Submitted:
01 June 2023
Posted:
02 June 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Evidence Acquisition
3. Artificial Intelligence Aided Diagnosis of RCC Subtypes
3.1. RCC Diagnosis and Subtyping in Biopsy Specimens
3.2. RCC Diagnosis and Subtyping in Surgical Resection Specimens
4. Pathomics in Disease Prognosis
4.1. Cancer Grading
| Group | Aim | Number of patients |
Accuracy on the test set | External validation (N of patients) |
Accuracy on the external validation cohort | Algorithm |
|---|---|---|---|---|---|---|
| Yeh et al. [80] | RCC grading |
39 ccRCC | AUC: 0.97 | N.A. | N.A. | SVM |
|
Holdbrook et al. [82] |
1) RCC grading, 2) survival prediction |
59 ccRCC |
1) F-score: 0.78 – 0.83 grade prediction 2) High degree of correlation (R = 0.59) with a multigene score |
N.A. |
N.A. |
DNN – features concatenation |
|
Tian et al. [84] |
1) RCC grading, 2) survival prediction |
395 ccRCC |
1) 84.6% sensitivity and 81.3% specificity grade prediction 2) predicted grade associated with overall survival (HR: 2.05; 95% CI 1.21-3.47) |
N.A. |
N.A. |
DNN - LASSO model |
4.2. Molecular-Morphological Connections and AI-Based Therapy Response Prediction
| Group | Aim | Number of patients | Accuracy on the test set | External validation (N of patients) |
Accuracy on the external validation cohort | Algorithm |
|---|---|---|---|---|---|---|
| Marostica et al. [61] | 1) RCC diagnosis 2) RCC subtyping, 3) CNAs identification 4) RCC survival prediction 5) tumor mutation burden prediction |
1) & 2) 537 ccRCC, 288 pRCC, 103 chRCC 3) 528 ccRCC, 288 pRCC, 66 chRCC 4) 269 stage I ccRCC 5) 302 ccRCC |
1)AUC: 0.990 ccRCC, 1.00 pRCC, 0.9998 chRCC 2) AUC: 0.953 3) ccRCC KRAS CNA: AUC=0.724; pRCC somatic mutations: AUC: 0.419 – 0.684; 4) short vs. Long-term survivors log-rank test P = 0.02, n = 269 5) Spearman correlation coefficient: 0.419 |
1)&2) 841 ccRCC, 41 pRCC, 31 chRCC | 1) 0.964 – 0.985 ccRCC; 2) 0.782 – 0.993 |
DCNN |
|
Go et al. [99] |
RCC VEGFR-TKI response classifier; survival prediction |
101 m-ccRCC |
Apparent accuracy of the model: 87.5%; C-index = 0.7001 for PFS; C-index of 0.6552 for OS |
N.A. |
N.A. |
SVM |
| Ing et al. [101] | 1) RCC vascular phenotypes; 2) survival prediction; 3) identification of prognostic gene signature 4) prediction models |
1); 2) & 3)64 ccRCC 4) 301 ccRCC |
1) AUC = 0.79; 2) log-rank p = 0.019, HR = 2.4 3) Wilcoxon rank-sum test p < 0.0511 4) C-Index: Stage = 0.7, Stage + 14VF = 0.74, Stage + 14GT = 0.74 |
N.A. |
N.A. | 1) SVM; Random Forest classifier 2) correlation analysis and information gain 3) two generalized linear models with elastic net regularization |
|
Zheng et al. [107] |
RCC methylation profile |
326 RCC (also tested on glioma) |
average AUC and F1 score higher than 0.6 |
N.A. |
N.A. |
Classic ML, FCNN |
4.3. Prognosis Prediction Models Based on Computational Pathology
| Group | Aim | Number of patients |
Accuracy on the test set | External validation (N of patients) |
Accuracy on the external validation cohort | Algorithm |
|---|---|---|---|---|---|---|
| Ning et al. [122] | RCC prognosis prediction |
209 ccRCC |
Mean C-index = 0.832 (0.761–0.903) |
N.A. | N.A. | CNN; BFPS algorithm for feature selection |
|
Cheng et al. [121] Schulz et al. [123] |
RCC prognosis prediction RCC prognosis prediction |
410 ccRCC 248 ccRCC |
log-rank test P values<0.05 Mean C-index of 0.7791 and a mean accuracy of 83.43%. (prognosis prediction) |
N.A. 18 ccRCC |
N.A. Mean C-index reached 0.799 ± 0.060 with a maximum of 0.8662. Accuracy averaged at 79.17% ± 9.8% with a maximum of 94.44%. |
lmQCM – gene coexpression and analysis; ML – LASSO-Cox model for prognosis prediction CNN consisting of one individual 18-layer residual network (ResNet) per image modality and a dense layer for genomic data |
5. Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Capitanio U, Bensalah K, Bex A, Boorjian SA, Bray F, Coleman J; et al. Epidemiology of Renal Cell Carcinoma. Eur Urol 2019, 75, 74. [Google Scholar] [CrossRef] [PubMed]
- Garfield K, LaGrange CA. Renal Cell Cancer. Renal Cell Cancer. StatPearls 2022.
- Bukavina L, Bensalah K, Bray F, Carlo M, Challacombe B, Karam JA; et al. Epidemiology of Renal Cell Carcinoma: 2022 Update. Eur Urol 2022, 82, 529–542. [Google Scholar] [CrossRef] [PubMed]
- Moch H, Cubilla AL, Humphrey PA, Reuter VE, Ulbright TM. The 2016 WHO Classification of Tumours of the Urinary System and Male Genital Organs-Part A: Renal, Penile, and Testicular Tumours. Eur Urol 2016, 70, 93–105. [CrossRef]
- Cimadamore A, Caliò A, Marandino L, Marletta S, Franzese C, Schips L; et al. Hot topics in renal cancer pathology: Implications for clinical management. Expert Rev Anticancer Ther 2022, 22, 1275–1287. [Google Scholar] [CrossRef]
- Fuhrman SA, Lasky LC, Limas C. Prognostic significance of morphologic parameters in renal cell carcinoma. Am J Surg Pathol 1982, 6, 655–663. [Google Scholar] [CrossRef]
- Zhang L, Zha Z, Qu W, Zhao H, Yuan J, Feng Y; et al. Tumor necrosis as a prognostic variable for the clinical outcome in patients with renal cell carcinoma: A systematic review and meta-analysis. BMC Cancer 2018, 18. [CrossRef]
- Sun M, Shariat SF, Cheng C, Ficarra V, Murai M, Oudard S; et al. Prognostic factors and predictive models in renal cell carcinoma: A contemporary review. Eur Urol 2011, 60, 644–661. [Google Scholar] [CrossRef] [PubMed]
- Hora M, Albiges L, Bedke J, Campi R, Capitanio U, Giles RH; et al. European Association of Urology Guidelines Panel on Renal Cell Carcinoma Update on the New World Health Organization Classification of Kidney Tumours 2022, The Urologist’s Point of View. Eur Urol 2023, 83, 97–100. [Google Scholar] [CrossRef]
- Baidoshvili A, Bucur A, van Leeuwen J, van der Laak J, Kluin P, van Diest PJ. Evaluating the benefits of digital pathology implementation: Time savings in laboratory logistics. Histopathology 2018, 73, 784–794. [Google Scholar] [CrossRef]
- Shmatko A, Ghaffari Laleh N, Gerstung M, Kather JN. Artificial intelligence in histopathology: Enhancing cancer research and clinical oncology. Nat Cancer 2022, 3, 1026–1038. [Google Scholar] [CrossRef]
- Komura D, Ishikawa S. Machine Learning Methods for Histopathological Image Analysis. Comput Struct Biotechnol J 2018, 16, 34–42. [Google Scholar] [CrossRef]
- Lecun Y, Bengio Y, Hinton G. Deep learning. Nature 2015 521, 7553 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Roussel E, Capitanio U, Kutikov A, Oosterwijk E, Pedrosa I, Rowe SP; et al. Novel Imaging Methods for Renal Mass Characterization: A Collaborative Review. Eur Urol 2022, 81, 476–488. [Google Scholar] [CrossRef]
- Bera K, Schalper KA, Rimm DL, Velcheti V, Madabhushi A. Artificial intelligence in digital pathology - new tools for diagnosis and precision oncology. Nat Rev Clin Oncol 2019, 16, 703–715. [Google Scholar] [CrossRef] [PubMed]
- Niazi MKK, Parwani A v. , Gurcan MN. Digital pathology and artificial intelligence. Lancet Oncol 2019, 20, e253–61. [Google Scholar] [CrossRef] [PubMed]
- Colling R, Pitman H, Oien K, Rajpoot N, Macklin P, Bachtiar V; et al. Artificial intelligence in digital pathology: A roadmap to routine use in clinical practice. J Pathol 2019, 249, 143–150. [Google Scholar] [CrossRef] [PubMed]
- Volpe A, Patard JJ. Prognostic factors in renal cell carcinoma. World J Urol 2010, 28, 319–327. [Google Scholar] [CrossRef]
- Tucker MD, Rini BI. Predicting Response to Immunotherapy in Metastatic Renal Cell Carcinoma. Cancers (Basel) 2020, 12, 1–20. [Google Scholar] [CrossRef]
- Wang D, Khosla A, Gargeya R, Irshad H, Beck AH, Israel B. Deep Learning for Identifying Metastatic Breast Cancer 2016. [CrossRef]
- Hayashi Y. Black Box Nature of Deep Learning for Digital Pathology: Beyond Quantitative to Qualitative Algorithmic Performances. Lecture Notes in Computer Science (Including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics) 2020, 12090 LNCS:95–101. [CrossRef]
- Tougui I, Jilbab A, Mhamdi J el. Impact of the Choice of Cross-Validation Techniques on the Results of Machine Learning-Based Diagnostic Applications. Healthc Inform Res 2021, 27, 189–199. [Google Scholar] [CrossRef]
- Cabitza F, Campagner A, Soares F, García de Guadiana-Romualdo L, Challa F, Sulejmani A; et al. The importance of being external. methodological insights for the external validation of machine learning models in medicine. Comput Methods Programs Biomed 2021, 208, 106288. [Google Scholar] [CrossRef]
- Creighton CJ, Morgan M, Gunaratne PH, Wheeler DA, Gibbs RA, Robertson G; et al. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 2013 499, 7456 2013, 499, 43–49. [Google Scholar] [CrossRef]
- Krajewski KM, Pedrosa I. Imaging Advances in the Management of Kidney Cancer. Journal of Clinical Oncology 2018, 36, 3582. [Google Scholar] [CrossRef] [PubMed]
- Roussel E, Campi R, Amparore D, Bertolo R, Carbonara U, Erdem S; et al. Expanding the Role of Ultrasound for the Characterization of Renal Masses. Expanding the Role of Ultrasound for the Characterization of Renal Masses. J Clin Med 2022, 11. [Google Scholar] [CrossRef]
- Shuch B, Hofmann JN, Merino MJ, Nix JW, Vourganti S, Linehan WM; et al. Pathologic validation of renal cell carcinoma histology in the Surveillance, Epidemiology, and End Results program. Urol Oncol 2014, 32, 23–e9. [Google Scholar] [CrossRef]
- Al-Aynati M, Chen V, Salama S, Shuhaibar H, Treleaven D, Vincic L. Interobserver and Intraobserver Variability Using the Fuhrman Grading System for Renal Cell Carcinoma. Arch Pathol Lab Med 2003, 127, 593–596. [Google Scholar] [CrossRef]
- Williamson SR, Rao P, Hes O, Epstein JI, Smith SC, Picken MM; et al. Challenges in pathologic staging of renal cell carcinoma: A study of interobserver variability among urologic pathologists. American Journal of Surgical Pathology 2018, 42, 1253–1261. [Google Scholar] [CrossRef]
- Gavrielides MA, Gallas BD, Lenz P, Badano A, Hewitt SM. Observer variability in the interpretation of HER2/neu immunohistochemical expression with unaided and computer-aided digital microscopy. Arch Pathol Lab Med 2011, 135, 233–242. [Google Scholar] [CrossRef] [PubMed]
- Ficarra V, Martignoni G, Galfano A, Novara G, Gobbo S, Brunelli M; et al. Prognostic role of the histologic subtypes of renal cell carcinoma after slide revision. Eur Urol 2006, 50, 786–794. [Google Scholar] [CrossRef]
- Multicenter determination of optimal interobserver agreement using the Fuhrman grading system for renal cell carcinoma - Lang - 2005 - Cancer - Wiley Online Library n.d. https://acsjournals.onlinelibrary.wiley.com/doi/full/10.1002/cncr.20812 (accessed February 1, 2023). 1 February.
- Smaldone MC, Egleston B, Hollingsworth JM, Hollenbeck BK, Miller DC, Morgan TM; et al. Understanding Treatment Disconnect and Mortality Trends in Renal Cell Carcinoma Using Tumor Registry Data. Med Care 2017, 55, 398–404. [Google Scholar] [CrossRef]
- Kutikov A, Smaldone MC, Egleston BL, Manley BJ, Canter DJ, Simhan J; et al. Anatomic Features of Enhancing Renal Masses Predict Malignant and High-Grade Pathology: A Preoperative Nomogram Using the RENAL Nephrometry Score. Eur Urol 2011, 60, 241. [Google Scholar] [CrossRef]
- Pierorazio PM, Patel HD, Johnson MH, Sozio SM, Sharma R, Iyoha E; et al. Distinguishing malignant and benign renal masses with composite models and nomograms: A systematic review and meta-analysis of clinically localized renal masses suspicious for malignancy. Cancer 2016, 122, 3267–3276. [Google Scholar] [CrossRef]
- Joshi S, Kutikov A. Understanding Mutational Drivers of Risk: An Important Step Toward Personalized Care for Patients with Renal Cell Carcinoma. Eur Urol Focus 2017, 3, 428–429. [Google Scholar] [CrossRef] [PubMed]
- Nguyen MM, Gill IS, Ellison LM. The evolving presentation of renal carcinoma in the United States: Trends from the Surveillance, Epidemiology, and End Results program. J Urol 2006, 176, 2397–2400. [Google Scholar] [CrossRef] [PubMed]
- Sohlberg EM, Metzner TJ, Leppert JT. The Harms of Overdiagnosis and Overtreatment in Patients with Small Renal Masses: A Mini-review. Eur Urol Focus 2019, 5, 943–945. [Google Scholar] [CrossRef] [PubMed]
- Campi R, Stewart GD, Staehler M, Dabestani S, Kuczyk MA, Shuch BM; et al. Novel Liquid Biomarkers and Innovative Imaging for Kidney Cancer Diagnosis: What Can Be Implemented in Our Practice Today? A Systematic Review of the Literature. Eur Urol Oncol 2021, 4, 22–41. [Google Scholar] [CrossRef]
- Warren H, Palumbo C, Caliò A, Tran MGB, Campi R, Courcier J; et al. World J Urol 2023, 1–2. [CrossRef]
- Kutikov A, Smaldone MC, Uzzo RG, Haifler M, Bratslavsky G, Leibovich BC. Renal Mass Biopsy: Always, Sometimes, or Never? Eur Urol 2016, 70, 403–406. [CrossRef]
- Lane BR, Samplaski MK, Herts BR, Zhou M, Novick AC, Campbell SC. Renal mass biopsy--a renaissance? J Urol 2008, 179, 20–27. [Google Scholar] [CrossRef]
- Marconi L, Dabestani S, Lam TB, Hofmann F, Stewart F, Norrie J; et al. Systematic Review and Meta-analysis of Diagnostic Accuracy of Percutaneous Renal Tumour Biopsy. Eur Urol 2016, 69, 660–673. [Google Scholar] [CrossRef]
- Evans AJ, Delahunt B, Srigley JR. Issues and challenges associated with classifying neoplasms in percutaneous needle biopsies of incidentally found small renal masses. Semin Diagn Pathol 2015, 32, 184–195. [Google Scholar] [CrossRef]
- Kümmerlin I, ten Kate F, Smedts F, Horn T, Algaba F, Trias I; et al. Core biopsies of renal tumors: A study on diagnostic accuracy, interobserver, and intraobserver variability. Eur Urol 2008, 53, 1219–1227. [Google Scholar] [CrossRef] [PubMed]
- Elmore JG, Longton GM, Carney PA, Geller BM, Onega T, Tosteson ANA; et al. Diagnostic concordance among pathologists interpreting breast biopsy specimens. JAMA 2015, 313, 1122–1132. [Google Scholar] [CrossRef]
- Elmore JG, Barnhill RL, Elder DE, Longton GM, Pepe MS, Reisch LM; et al. Pathologists’ diagnosis of invasive melanoma and melanocytic proliferations: Observer accuracy and reproducibility study. BMJ 2017, 357. [CrossRef]
- Shah MD, Parwani A v. , Zynger DL. Impact of the Pathologist on Prostate Biopsy Diagnosis and Immunohistochemical Stain Usage Within a Single Institution. Am J Clin Pathol 2017, 148, 494–501. [Google Scholar] [CrossRef] [PubMed]
- Fenstermaker M, Tomlins SA, Singh K, Wiens J, Morgan TM. Development and Validation of a Deep-learning Model to Assist With Renal Cell Carcinoma Histopathologic Interpretation. Urology 2020, 144, 152–157. [Google Scholar] [CrossRef]
- van Oostenbrugge TJ, Fütterer JJ, Mulders PFA. Diagnostic Imaging for Solid Renal Tumors: A Pictorial Review. Kidney Cancer 2018, 2, 79–93. [Google Scholar] [CrossRef]
- Williams GM, Lynch DT. Renal Oncocytoma. Renal Oncocytoma. StatPearls 2022.
- Leone AR, Kidd LC, Diorio GJ, Zargar-Shoshtari K, Sharma P, Sexton WJ; et al. Bilateral benign renal oncocytomas and the role of renal biopsy: Single institution review. BMC Urol 2017, 17, 1–6. [Google Scholar] [CrossRef]
- Zhu M, Ren B, Richards R, Suriawinata M, Tomita N, Hassanpour S. Development and evaluation of a deep neural network for histologic classification of renal cell carcinoma on biopsy and surgical resection slides. Development and evaluation of a deep neural network for histologic classification of renal cell carcinoma on biopsy and surgical resection slides. Sci Rep 2021, 11. [Google Scholar] [CrossRef]
- Volpe A, Mattar K, Finelli A, Kachura JR, Evans AJ, Geddie WR; et al. Contemporary results of percutaneous biopsy of 100 small renal masses: A single center experience. J Urol 2008, 180, 2333–2337. [Google Scholar] [CrossRef]
- Wang R, Wolf JS, Wood DP, Higgins EJ, Hafez KS. Accuracy of Percutaneous Core Biopsy in Management of Small Renal Masses. Urology 2009, 73, 586–590. [Google Scholar] [CrossRef]
- Barwari K, de La Rosette JJ, Laguna MP. The penetration of renal mass biopsy in daily practice: A survey among urologists. J Endourol 2012, 26, 737–747. [Google Scholar] [CrossRef]
- Escudier, B. Emerging immunotherapies for renal cell carcinoma. Annals of Oncology 2012, 23, viii35–40. [Google Scholar] [CrossRef] [PubMed]
- Bertolo R, Pecoraro A, Carbonara U, Amparore D, Diana P, Muselaers S; et al. Resection Techniques During Robotic Partial Nephrectomy: A Systematic Review. Eur Urol Open Sci 2023, 52, 7–21. [Google Scholar] [CrossRef] [PubMed]
- Tabibu S, Vinod PK, Jawahar C v. Pan-Renal Cell Carcinoma classification and survival prediction from histopathology images using deep learning. Sci Rep 2019, 9. [CrossRef]
- Chen S, Zhang N, Jiang L, Gao F, Shao J, Wang T; et al. Clinical use of a machine learning histopathological image signature in diagnosis and survival prediction of clear cell renal cell carcinoma. Int J Cancer 2021, 148, 780–790. [Google Scholar] [CrossRef]
- Marostica E, Barber R, Denize T, Kohane IS, Signoretti S, Golden JA; et al. Development of a Histopathology Informatics Pipeline for Classification and Prediction of Clinical Outcomes in Subtypes of Renal Cell Carcinoma. Clin Cancer Res 2021, 27, 2868–2878. [Google Scholar] [CrossRef]
- Pathology Outlines - WHO classification n.d. https://www.pathologyoutlines.com/topic/kidneytumorWHOclass.html (accessed January 24, 2023). 24 January.
- Cimadamore A, Cheng L, Scarpelli M, Massari F, Mollica V, Santoni M; et al. Towards a new WHO classification of renal cell tumor: What the clinician needs to know—A narrative review. Transl Androl Urol 2021, 10, 1506. [Google Scholar] [CrossRef] [PubMed]
- Moch H, Cubilla AL, Humphrey PA, Reuter VE, Ulbright TM. The 2016 WHO Classification of Tumours of the Urinary System and Male Genital Organs-Part A: Renal, Penile, and Testicular Tumours. Eur Urol 2016, 70, 93–105. [CrossRef]
- Weng S, DiNatale RG, Silagy A, Mano R, Attalla K, Kashani M; et al. The Clinicopathologic and Molecular Landscape of Clear Cell Papillary Renal Cell Carcinoma: Implications in Diagnosis and Management. Eur Urol 2021, 79, 468–477. [Google Scholar] [CrossRef]
- Williamson SR, Eble JN, Cheng L, Grignon DJ. Clear cell papillary renal cell carcinoma: Differential diagnosis and extended immunohistochemical profile. Modern Pathology 2013, 26, 697–708. [Google Scholar] [CrossRef]
- Abdeltawab HA, Khalifa FA, Ghazal MA, Cheng L, El-Baz AS, Gondim DD. A deep learning framework for automated classification of histopathological kidney whole-slide images. J Pathol Inform 2022, 13, 100093. [Google Scholar] [CrossRef]
- Faust K, Roohi A, Leon AJ, Leroux E, Dent A, Evans AJ; et al. Unsupervised Resolution of Histomorphologic Heterogeneity in Renal Cell Carcinoma Using a Brain Tumor-Educated Neural Network. JCO Clin Cancer Inform 2020, 4, 811–821. [Google Scholar] [CrossRef]
- Renal Cell Carcinoma EAU Guidelines on 2022.
- Gelb, AB. Gelb AB. C O M M U N I C A T I O N Union Internationale Contre le Cancer (UICC) and the American Joint Renal Cell Carcinoma Committee on Cancer (AJCC) Current Prognostic Factors BACKGROUND. Renal cell carcinomas include several distinct entities with a range n.d. [CrossRef]
- Beksac AT, Paulucci DJ, Blum KA, Yadav SS, Sfakianos JP, Badani KK. Heterogeneity in renal cell carcinoma. Urologic Oncology: Seminars and Original Investigations 2017, 35, 507–515. [Google Scholar] [CrossRef]
- Dall’Oglio MF, Ribeiro-Filho LA, Antunes AA, Crippa A, Nesrallah L, Gonçalves PD; et al. Microvascular Tumor Invasion, Tumor Size and Fuhrman Grade: A Pathological Triad for Prognostic Evaluation of Renal Cell Carcinoma. J Urol 2007, 178, 425–428. [Google Scholar] [CrossRef]
- Tsui KH, Shvarts O, Smith RB, Figlin RA, Dekernion JB, Belldegrun A. PROGNOSTIC INDICATORS FOR RENAL CELL CARCINOMA: A MULTIVARIATE ANALYSIS OF 643 PATIENTS USING THE REVISED 1997 TNM STAGING CRITERIA. J Urol 2000, 163, 1090–1095. [Google Scholar] [CrossRef]
- Ficarra V, Righetti R, Pillonia S, D’amico A, Maffei N, Novella G; et al. Prognostic Factors in Patients with Renal Cell Carcinoma: Retrospective Analysis of 675 Cases. Eur Urol 2002, 41, 190–198. [Google Scholar] [CrossRef]
- Scopus preview - Scopus - Document details - Prognostic significance of morphologic parameters in renal cell carcinoma n.d. https://www.scopus.com/record/display.uri?eid=2-s2.0-2642552183&origin=inward&txGid=18f4bff1afabc920febe75bb222fbbab (accessed January 18, 2023).
- Prognostic value of nuclear grade of renal cell carcinoma n.d. https://acsjournals.onlinelibrary.wiley.com/doi/epdf/10.1002/1097-0142(19951215)76, 12%3C2543, :AID-CNCR2820761221%3E3.0.CO;2-S?src=getftr (accessed January 18, 2023).
- Intraobserver and Interobserver Variability of Fuhrman and Modified Fuhrman Grading Systems for Conventional Renal Cell Carcinoma - Bektas - 2009 - The Kaohsiung Journal of Medical Sciences - Wiley Online Library n.d. https://onlinelibrary.wiley.com/doi/abs/10.1016/S1607-551X(09)70562-5 (accessed January 18, 2023).
- Al-Aynati M, Chen V, Salama S, Shuhaibar H, Treleaven D, Vincic L. Interobserver and Intraobserver Variability Using the Fuhrman Grading System for Renal Cell Carcinoma. Arch Pathol Lab Med 2003, 127, 593–596. [Google Scholar] [CrossRef] [PubMed]
- Multicenter determination of optimal interobserver agreement using the Fuhrman grading system for renal cell carcinoma n.d. https://acsjournals.onlinelibrary.wiley.com/doi/epdf/10.1002/cncr.20812?src=getftr (accessed January 18, 2023).
- Yeh F-C, Parwani A V. , Pantanowitz L, Ho C. Automated grading of renal cell carcinoma using whole slide imaging. J Pathol Inform 2014, 5, 23. [Google Scholar] [CrossRef] [PubMed]
- Paner GP, Stadler WM, Hansel DE, Montironi R, Lin DW, Amin MB. Updates in the Eighth Edition of the Tumor-Node-Metastasis Staging Classification for Urologic Cancers. Eur Urol 2018, 73, 560–569. [Google Scholar] [CrossRef]
- Holdbrook DA, Singh M, Choudhury Y, Kalaw EM, Koh V, Tan HS; et al. Automated Renal Cancer Grading Using Nuclear Pleomorphic Patterns. JCO Clin Cancer Inform 2018, 2, 1–12. [Google Scholar] [CrossRef]
- Qayyum T, McArdle P, Orange C, Seywright M, Horgan P, Oades G; et al. Reclassification of the Fuhrman grading system in renal cell carcinoma-does it make a difference? Springerplus 2013, 2, 1–4. [CrossRef]
- Tian K, Rubadue CA, Lin DI, Veta M, Pyle ME, Irshad H; et al. Automated clear cell renal carcinoma grade classification with prognostic significance. Automated clear cell renal carcinoma grade classification with prognostic significance. PLoS ONE 2019, 14. [Google Scholar] [CrossRef]
- Song J, Xiao L, Lian Z. Contour-Seed Pairs Learning-Based Framework for Simultaneously Detecting and Segmenting Various Overlapping Cells/Nuclei in Microscopy Images. IEEE Trans Image Process 2018, 27, 5759–5774. [Google Scholar] [CrossRef] [PubMed]
- Role of VHL gene mutation in human renal cell carcinoma | SpringerLink n.d. https://link.springer.com/article/10.1007/s13277-011-0257-3 (accessed January 18, 2023). 18 January.
- Nogueira M, Kim HL. Molecular markers for predicting prognosis of renal cell carcinoma. Urologic Oncology: Seminars and Original Investigations 2008, 26, 113–124. [Google Scholar] [CrossRef] [PubMed]
- Roussel E, Beuselinck B, Albersen M. Tailoring treatment in metastatic renal cell carcinoma. Nat Rev Urol 2022, 19, 455–456. [Google Scholar] [CrossRef]
- Funakoshi T, Lee CH, Hsieh JJ. A systematic review of predictive and prognostic biomarkers for VEGF-targeted therapy in renal cell carcinoma. Cancer Treat Rev 2014, 40, 533–547. [Google Scholar] [CrossRef]
- Rodriguez-Vida A, Strijbos M, Hutson T. Predictive and prognostic biomarkers of targeted agents and modern immunotherapy in renal cell carcinoma. ESMO Open 2016, 1, e000013. [Google Scholar] [CrossRef]
- Motzer RJ, Robbins PB, Powles T, Albiges L, Haanen JB, Larkin J; et al. Avelumab plus axitinib versus sunitinib in advanced renal cell carcinoma: Biomarker analysis of the phase 3 JAVELIN Renal 101 trial. Nat Med 2020, 26, 1733. [Google Scholar] [CrossRef]
- Schimmel H, Zegers I, Emons H. Standardization of protein biomarker measurements: Is it feasible? 2010. [CrossRef]
- Mayeux, R. Biomarkers: Potential Uses and Limitations. NeuroRx 2004, 1, 182. [Google Scholar] [CrossRef]
- Singh NP, Bapi RS, Vinod PK. Machine learning models to predict the progression from early to late stages of papillary renal cell carcinoma. Comput Biol Med 2018, 100, 92–99. [Google Scholar] [CrossRef]
- Bhalla S, Chaudhary K, Kumar R, Sehgal M, Kaur H, Sharma S; et al. Gene expression-based biomarkers for discriminating early and late stage of clear cell renal cancer. Scientific Reports 2017 7, 1 2017, 7, 1–13. [Google Scholar] [CrossRef]
- Fernandes FG, Silveira HCS, Júnior JNA, da Silveira RA, Zucca LE, Cárcano FM; et al. Somatic Copy Number Alterations and Associated Genes in Clear-Cell Renal-Cell Carcinoma in Brazilian Patients. Int J Mol Sci 2021, 22, 1–14. [Google Scholar] [CrossRef]
- D’Avella C, Abbosh P, Pal SK, Geynisman DM. Mutations in renal cell carcinoma. Urologic Oncology: Seminars and Original Investigations 2020, 38, 763–773. [Google Scholar] [CrossRef]
- Havel JJ, Chowell D, Chan TA. The evolving landscape of biomarkers for checkpoint inhibitor immunotherapy. Nature Reviews Cancer 2019 19, 3 2019, 19, 133–150. [Google Scholar] [CrossRef]
- Go H, Kang MJ, Kim PJ, Lee JL, Park JY, Park JM; et al. Development of Response Classifier for Vascular Endothelial Growth Factor Receptor (VEGFR)-Tyrosine Kinase Inhibitor (TKI) in Metastatic Renal Cell Carcinoma. Pathol Oncol Res 2019, 25, 51–58. [Google Scholar] [CrossRef]
- Padmanabhan RK, Somasundar VH, Griffith SD, Zhu J, Samoyedny D, Tan KS; et al. An Active Learning Approach for Rapid Characterization of Endothelial Cells in Human Tumors. PLoS ONE 2014, 9, e90495. [Google Scholar] [CrossRef]
- Ing N, Huang F, Conley A, You S, Ma Z, Klimov S; et al. A novel machine learning approach reveals latent vascular phenotypes predictive of renal cancer outcome. A novel machine learning approach reveals latent vascular phenotypes predictive of renal cancer outcome. Sci Rep 2017, 7. [Google Scholar] [CrossRef]
- Herman JG, Latif F, Weng Y, Lerman MI, Zbar B, Liu S; et al. Silencing of the VHL tumor-suppressor gene by DNA methylation in renal carcinoma. Proc Natl Acad Sci U S A 1994, 91, 9700. [Google Scholar] [CrossRef] [PubMed]
- Yamana K, Ohashi R, Tomita Y. Contemporary Drug Therapy for Renal Cell Carcinoma— Evidence Accumulation and Histological Implications in Treatment Strategy. Biomedicines 2022, 10, 2840. [Google Scholar] [CrossRef]
- Zhu L, Wang J, Kong W, Huang J, Dong B, Huang Y; et al. LSD1 inhibition suppresses the growth of clear cell renal cell carcinoma via upregulating P21 signaling. Acta Pharm Sin B 2019, 9, 324–334. [Google Scholar] [CrossRef]
- Chen W, Zhang H, Chen Z, Jiang H, Liao L, Fan S; et al. Development and evaluation of a novel series of Nitroxoline-derived BET inhibitors with antitumor activity in renal cell carcinoma. Oncogenesis 2018 7, 11 2018, 7, 1–11. [Google Scholar] [CrossRef]
- Joosten SC, Smits KM, Aarts MJ, Melotte V, Koch A, Tjan-Heijnen VC; et al. Epigenetics in renal cell cancer: Mechanisms and clinical applications. Nature Reviews Urology 2018 15, 7 2018, 15, 430–451. [Google Scholar] [CrossRef]
- Zheng H, Momeni A, Cedoz PL, Vogel H, Gevaert O. Whole slide images reflect DNA methylation patterns of human tumors. Whole slide images reflect DNA methylation patterns of human tumors. NPJ Genom Med 2020, 5. [Google Scholar] [CrossRef]
- Singh NP, Vinod PK. Integrative analysis of DNA methylation and gene expression in papillary renal cell carcinoma. Mol Genet Genomics 2020, 295, 807–824. [Google Scholar] [CrossRef]
- Guida A, le Teuff G, Alves C, Colomba E, di Nunno V, Derosa L; et al. Identification of international metastatic renal cell carcinoma database consortium (IMDC) intermediate-risk subgroups in patients with metastatic clear-cell renal cell carcinoma. Oncotarget 2020, 11, 4582–4592. [Google Scholar] [CrossRef]
- Zigeuner R, Hutterer G, Chromecki T, Imamovic A, Kampel-Kettner K, Rehak P; et al. External validation of the Mayo Clinic stage, size, grade, and necrosis (SSIGN) score for clear-cell renal cell carcinoma in a single European centre applying routine pathology. Eur Urol 2010, 57, 102–111. [Google Scholar] [CrossRef] [PubMed]
- Prediction of progression after radical nephrectomy for patients with clear cell renal cell carcinoma: A stratification tool for prospective clinical trials - PubMed n.d. https://pubmed.ncbi.nlm.nih.gov/12655523/ (accessed March 5, 2023).
- Leibovich BC, Blute ML, Cheville JC, Lohse CM, Frank I, Kwon ED; et al. Prediction of progression after radical nephrectomy for patients with clear cell renal cell carcinoma: A stratification tool for prospective clinical trials. Cancer 2003, 97, 1663–1671. [Google Scholar] [CrossRef] [PubMed]
- Zisman A, Pantuck AJ, Dorey F, Said JW, Shvarts O, Quintana D; et al. Improved prognostication of renal cell carcinoma using an integrated staging system. J Clin Oncol 2001, 19, 1649–1657. [Google Scholar] [CrossRef]
- Lubbock ALR, Stewart GD, O’Mahony FC, Laird A, Mullen P, O’Donnell M; et al. Overcoming intratumoural heterogeneity for reproducible molecular risk stratification: A case study in advanced kidney cancer. BMC Med 2017, 15, 1–12. [Google Scholar] [CrossRef]
- Heng DYC, Xie W, Regan MM, Harshman LC, Bjarnason GA, Vaishampayan UN; et al. External validation and comparison with other models of the International Metastatic Renal-Cell Carcinoma Database Consortium prognostic model: A population-based study. Lancet Oncol 2013, 14, 141–148. [Google Scholar] [CrossRef]
- Erdem S, Capitanio U, Campi R, Mir MC, Roussel E, Pavan N; et al. External validation of the VENUSS prognostic model to predict recurrence after surgery in non-metastatic papillary renal cell carcinoma: A multi-institutional analysis. Urol Oncol 2022, 40, 198–e9. [Google Scholar] [CrossRef]
- di Nunno V, Mollica V, Schiavina R, Nobili E, Fiorentino M, Brunocilla E; et al. Improving IMDC Prognostic Prediction Through Evaluation of Initial Site of Metastasis in Patients With Metastatic Renal Cell Carcinoma. Clin Genitourin Cancer 2020, 18, e83–90. [Google Scholar] [CrossRef]
- Huang SC, Pareek A, Seyyedi S, Banerjee I, Lungren MP. Fusion of medical imaging and electronic health records using deep learning: A systematic review and implementation guidelines. Npj Digital Medicine 2020 3, 1 2020, 3, 1–9. [Google Scholar] [CrossRef]
- Wessels F, Schmitt M, Krieghoff-Henning E, Kather JN, Nientiedt M, Kriegmair MC; et al. Deep learning can predict survival directly from histology in clear cell renal cell carcinoma. Deep learning can predict survival directly from histology in clear cell renal cell carcinoma. PLoS ONE 2022, 17. [Google Scholar] [CrossRef]
- Chen S, Jiang L, Gao F, Zhang E, Wang T, Zhang N; et al. Machine learning-based pathomics signature could act as a novel prognostic marker for patients with clear cell renal cell carcinoma. Br J Cancer 2022, 126, 771–777. [Google Scholar] [CrossRef] [PubMed]
- Cheng J, Zhang J, Han Y, Wang X, Ye X, Meng Y; et al. Integrative Analysis of Histopathological Images and Genomic Data Predicts Clear Cell Renal Cell Carcinoma Prognosis. Cancer Res 2017, 77, e91–100. [Google Scholar] [CrossRef]
- Ning Z, Ning Z, Pan W, Pan W, Chen Y, Xiao Q; et al. Integrative analysis of cross-modal features for the prognosis prediction of clear cell renal cell carcinoma. Bioinformatics 2020, 36, 2888–2895. [Google Scholar] [CrossRef] [PubMed]
- Schulz S, Woerl AC, Jungmann F, Glasner C, Stenzel P, Strobl S; et al. Multimodal Deep Learning for Prognosis Prediction in Renal Cancer. Multimodal Deep Learning for Prognosis Prediction in Renal Cancer. Front Oncol 2021, 11. [Google Scholar] [CrossRef]
- Khene ZE, Kutikov A, Campi R. Machine learning in renal cell carcinoma research: The promise and pitfalls of ‘renal-izing’ the potential of artificial intelligence. BJU Int 2023. [CrossRef]
- Wu Z, Carbonara U, Campi R. Re: Criteria for the Translation of Radiomics into Clinically Useful Tests. Eur Urol 2023, 0. [CrossRef]
- Shortliffe EH, Sepúlveda MJ. Clinical Decision Support in the Era of Artificial Intelligence. JAMA 2018, 320, 2199–2200. [Google Scholar] [CrossRef] [PubMed]
- Durán JM, Jongsma KR. Who is afraid of black box algorithms? On the epistemological and ethical basis of trust in medical AI. J Med Ethics 2021, 47, 329–335. [Google Scholar] [CrossRef]
- Teo YY (Alan), Danilevsky A, Shomron N. Overcoming Interpretability in Deep Learning Cancer Classification. Methods in Molecular Biology 2021, 2243, 297–309. [Google Scholar] [CrossRef]
- Das S, Moore T, Wong WK, Stumpf S, Oberst I, McIntosh K; et al. End-user feature labeling: Supervised and semi-supervised approaches based on locally-weighted logistic regression. Artif Intell 2013, 204, 56–74. [Google Scholar] [CrossRef]
- Krzywinski M, Altman N. Points of significance: Power and sample size. Nat Methods 2013, 10, 1139–1140. [Google Scholar] [CrossRef]
- Button KS, Ioannidis JPA, Mokrysz C, Nosek BA, Flint J, Robinson ESJ; et al. Power failure: Why small sample size undermines the reliability of neuroscience. Nature Reviews Neuroscience 2013 14, 5 2013, 14, 365–376. [Google Scholar] [CrossRef]
- Wang, L. Heterogeneous Data and Big Data Analytics. Automatic Control and Information Sciences 2017, 3, 8–15. [Google Scholar] [CrossRef]
- Borodinov N, Neumayer S, Kalinin S v., Ovchinnikova OS, Vasudevan RK, Jesse S. Deep neural networks for understanding noisy data applied to physical property extraction in scanning probe microscopy. NPJ Comput Mater 2019, 5. [CrossRef]
- Goh WW bin, Wong L. Dealing with Confounders in Omics Analysis. Trends Biotechnol 2018, 36, 488–498. [Google Scholar] [CrossRef] [PubMed]
- Deep Learning n.d. https://mitpress.mit.edu/9780262035613/deep-learning/ (accessed April 23, 2023). 23 April.
- Veta M, Heng YJ, Stathonikos N, Bejnordi BE, Beca F, Wollmann T; et al. Predicting breast tumor proliferation from whole-slide images: The TUPAC16 challenge. Med Image Anal 2019, 54, 111–121. [Google Scholar] [CrossRef] [PubMed]
- Sirinukunwattana K, Pluim JPW, Chen H, Qi X, Heng PA, Guo YB; et al. Gland Segmentation in Colon Histology Images: The GlaS Challenge Contest. Med Image Anal 2016, 35, 489–502. [Google Scholar] [CrossRef]
- Yagi, Y. Color standardization and optimization in Whole Slide Imaging. Diagn Pathol 2011, 6, S15. [Google Scholar] [CrossRef]
- Tellez D, Litjens G, Bándi P, Bulten W, Bokhorst JM, Ciompi F; et al. Quantifying the effects of data augmentation and stain color normalization in convolutional neural networks for computational pathology. Med Image Anal 2019, 58, 101544. [Google Scholar] [CrossRef]
- Ho SY, Phua K, Wong L, bin Goh WW. Extensions of the External Validation for Checking Learned Model Interpretability and Generalizability. Patterns 2020, 1, 100129. [Google Scholar] [CrossRef] [PubMed]
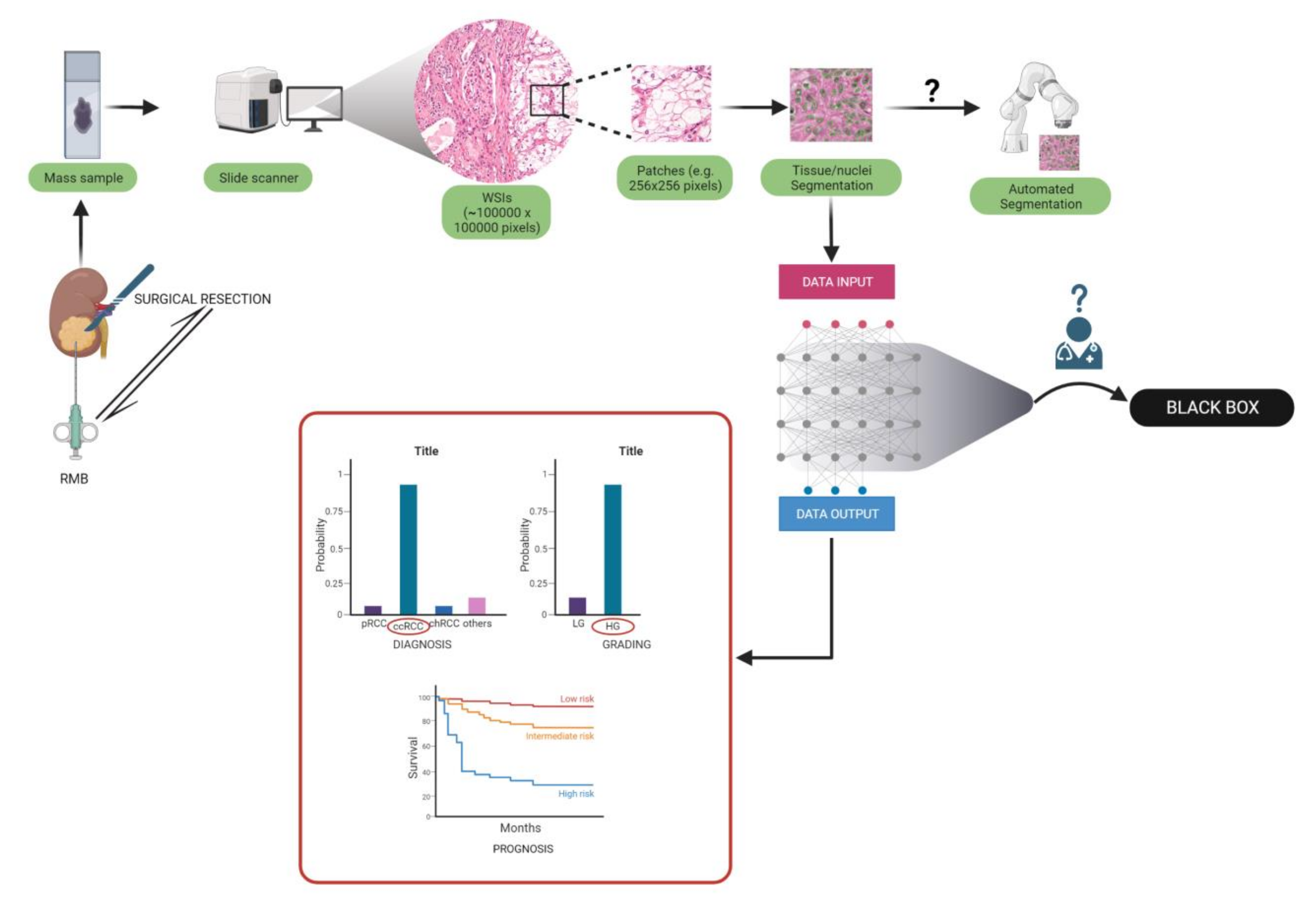
| Group | Aim | Number of patients |
Accuracy on the test set | External validation (N of patients) |
Accuracy on the external validation cohort | Algorithm |
|---|---|---|---|---|---|---|
| Fenstermaker et al. [49] | 1) RCC diagnosis, 2) subtyping, 3) grading |
15 ccRCC 15 pRCC 12 chRCC |
1) 99.1%; 2) 97.5%; 3) 98.4% |
N.A. | N.A. | CNN |
|
Zhu et al. [53] |
RCC subtyping |
486 SR (30 NT, 27 RO, 38 chRCC, 310 ccRCC, 81 pRCC), 79 RMB (24 RO, 34 ccRCC, 21 pRCC) |
1) 97% on SRS, 2) 97% on RMB |
0 RO 109 ChRCC 505 ccRCC 294 pRCC: |
95% accuracy (only SRs) |
DNN |
|
Chen et al. [60] |
1) RCC diagnosis, 2) subtyping, 3) survival prediction |
1) & 2) 362 NT, 362ccRCC, 128pRCC, 84chRCC 3) 283ccRCC |
1) 94.5% vs. NT 2) 97% vs. pRCC and chRCC 3) 88.8%, 90.0%, 89.6% in 1-3-5 y DFS |
1) & 2) 150 NP 150 ccRCC 52 pRCC 84 chRCC 3) 120ccRCC |
1)87.6% vs. NP 2)81.4% vs. pRCC and chRCC 3) 72.0%, 80.9%, 85.9% in 1-3-5 y DFS |
CNN |
| Tabibu et al. [59] | 1) RCC diagnosis, 2) subtyping, |
509 NT 1027 ccRCC 303 pRCC 254 chRCC |
1)93.9% ccRCC vs. NP 87.34% chRCC vs. NP 2)92.16% subtyping |
N.A. | N.A. | CNN (Resnet 18 and 34 architecture based); DAG-SVM on top of CNN for subtyping |
|
Abdeltawab et al. [67] |
RCC subtyping |
27 ccRCC 14 ccpRCC |
91% in ccpRCC |
10 ccRCC. |
90% in ccRCC |
CNN |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





