1. Introduction
Genus
Tulipa L. includes a large number of species, both diverse in their morphological characteristics, and morphologically similar and difficult to identify. The number of species in the genus differs according to different researchers, currently there are from 76 species [
1] to 87 species [
2]. In some cases, it is difficult to accurately identify closely related species of the
Tulipa L. In this regard, difficulties arise in determining the types of tulip entering the botanical collections.
In order to clarify the species, as well as to study the polymorphism of plants within a taxon, in addition to analyzing morphological features, it may be useful to use molecular markers, which are currently widely used in taxonomic studies and for studying biodiversity. Previously, the study of DNA polymorphism in varieties and species, as well as tulip populations, was carried out using AFLP markers [
3,
4,
5]. Later, microsatellite sequences (SSR), such as EST-SSR and NBS-LRR markers [
6,
7], as well as intermicrosatellite sequences (Inter Simple Sequence Repeat—ISSR), were used to study genetic diversity, population structure and phylogenetic studies in the genus Tulipa, and the latter were used to study intra- and interspecific polymorphism [
8,
9,
10,
11].
T. tarda is widespread in horticulture and is described in detail in a number of sources, the species grows naturally in Central Asia [
12,
13,
14,
15,
16], etc. In the description of
T. tarda, researchers mention that the perianth can be not only white-yellow with white tips of the perianth leaves, but also completely yellow, which is much rarer [
13,
14]. The following description is given for
T. urumiensis in the literature: a species from the subgenus
Eriostemones, close to
T. australis, grows in Northwestern Iran, is difficult to cultivate. It differs in dwarf sizes: the height of the plant is 5–8 cm, and most of the stem is underground, the height of the flower is 3–4 cm, the length of the lower leaf is up to 12 cm, the leaves (2–3) are close directed upwards, the flower is elongated-bell-shaped, yellow [
13].
Tulipa urumiensis also describe in the International Register of Names of Tulips of the Royal Association of Bulb Producers KAVB [
15]: origin—northwestern Iran; Lake Rezai (Urmia), perianth is cleaer yellow, outer segments are pigmented on reverse with olive and red, anthers are yellow, multi-flowered, height 10 cm. Zonneveld [
2] combined both species under the name T. tarda, while Christenhusz et al. [
1], on the contrary, suggested using the name T. urumiensis for the combined species.
The purpose of this work was to study the complex of morphological parameters, as well as DNA polymorphism in the composition of ISSR fragments and to clarify the species belonging of tulip plants from two subgenera Tulipa L. и Eriostemones (Boiss.) Hall.
2. Materials and Methods
The following species from the subgenus were included in this study:
Tulipa vvedenskyi Z. Botsch., Tulipa kaufmanniana Regel, Tulipa bifloriformis Vved., Tulipa turkestanica Regel., Tulipa biflora Pall., Tulipa tarda Stapf., Tulipa urumiensis Stapf.
Table 1 provides a description of the samples taken for the study and the source of the origin of the material. To study the morphological features and composition of ISSR fragments, plants from the collection of tulips of the Department of Ornamental Plants of the Main Botanical Garden Russian Academy of Sciences (GBS RAS), Moscow, Russia were taken. The study included both plants grown from seeds and vegetative clones. The plants for the study were taken in the flowering phase. We studied 2–3 plants for vegetative clones and 5–8 plants for samples grown from seeds. Species identification was carried out using the key [
12,
13] and morphological descriptions [
16,
17,
18], and plants taken for study were compared with herbarium samples stored in the Herbarium of the Main Botanical Garden RAS (MHA).
The study of morphological features. Morphological signs were studied from herbarium samples collected at the experimental site of the Laboratory of Plant Physiology and Immunity and the Laboratory of Ornamental Plants of the GBS RAS. The following signs were studied: plant height; length, color and pubescence of the stem, length; width, color, fringing and pubescence of the leaf; number of flowers; length, width, color of the outer and inner tepals; color of anthers and staminate filaments, pubescence at the base of the staminate filaments. The selected features are used to describe a species of Tulipa, as well as when compiling keys for determining the types of tulip.
Study of the composition of molecular markers. DNA was isolated from fresh leaves using CTAB method [
19]. The conditions of PCR were described in detail earlier [
20]. For ISSR-PCR, primers synthesized and purified in PAAG by Syntol Ltd. (Moscow, Russia) were used. 10 primers were selected for PCR, their description is given in
Table 2. Each amplicon, which was visualized as a strip in an electrophoretic agarose gel, was considered as a counting feature and taken into account as a binary code (1/0). Obscure bands were considered missing.
Statistical processing of the results. Based on the results obtained, two matrices were compiled: one for morphological features, the second for molecular genetic data. Next, cluster analysis and data processing was carried out by the method of main coordinates in the PAST program [
21]. To assess the stability of the resulting dendrogram, a bootstrap analysis was performed with 1000 replicas. For morphological data, the matrix included quantitative and qualitative features, cluster analysis was performed using the Gover distance. The presence/absence matrix of ISSR fragments was analyzed by cluster analysis using the Jacquard similarity measure.
3. Results and Discussion
3.1. Analysis of Morphological Features
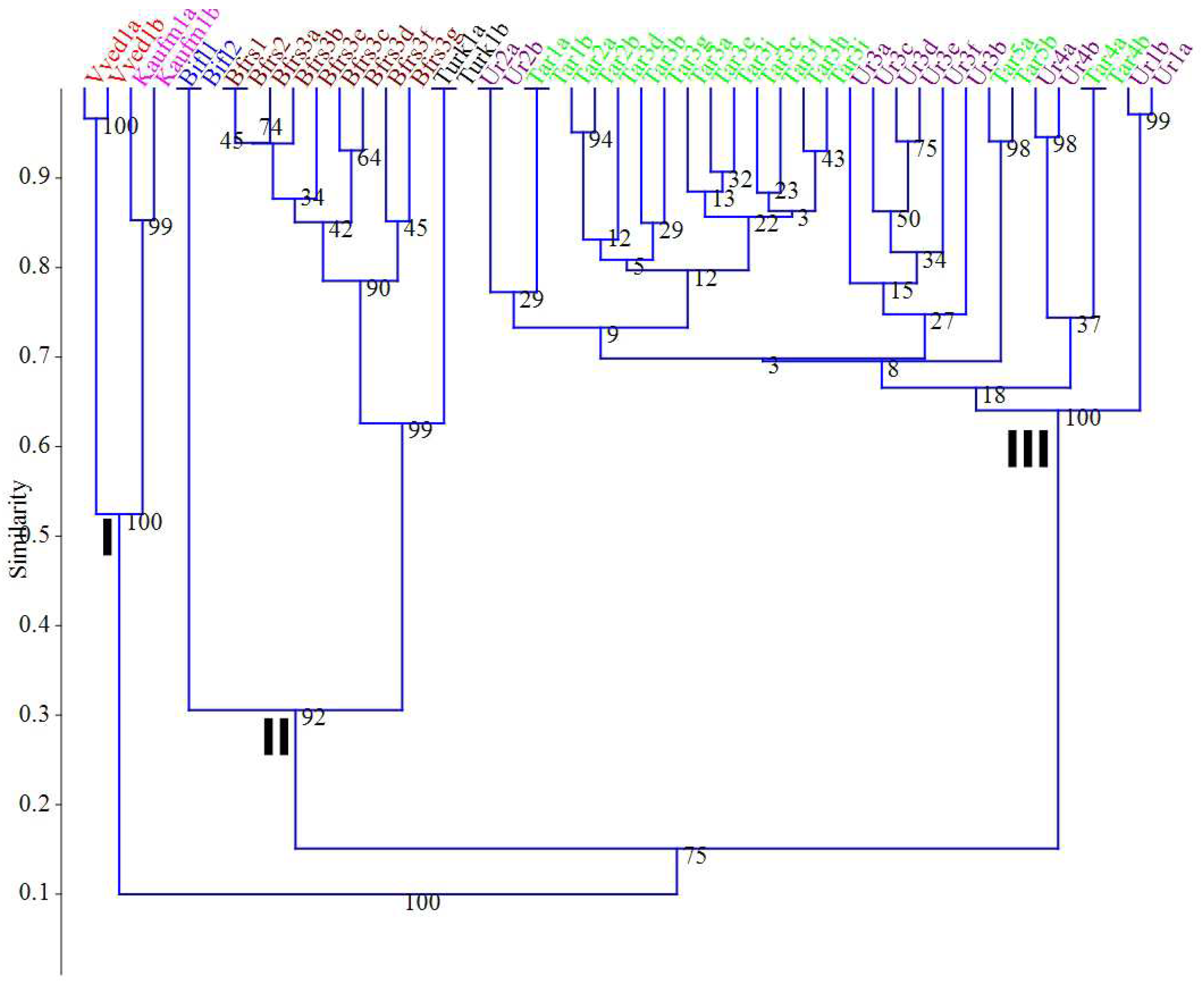
The study of morphological features showed that two subgenera
Tulipa L. and Eriostemones Boiss. They are clearly separated from each other, but the results obtained do not always allow us to distinguish species according to their systematic position. So, as a result of cluster analysis, the samples were distributed as follows (
Figure 1).
Plants from the subgenus Tulipa formed a separate cluster I, and the neighboring cluster II—plants from the subgenus Eriostemones. Cluster III also consists of representatives of Eriostemones, but samples were grouped rather randomly and not according to their species and systematic position. The bootstrap support for large clusters is low. At the same time, the samples of Tulipa bifloriformis and T. turkestanica formed a single subcluster in subcluster III with a high similarity measure and a bootstrap support coefficient of 93%. This may indicate that these samples belong to the same species. The analysis of morphological features by the method of principal coordinates (PCoA) showed similar results (
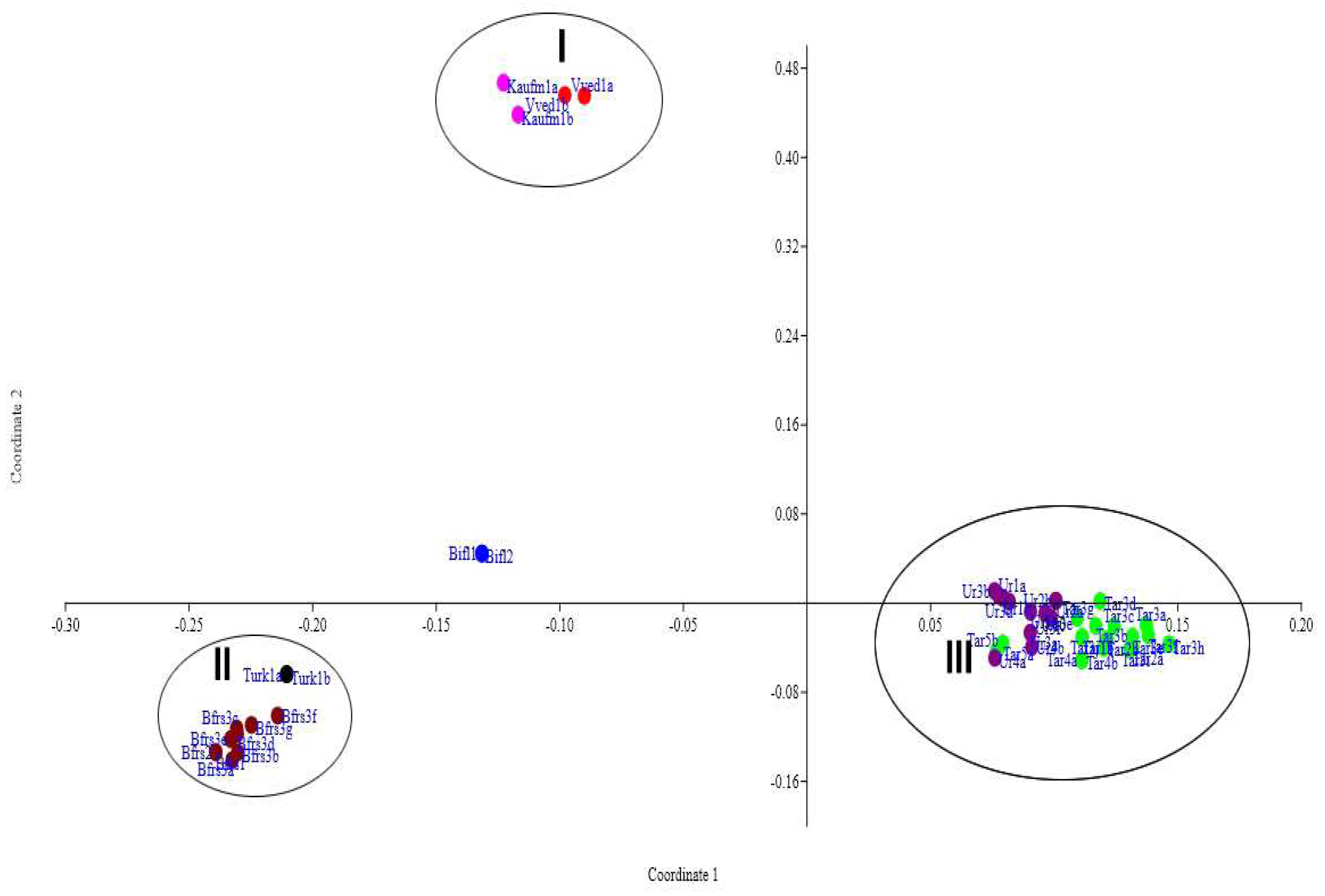
Figure 2).
The method of main coordinates also failed to separate the studied samples according to species, but all plants were divided into two clouds corresponding to the subgenera Tulipa and Eriostemones. It is not possible to distinguish species within a subgenus by the complex of selected morphological features of PCoA. Thus, the results of the analysis of only morphological features using cluster analysis and the method of principal coordinates not being informative in this case.
Khaleghi et al. analyzing the complex of morphological features by the method of principal components also obtained the separation of samples according to the subgenera to which they belonged [
22]. In turn, Dekhkanov. et al. carried out cladistic analysis of a complex of morphological features of 48 tulip species from Central Asia and obtained a tree [
23], which (despite low bootstrap support) agrees mainly with the molecular data of Zonneveld [
2] and Christenhusz et al. [
1]. In this study, the species were mostly divided not only into subgenera, but also into sections [
23].
3.2. Analysis of Molecular Data
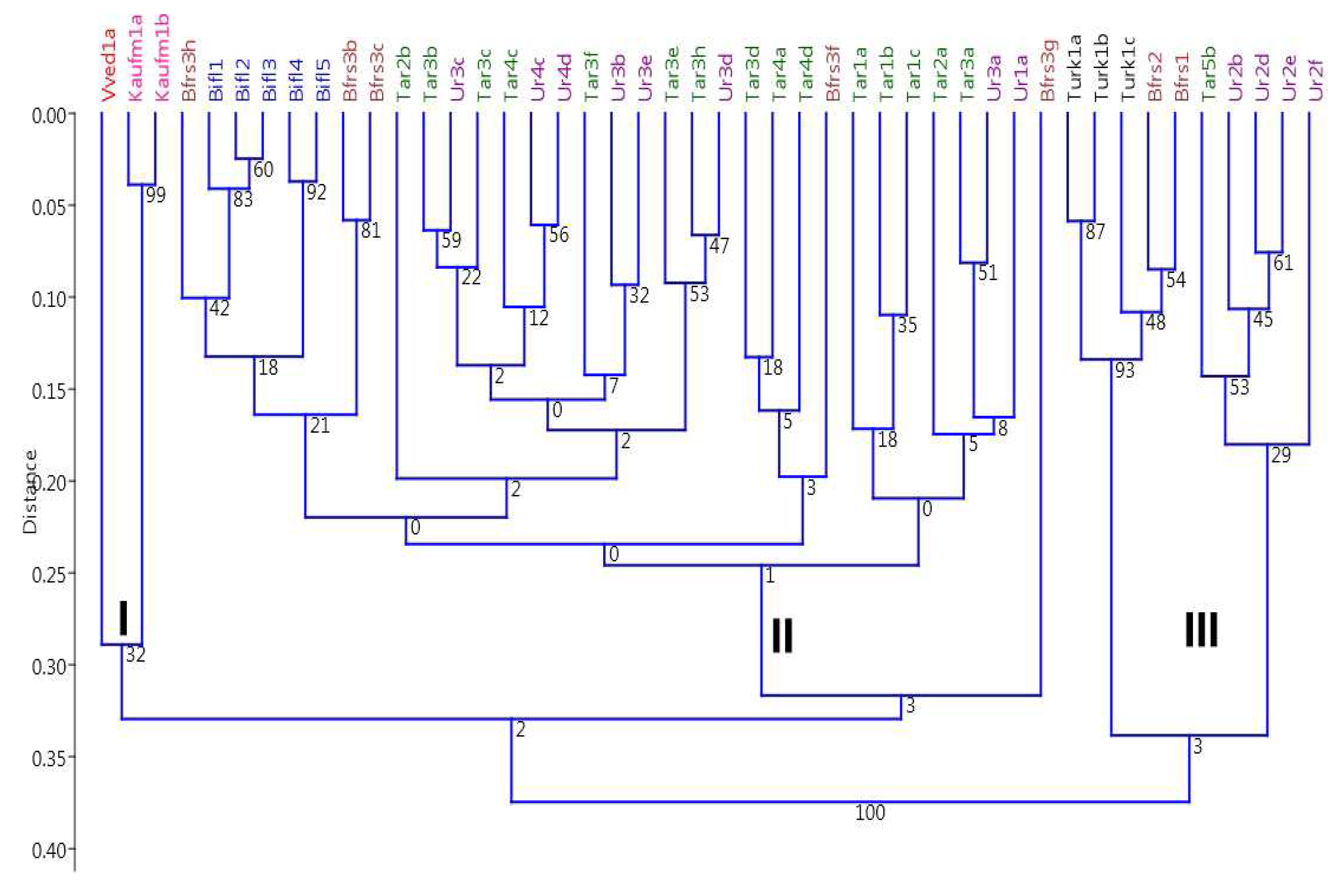
As a result of PCR, we obtained 102 polymorphic fragments. The data of the cluster analysis of the obtained matrix are presented in
Figure 3. In this case, a clearer distribution of samples was observed according to their taxonomic position. The specimens were also divided into two main groups according to two subgenera:
Tulipa and
Eriostemones. Plants grown from seeds were distinguished by significant genetic diversity, while those propagated vegetatively represented one clone and had the same composition of ISSR fragments. Two species from the subgenus
Tulipa,
T. vvedenskyi and
T. kaufmanniana formed one cluster with a high similarity measure and 99% bootstrap support. Within the second group, corresponding to the subgenus
Eriostemones, the samples were grouped into cluster II and III.
Within cluster II, T. biflora plants formed a subcluster sister to the subcluster uniting T. turkestanica Regel with T. bifloriformis Vved. Since, according to the results of ISSR-PCR analysis, the above species are combined into one cluster (II) with a high degree of bootstrap support (92), this may indicate their closer relationship with each other than with T. tarda (cluster III) The T. tarda and the T. urumiensis also formed a common subcluster, isolated from the other Eriostemones species. In this group, the samples of T. tarda and T. urumiensis from different sources were practically inseparable and had a high degree of similarity. Thus, a significant similarity in the composition of ISSR fragments indicates that the plants received as T. urumiensis are actually the yellow-flowered form of T. tarda. Based on the above, it can be concluded that the studied types of tulips have a polymorphism of ISSR markers, which allows for a more thorough identification of varieties.
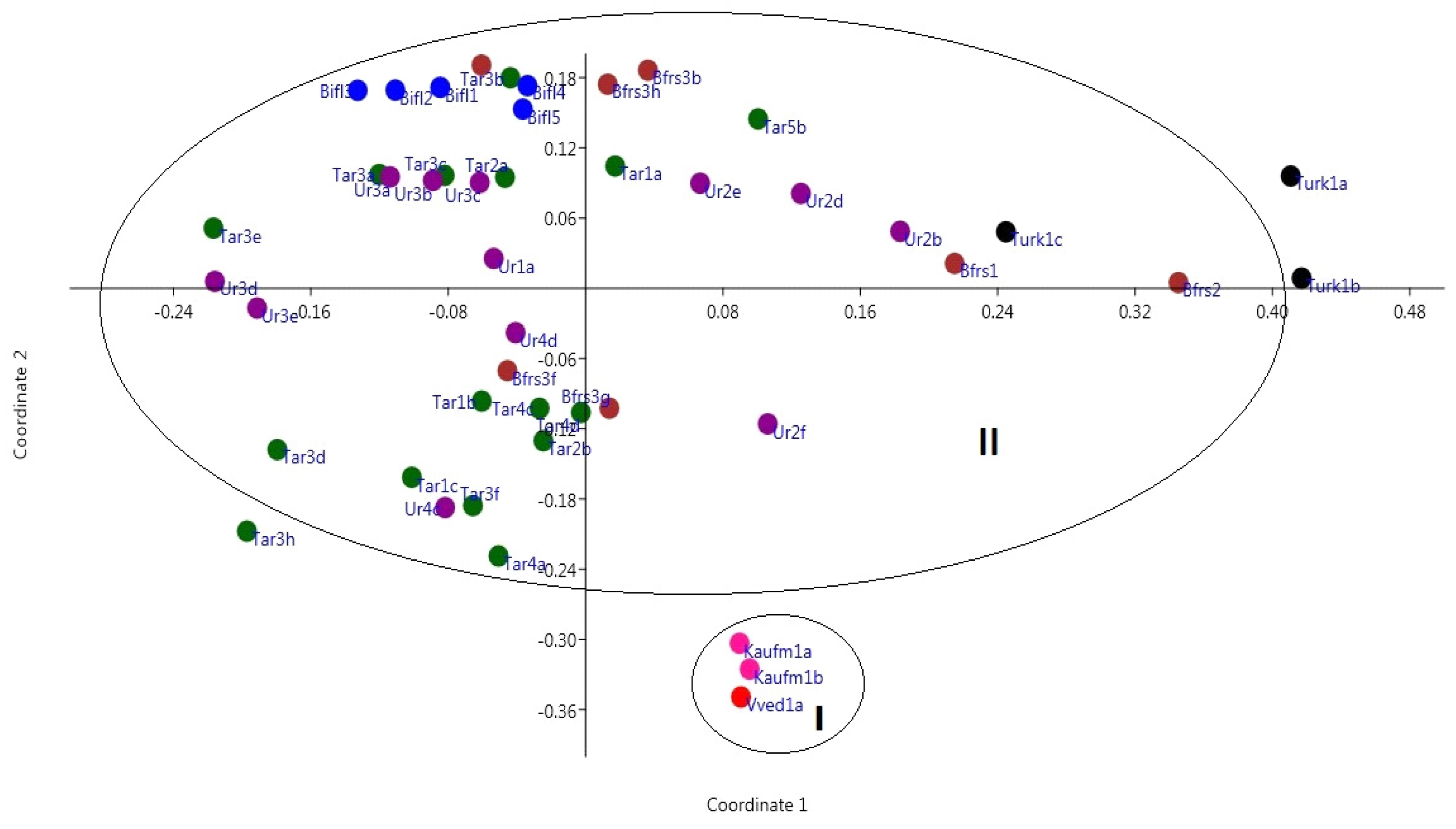
As a result of statistical processing of data from fragment analysis by the method of principal coordinates, a more significant picture was obtained than when analyzing morphological data by this method. In
Figure 4, you can see three well-separated clouds, in addition,
T. biflora samples occupied a separate position. The first cloud was formed by
T. vvedenskyi and
T. kaufmanniana, the second by
Tulipa bifloriformis and
T. turkestanica, the third by
T. tarda and
T. urumiensis.
The samples of
T. tarda and
T. urumiensis formed a single group both in the analysis of morphological features and molecular data, and also when we determined by the key, all were assigned to
T. tarda. In our experience, fully formed flowering plants of
T. urumiensis have an average plant height of 20–21 cm (sometimes up to 31 cm, in
T. tarda it is 18–20 cm), leaves are usually 5 (in
T. tarda—5–6), the length of the lower leaf is 19–21 cm (in
T. tarda—21–28 cm). Thus,
T. urumiensis plants are significantly larger than the plants described in the literature [
13,
15] and morphologically similar to
T. tarda.
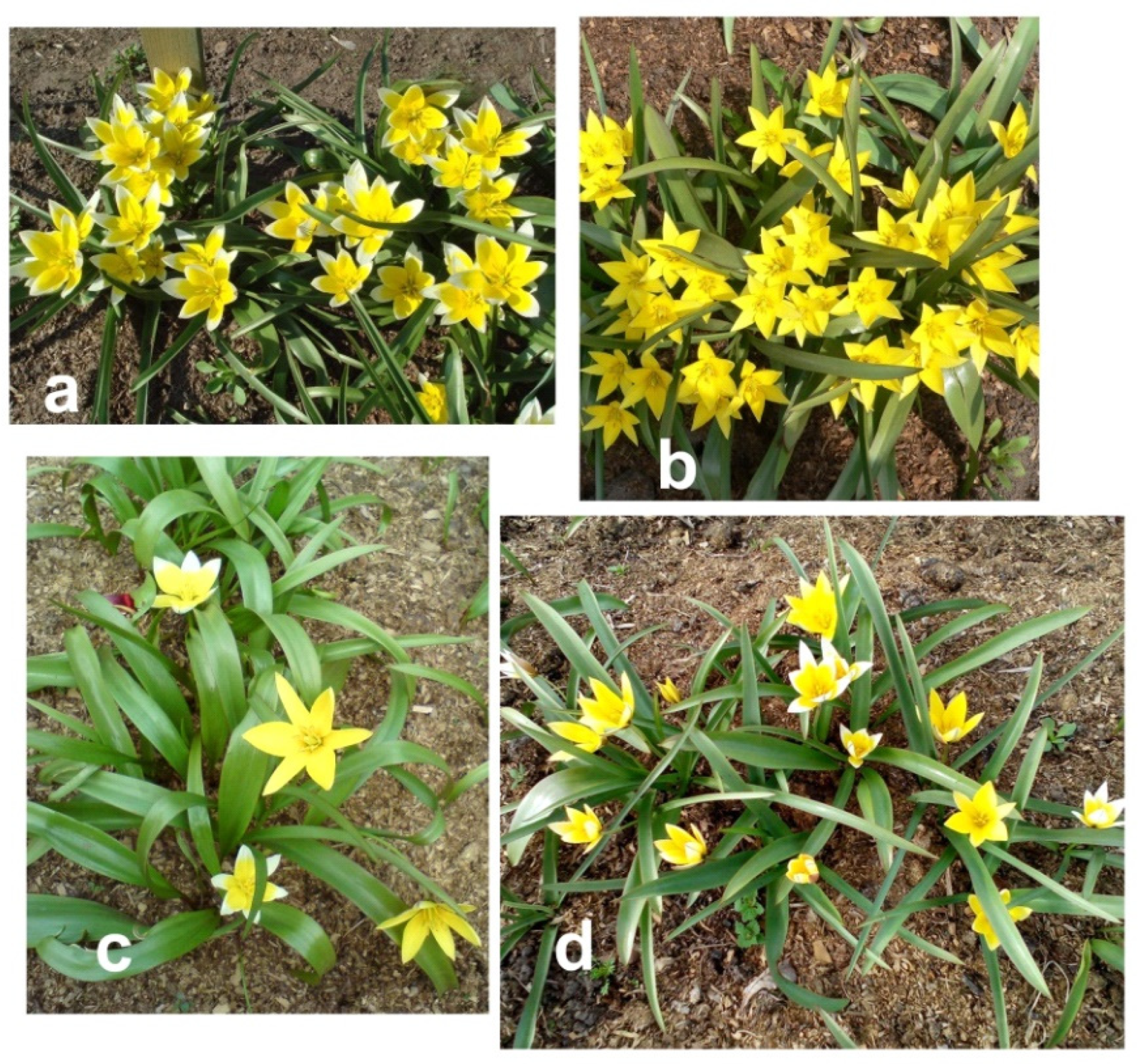
Summing up the data obtained, it can be concluded that the samples obtained from different sources as the
Tulipa urumiensis Stapf are in fact
T. tarda Stapf., i.e., its yellow-flowered form. This is evidenced also by the data of the analysis of ISSR fragments and morphological features of plants Ur 1–3 and Tar 1–3, which were grown from seeds obtained from free pollination of yellow-flowered plants
T. urumiensis and white-flowered plants
T. tarda, respectively. Among the plants from both clones, there were both plants with yellow and white flowers, otherwise having signs of
T. tarda (
Figure 5). Recently, Christenhusz and Wilson proposed to classify cultivated
Tulipa urumiensis plants as
T. tarda [
24], which in our opinion is more correct, since the morphological features of all
T. urumiensis specimens correspond to
T. tarda and are supported by the results of molecular genetic studies. The species
Tulipa urumiensis Stapf was described from a plant grown in the Van Tubergen nursery from a bulb, which is known to have been brought from Urmia (Iran) and this species has never been rediscovered in Iran, it may have died out there or the bulb was mistakenly labeled when shipped to the Netherlands [
24].
As a result of our research, we found that the plants received as the
T. turkestanika Regel and
T. bifloriformis Vved. with a high degree of probability, they belong to the same species—
T. bifloriformis Vved and are two clones of this species, because they are similar in composition of ISSR-markers and morphologically. According to Zonneveld [
2],
T. bifloriformis is synonymous with
T. turkestanika, but in all likelihood, this conclusion applies only to those cultivated clones that come from European sources. To determine the boundaries of these two species, an additional study of the complex of morphological features and polymorphism of plant DNA from natural habitats is necessary.
Thus, a comprehensive analysis of morphological features and ISSR markers in the presence of a sufficient number of plants makes it possible to more fully analyze samples and clarify the species of tulip plants.
Author Contributions
methodology, analysis of results, writing of the article—Semenova M.V.; data collection—Olekhnovich L.S.; data processing and visualization—Enina O.L. All authors have read the published version of the manuscript and agreed with it.
Funding
This research was funded by Institutional research project № 122042700002-6 of the Tsitsin Main Botanical Garden of the RAS.
Data Availability Statement
Not applicable.
Acknowledgments
The authors express their sincere gratitude N.N. Danilina for the plant material and the staff and I.A. Shantzer, head of the Main Botanical Garden Laboratory of Molecular Genetic Systematics for the opportunity to conduct molecular genetic research. We thank the Ministry of Science and Higher Education of the Russian Federation for the support of the Center of Collective Use “Herbarium MBG RAS”, grant 075-15-2021-678.
Conflicts of Interest
The authors declare that there is no conflict of interest.
References
- Christenhusz M.J.M, Govaerts R., Hall T., Borland K., Stranc P., Tuomisto A., ChaseM.W., Fay M.F. Tiptoe through the tulips - cultural history, molecular phylogenetics and classification of Tulipa (Liliaceae). Botanical Journal of the Linnean Society. 2013, 172 (3), 80-328. [CrossRef]
- Zonneveld B. J. M.The systematic value of nuclear genome size for ‘‘all’’ species of Tulipa L. (Liliaceae). Plant SystEvol. 2009, 281, 217-245.
- Bondrea I., Pamfil D., van Heusden S., van Tuyl J., Meijer-Dekens F., Bondrea M., Sestras R., Rusu A.R, Lucaci M., Patrascu B. I., Balteanu V.A.. AFLP as amodern technique for DNA fingerprinting and identification Tulipa cultivars. Bull. USAMV-CN. 2007, 63–64.
- Kutlunina N. A., Polezhaeva M. A., Permyakova M. V. Morphological and genetic (AFLP) analyzes of Tulipa biebersteiniana (Liliaceae) species. Genetics, 49(4), 2013, 461–471. [CrossRef]
- Morgil H., Levent Şık L., Erol O. 2017. Genetic diversity by AFLP analysis within Tulipa orphanidea L. (Liliaceae) populations in Manisa. Celal Bayar University J. of Sci., 2017, 13 (4), 913-917.
- Pourkhaloee A., Khosh-Khui M., Arens P., Salehi H., Razi H., Niazi A., Afsharifar A. van Tuyl J.. Genetic Diversity and Population Structure of Iranian tulips revealed by EST-SSR and NBS-LRR Markers. International Journal of Horticultural Science and Technology. 2017, 4 (2), 167-182. [CrossRef]
- Pourkhaloee A., Khosh-Khui M.,Arens P.,Salehi H.,Razi H., Niazi A., Afsharifar A. van Tuyl J.,. Molecular analysis of genetic diversity, population structure, and phylogeny of wild and cultivated tulips (Tulipa L.) by genic microsatellites. Horticulture, Environment, and Biotech. 2018, 59, 875–888. [CrossRef]
- Kiani M., Memariani F., Zarghami H. Molecular analysis of species of L. from Iran based on ISSR markers. Plant Syst Evol. 2012, 298, 1515–1522.
- Kashin A. S., Kritskaya T. A., Schanzer I. A. Genetic polymorphism of Tulipa gesneriana L. According to ISSR marking data. Genetics. 2016, 52 (10) 1134–1145.
- Kritskaya T.A., Kashin A.S., Schanzer I.A., Danilov V.A. Genetic differentiation of Tulipa suaveolens (Liliaceae) in the north-east of its range in the European part of Russia [in Russian] Botanical journal. 2018, 103(2), 187-200. [CrossRef]
- Kritskaya T.A., Kritsky A.A., Kashin A.S. Estimation of the level of intraspecific polymorphism of Tulip Schrenk based on ISSR markers. Problems of Botany of Southern Siberia and Mongolia. 2014, 13,125−127.
- Vvedenskiy A.I. Tulipa. In Flora of the USSR. [in Russian]. Komarov V.L. Ed. Publishing House Academy of Sciences of the USSR: Leningrad, USSR, 1935; Volume 4, pp. 320–364.
- Dekorativnye travyanistye rasteniya dlya otkrytogo grunta SSSR. Izd. «Nauka»: Leningrad, USSR, 1977; Volume 2, pp. 221-285.
- Baranova M.V. Monocotyledon Perennials in outdoor collection of Botanical Garden of Peter the Gtreat of the Komarov Botanical Institute RAS. Rostok: St. Petersburg, Russian Federation, 2013, 320 pp.
- Classified List and International Register of Tulip Names. editor J.van Scheepen. Published by Royal General Bulbgrowers’ Association KAVB. Hillegom, The Netherlands, 1996, 623. ISBN90-73350-02-6.
- Botschantzeva Z.P. Tulips. Morphology, Cytology and Byiology. (Russian Edn), Publishing House Uzbek SSR Academy of Sciences: Tashkent, USSR, 1962; 408 p.
- Sharipov A., Pratov U. Tyul’pany Srednej Azii (na uzbekskom i russkom yazykah). Uzbekiston millij enciklopediyasi: Tashkent, Uzbekistan,1997; 144 c.
- Ivaschenko A.A. Tulips and other bulbs plants of Kazakhstan. Printing House «Two capitals»: Almaty, Kazakhstan, 2005, 192 p.
- Doyle J.J. & Doyle J.L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15.
- Schanzer I.A., Semenova M.V., Shelepova O.V., Voronkova T.V. Genetic diversity and natural hybridization in populations of clonal plants of Mentha aquatica L. (Lamiaceae), Wulfenia. 2012, 19, 131–139.
- Hammer O., Harper D. A. T., Ryan P. D. PAST: Palaeontological Statistics software package for education and data analysis. Palaeontologia Electronica. 2001, 4(1, 4), S.9. http://palaeo-electronica.org/2001_1/past/issue1_01.htm.
- Khaleghi A., Khadivi A., Zonneveld B. J. M. Morphological variations among and within species of wild tulip (Tulipa L.) from Iran. Genet Resour Crop Evol. 2018, 65, 2241–2266.
- Dekhkonov D., Tojibaev K., Yusupov Z., Makhmudjanov D., Asatulloev T. Morphology of tulips (Tulipa, Liliaceae) in its primary centre of diversity. Plant Diversity of Central Asia, 2022, 1, 52–70. http://doi.org/10.54981/PCDA/vol1_iss1/a1.
- Christenhusz M. and Wilson B. Proposal to reject the name Tulipa urumiensis (Liliaceae). Taxon. 2022. 71(4):906-907. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).