Prodigiosin inhibits bacterial growth and virulence factors as a potential physiological response to interspecies competition. PLoS ONE 2021, 16. [Google Scholar] [CrossRef]
- Lin, C.; Jia, X.; Fang, Y.; Chen, L.; Zhang, H.; Lin, R.; Chen, J. Enhanced production of prodigiosin by Serratia marcescens FZSF02 in the form of pigment pellets. Electron J. Biotechnol. 2019, 40, 58–64. [Google Scholar] [CrossRef]
- Song, M.J.; Bae, J.; Lee, D.S.; Kim, C.H.; Kim, J.S.; Kim, S.W.; Hong, S.I. Purification and characterization of prodigiosin produced by integrated bioreactor from Serratia sp. KH-95. J. Biosci. Bioeng. 2006, 101, 157–161. [Google Scholar] [CrossRef]
- EFSA and ECDC (European Food Safety Authority and European Centre for Disease Prevention and Control.) The European Union One Health 2018 Zoonoses Report. EFSA Journal 2019, 17.
- Daly, D.F.M.; McSweeney, P.L.H.; Sheehan, J.J. Pink discolouration defect in commercial cheese: a review. Dairy Sci. Technol. 2012, 92, 439–453. [Google Scholar] [CrossRef]
- Sutthiwong, N.; Sukdee, P.; Lekhavat, S.; Dufossé, L. Identification of red pigments produced by cheese-ripening bacterial strains of Glutamicibacter arilaitensis using HPLC. Dairy 2021, 2, 396–409. [Google Scholar] [CrossRef]
- Flórez, A.B.; Álvarez-Martín, P.; López-Díaz, T.M.; Mayo, B. Microbiological characterisation of the traditional Spanish blue-veined Cabrales cheese: identification of dominant lactic acid bacteria. Eur. Food Res. Technol. 2006, 223, 503–508. [Google Scholar] [CrossRef]
- Tunca Koyun, M.; Sirin, S.; Aslim, B.; Taner, G.; Nigdelioglu Dolanbay, S. Characterization of prodigiosin pigment by Serratia marcescens and the evaluation of its bioactivities. Toxicol. In Vitro 2022, 82, 105368. [Google Scholar] [CrossRef]
- Adeolu, M.; Alnajar, S.; Naushad, S.; Gupta, R.S. Genome-based phylogeny and taxonomy of the ‘Enterobacteriales’: proposal for Enterobacterales ord. nov. divided into the families Enterobacteriaceae, Erwiniaceae fam. nov., Pectobacteriaceae fam. nov., Yersiniaceae fam. nov., Hafniaceae fam. nov., Morganellaceae fam. nov., and Budviciaceae fam. nov. Int. J. Syst. Evol. Microbiol. 2016, 12, 5575–5599. [Google Scholar] [CrossRef]
- Kamarudin, M.I.; Fox, L.K.; Gaskins, C.T.; Gay, J.M. Environmental reservoirs for Serratia marcescens intramammary infections in dairy cows. J. Am. Vet. Med. Assoc. 1996, 208, 555–558. [Google Scholar] [CrossRef]
- Friman, M.J.; Eklund, M.H.; Pitkälä, A.H.; Rajala-Schultz, P.J.; Rantala, M. Description of two Serratia marcescens associated mastitis outbreaks in Finnish dairy farms and a review of literature. Acta Vet. Scand. 2019, 61. [Google Scholar] [CrossRef]
- Quinn, L.; Ailsworth, M.; Matthews, E.; Kellams, A.; Shirley, D.A. Serratia marcescens colonization causing pink breast milk and pink diapers: a case report and literature review. Breastfeed Med. 2018, 13, 388–394. [Google Scholar] [CrossRef] [PubMed]
- Sudhakar, C.; Shobana, C.; Selvankumar, T.; Selvam, K. Prodigiosin production from Serratia marcescens strain CSK and their antioxidant, antibacterial, cytotoxic effect and in-silico study of caspase-3 apoptotic protein. Biotechnol. Appl. Biochem. 2021, 69, 1984–1997. [Google Scholar] [CrossRef] [PubMed]
- Woodhams, D.C.; LaBumbard, B.C.; Barnhart, K.L.; Becker, M.H.; Bletz, M.C.; Escobar, L.A.; Flechas, S.V.; Forman, M.E.; Iannetta, A.A.; Joyce, M.D.; Rabemananjara, F.; Gratwicke, B.; Vences, M.; Minbiole, K.P.C. Prodigiosin, violacein, and volatile organic compounds produced by widespread cutaneous bacteria of amphibians can inhibit two Batrachochytrium fungal pathogens. Microb. Ecol. 2018, 75, 1049–1062. [Google Scholar] [CrossRef]
- Araújo, R.G.; Zavala, N.R.; Castillo-Zacarías, C.; Barocio, M. E.; Hidalgo-Vázquez, E.; Parra-Arroyo, L.; Rodríguez-Hernández, J.A.; Martínez-Prado, M.A.; Sosa-Hernández, J.E.; Martínez-Ruiz, M.; Chen, W. N.; Barceló, D.; Iqbal, H.M.N.; Parra-Saldívar, R. Recent advances in prodigiosin as a bioactive compound in nanocomposite applications. Molecules (Basel, Switzerland) 2022, 27. [Google Scholar] [CrossRef]
- Li, P.; He, S.; Zhang, X.; Gao, Q.; Liu, Y.; Liu, L. Structures, biosynthesis, and bioactivities of prodiginine natural products. Appl. Microbiol. Biotechnol. 2022, 106, 7721–7735. [Google Scholar] [CrossRef]
- Baráti-Deák, B.; Da Costa Arruda, G.C.; Perjéssy, J.; Klupács, A.; Zalán, Z.; Mohácsi-Farkas, C.; Belák, Á. Inhibition of foodborne pathogenic bacteria by excreted metabolites of Serratia marcescens strains isolated from a dairy-producing environment. Microorganisms 2023, 11, 403. [Google Scholar] [CrossRef] [PubMed]
- Denkovskienė, E.; Paškevičius, Š.; Misiūnas, A.; Stočkūnaitė, B.; Starkevič, U.; Vitkauskienė, A.; Hahn-Löbmann, S.; Schulz, S.; Giritch, A.; Gleba, Y.; Ražanskienė, A. Broad and efficient control of Klebsiella pathogens by peptidoglycan-degrading and pore-forming bacteriocins klebicins. Sci. Rep. 2019, 9. [Google Scholar] [CrossRef]
- Jansen, R.; Sood, S.; Huch, V.; Kunze, B.; Stadler, M.; Müller, R. Pyrronazols, metabolites from the myxobacteria Nannocystis pusilla and N. exedens, are unusual chlorinated pyrone-oxazole-pyrroles. J. Nat. Prod. 2014, 77, 320–326. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, S.A.; Ayivi, R.D.; Zimmerman, T.; Siddiqui, S.A.; Altemimi, A.B.; Fidan, H.; Esatbeyoglu, T.; Bakhshayesh, R.V. Lactic acid bacteria as antimicrobial agents: food safety and microbial food spoilage prevention. Foods (Basel, Switzerland) 2021, 10. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements.; opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas.; methods.; instructions or products referred to in the content. |
 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





