Submitted:
03 June 2023
Posted:
05 June 2023
You are already at the latest version
Abstract

Keywords:
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Extraction of peanut oil body by aqueous enzymatic extraction
2.3. Screening of fatty acids
2.4. Optimization of fatty acid demulsification
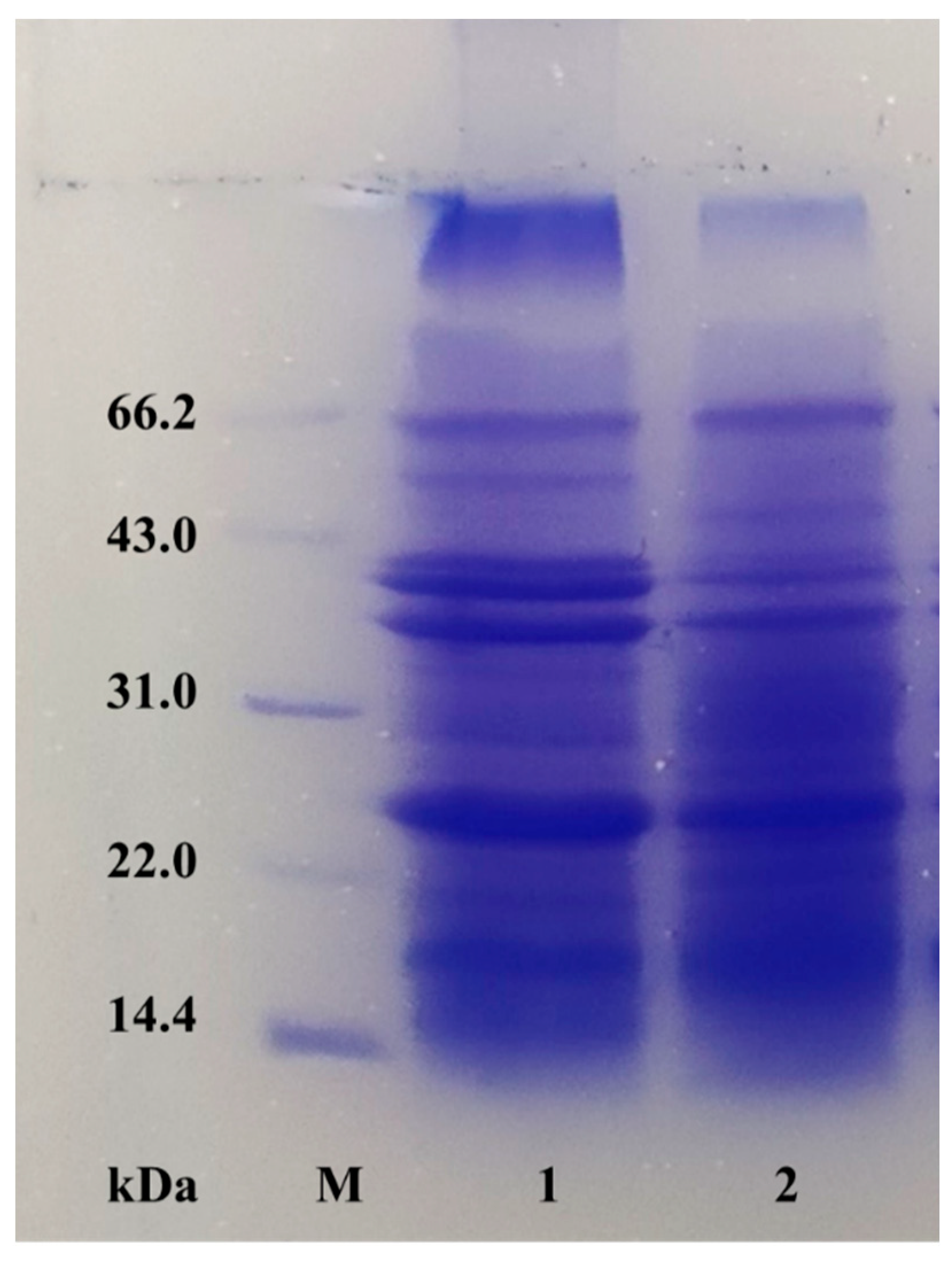
2.5. Sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE)
2.6. Physicochemical properties
2.7. Fatty acid composition
2.8. Determination of tocol (tocopherol and tocotrienol) contents
2.9. Oxidation stability
2.10. Statistical analysis
3. Results and Discussion
3.1. Fatty acid screening
3.2. Single-factor experiments
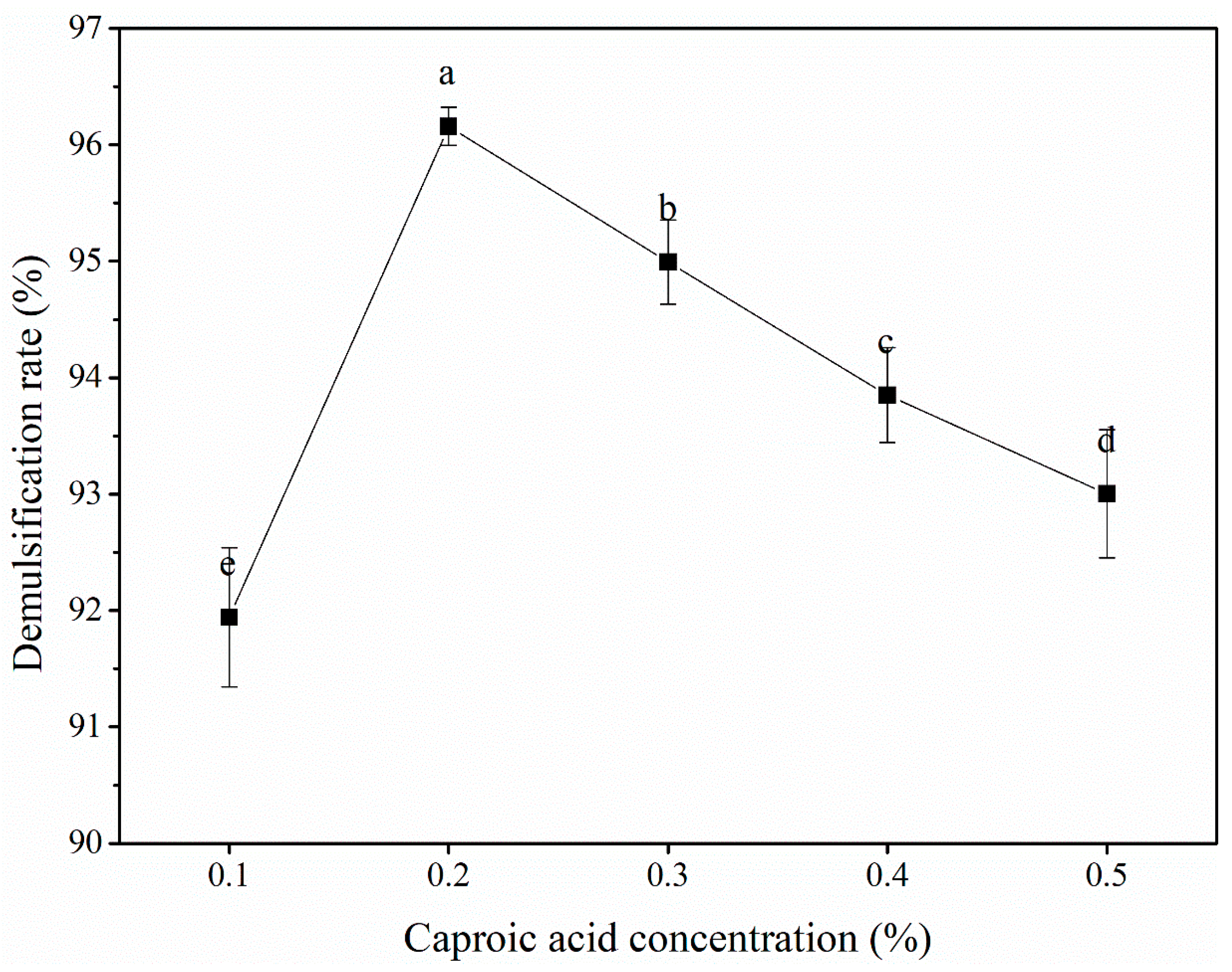
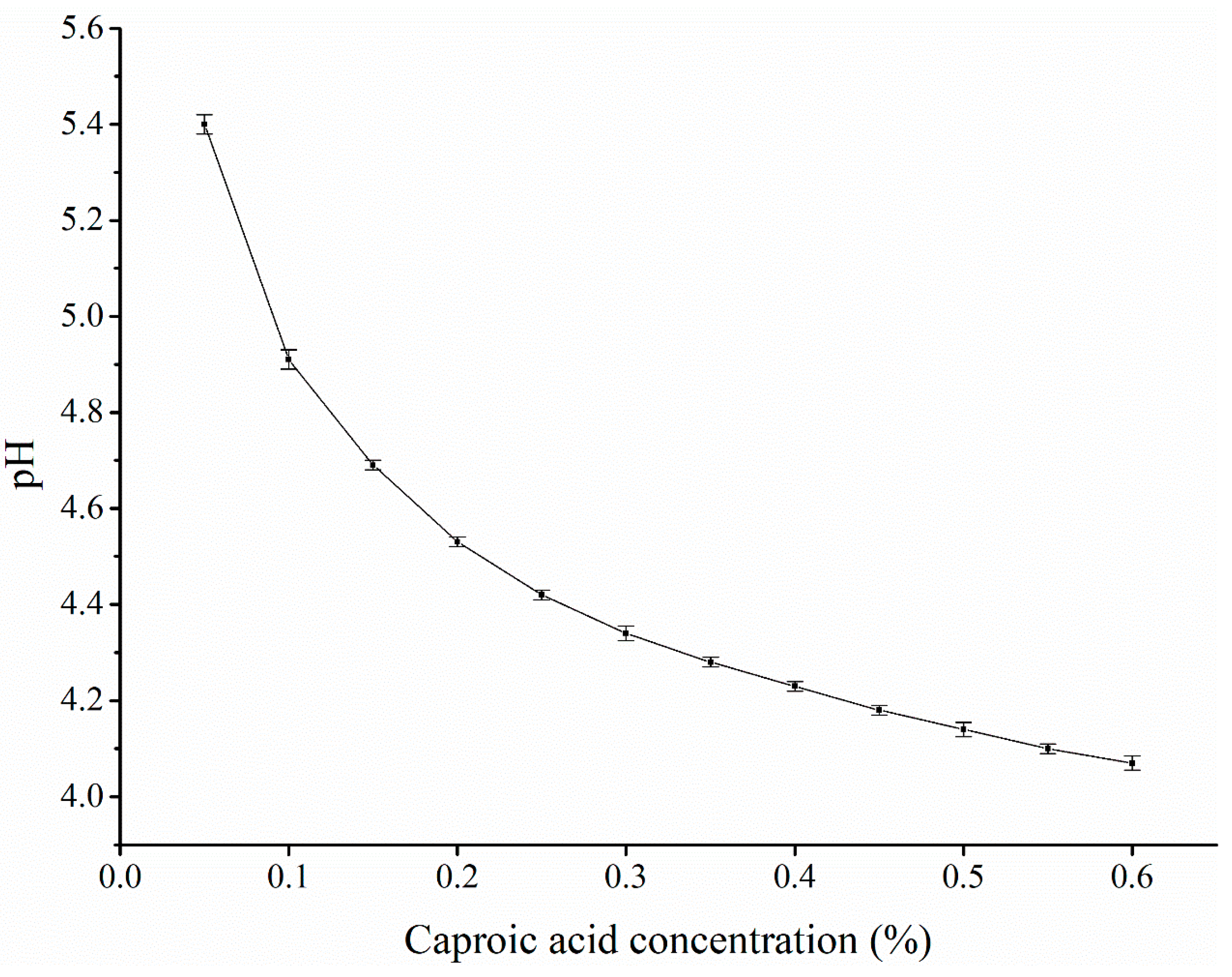
3.2.1. Caproic acid concentration
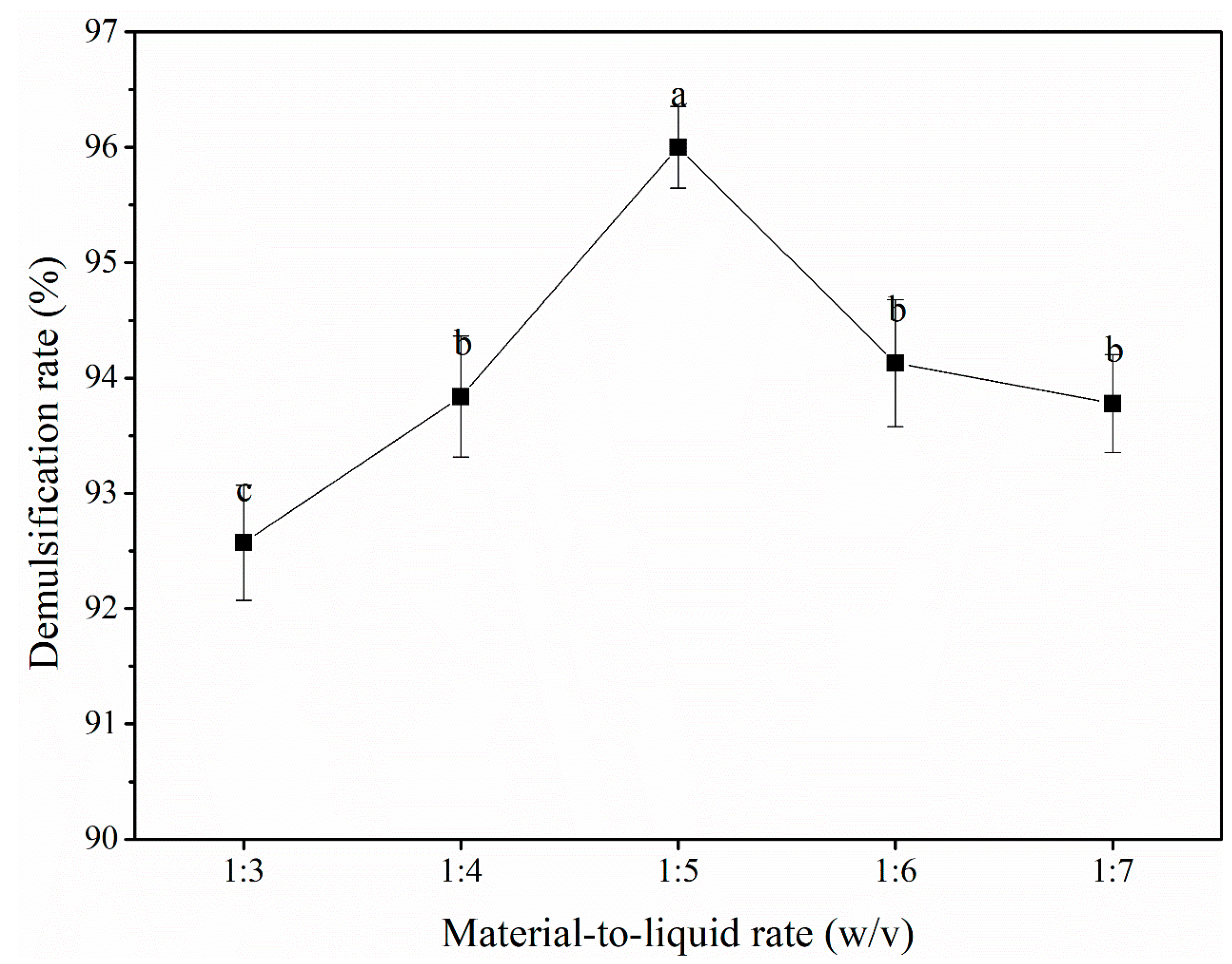
3.2.2. Material-to-liquid ratio
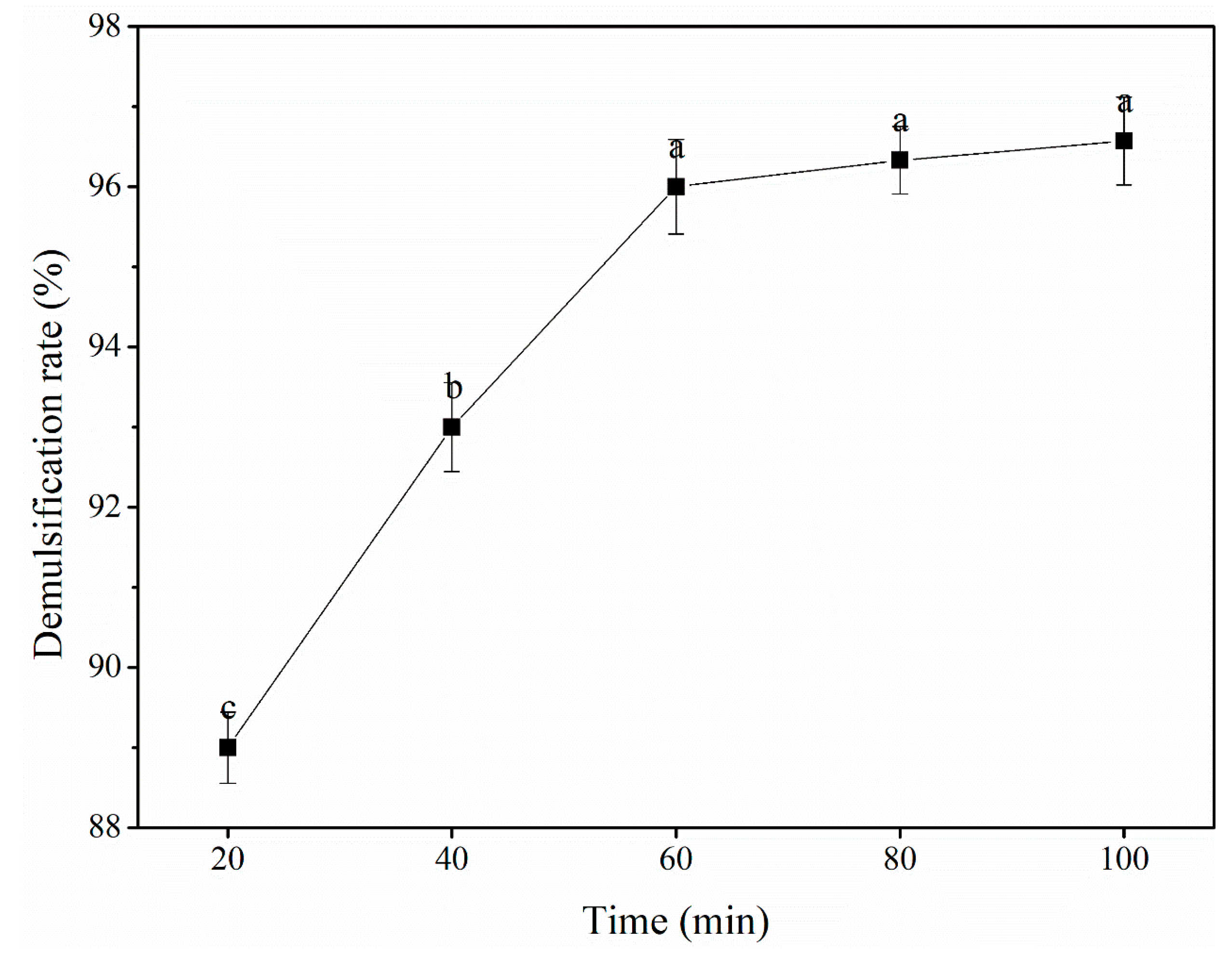
3.2.3. Reaction time
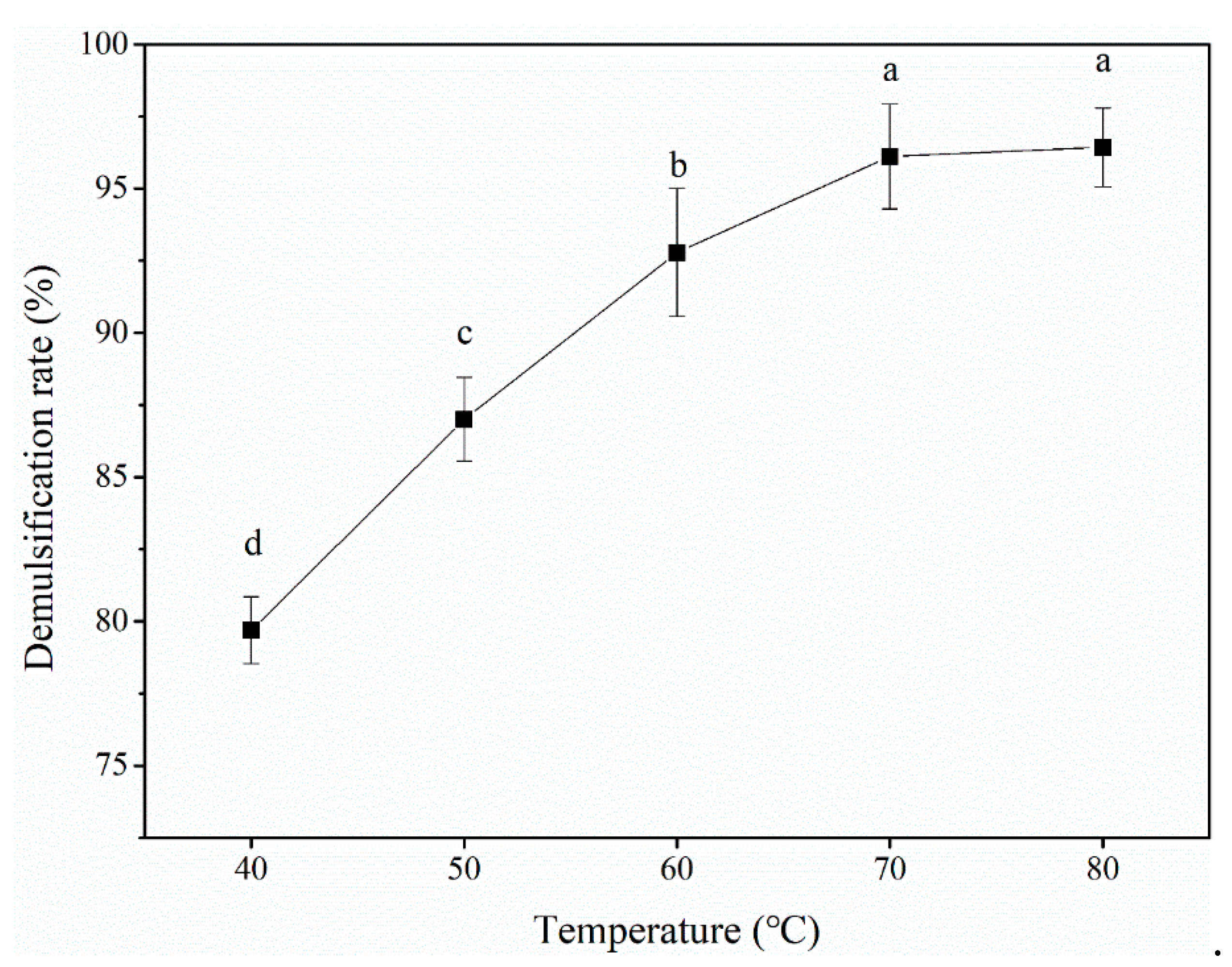
3.2.4. Temperature
3.3. RSM model development
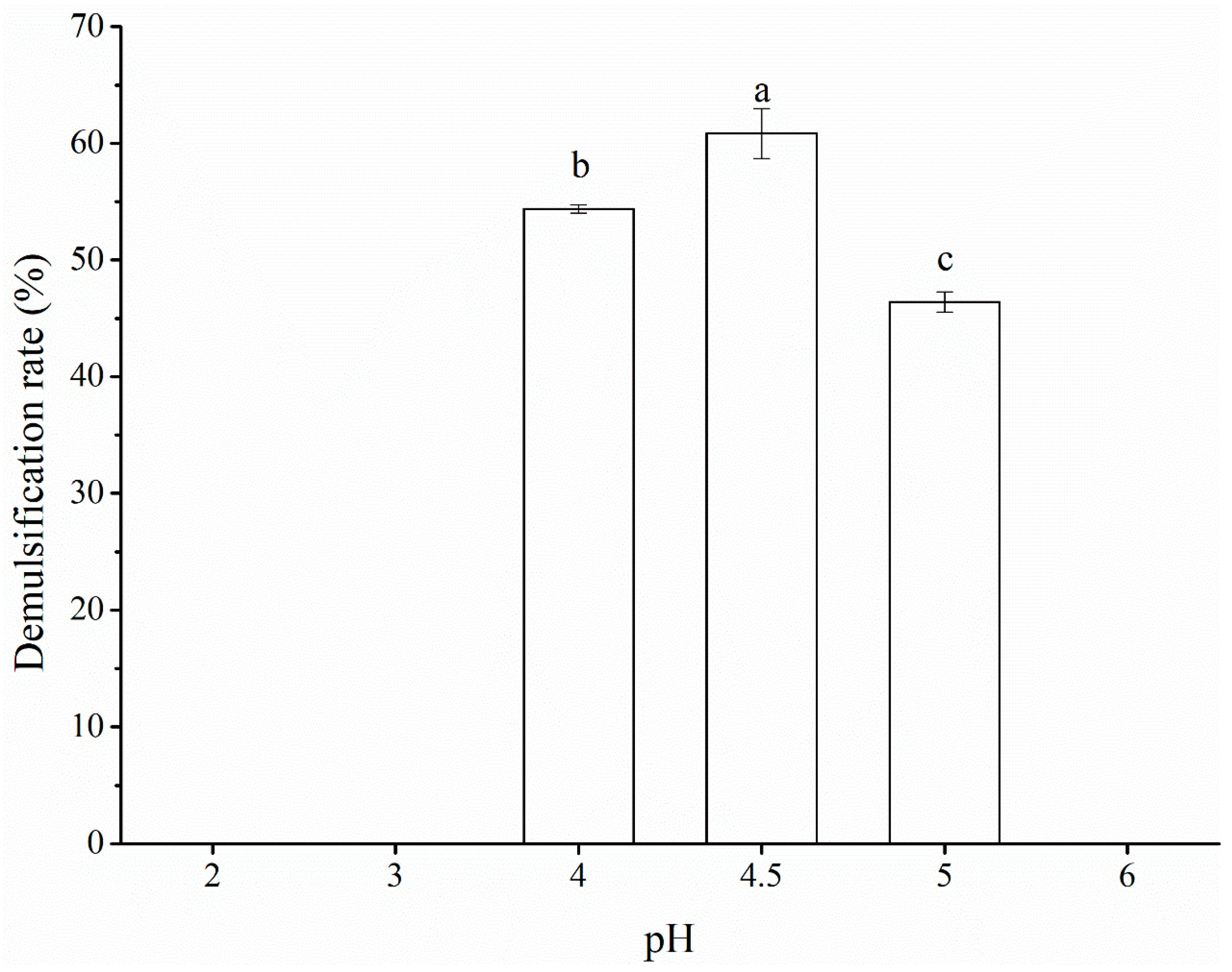
3.4. Determination of optimal conditions
3.5. Caproic acid demulsification mechanism
3.6. Physicochemical properties of caproic acid demulsified oil
3.7. Fatty acid composition
3.8. Total tocopherol and tocotrienol contents
3.9. Oxidation stability
4. Conclusions
Declaration of competing interests
Acknowledgments
References
- Bederska-Lojewska, D., Pieszka, M., Marzec, A., Rudzinska, M., Grygier, A., Siger, A., Cieslik-Boczula, K., Orczewska-Dudek, S., & Migdal, W. (2021). Physicochemical Properties, Fatty Acid Composition, Volatile Compounds of Blueberries, Cranberries, Raspberries, and Cuckooflower Seeds Obtained Using Sonication Method. Molecules, 26 (24).
- Bonku, R., & Yu, J. (2020). Health aspects of peanuts as an outcome of its chemical composition. Food Science and Human Wellness, 9 (1), 21-30.
- Capellini, M. C., Giacomini, V., Cuevas, M. S., & Rodrigues, C. E. C. (2017). Rice bran oil extraction using alcoholic solvents: Physicochemical characterization of oil and protein fraction functionality. Industrial Crops and Products, 104, 133-143.
- Chabrand, R. M., Kim, H.-J., Zhang, C., Glatz, C. E., & Jung, S. (2008). Destabilization of the emulsion formed during aqueous extraction of soybean oil. Journal of the American Oil Chemists Society, 85 (4), 383-390.
- Chen, L., Li, R., Ren, X., & Liu, T. (2016). Improved aqueous extraction of microalgal lipid by combined enzymatic and thermal lysis from wet biomass of Nannochloropsis oceanica. Bioresource Technology, 214, 138-143.
- Chiacchierini, E., Mele, G., Restuccia, D., & Vinci, G. (2007). Impact evaluation of innovative and sustainable extraction technologies on olive oil quality. Trends in Food Science & Technology, 18 (6), 299-305.
- Dash, D. R. , Pathak, S. S., & Pradhan, R. C. (2021). Extraction of oil from Terminalia chebula kernel by using ultrasound technology: Influence of process parameters on extraction kinetics. Industrial Crops and Products, 171.
- Diaz-Suarez, P. , Rosales-Quintero, A., Fernandez-Lafuente, R., Pola-Sanchez, E., Hernandez-Cruz, M. C., Ovando-Chacon, S. L., Rodrigues, R. C., & Tacias-Pascacio, V. G. (2021). Aqueous enzymatic extraction of Ricinus communis seeds oil using Viscozyme L. Industrial Crops and Products, 170.
- Dybowska, B. E., & Krupa-Kozak, U. (2020). Stability of oil-in-water emulsions as influenced by thermal treatment of whey protein dispersions or emulsions. International Journal of Dairy Technology, 73 (3), 513-520.
- Fang, X., Fei, X., Sun, H., & Jin, Y. (2016). Aqueous enzymatic extraction and demulsification of camellia seed oil (Camellia oleifera Abel.) and the oil's physicochemical properties. European Journal of Lipid Science and Technology, 118 (2), 244-251.
- Folch, J., Lees, M., & Sloane Stanley, G. H. (1957). A simple method for the isolation and purification of total lipides from animal tissues. The Journal of biological chemistry, 226 (1), 497-509.
- Fozo, E. M., & Quivey, R. G., Jr. (2004). Shifts in the membrane fatty acid profile of Streptococcus mutans enhance survival in acidic environments. Applied and Environmental Microbiology, 70 (2), 929-936.
- Gao, Y., Liu, C., Yao, F., & Chen, F. (2021). Aqueous enzymatic extraction of peanut oil body and protein and evaluation of its physicochemical and functional properties. International Journal of Food Engineering, 17 (11), 897-908.
- Gong, A. n., Shi, A.-m., Liu, H.-z., Yu, H.-w., Liu, L., Lin, W.-j., & Wang, Q. (2018). Relationship of chemical properties of different peanut varieties to peanut butter storage stability. Journal of Integrative Agriculture, 17 (5), 1003-1010.
- Hu, B. , Li, Y., Song, J., Li, H., Zhou, Q., Li, C., Zhang, Z., Liu, Y., Liu, A., Zhang, Q., Liu, S., & Luo, Q. (2020). Oil extraction from tiger nut (Cyperus esculentus L. ) using the combination of microwave-ultrasonic assisted aqueous enzymatic method - design, optimization and quality evaluation. Journal of Chromatography A, 1627.
- Ji, J., Liu, Y., Shi, L., Wang, N., & Wang, X. (2019). Effect of roasting treatment on the chemical composition of sesame oil. Lwt-Food Science and Technology, 101, 191-200.
- Ji, J. , Liu, Y., & Wang, D. (2020). Comparison of de-skin pretreatment and oil extraction on aflatoxins, phthalate esters, and polycyclic aromatic hydrocarbons in peanut oil. Food Control, 118.
- Ji, L., Yuan-Gang, Z., Meng, L., Cheng-Bo, G., Chun-Jian, Z., Thomas, E., & Yu-Jie, F. (2013). Aqueous enzymatic process assisted by microwave extraction of oil from yellow horn (Xanthoceras sorbifolia Bunge.) seed kernels and its quality evaluation. Food Chemistry, 138 (4), 2152-2158.
- Jiang, F., Yuan, L., Shu, N., Wang, W., Liu, Y., & Xu, Y.-J. (2020). Foodomics Revealed the Effects of Extract Methods on the Composition and Nutrition of Peanut Oil. Journal of Agricultural and Food Chemistry, 68 (4), 1147-1156.
- Jiang, L. H., Hua, D., Wang, Z., & Xu, S. Y. (2010). Aqueous enzymatic extraction of peanut oil and protein hydrolysates. Food and Bioproducts Processing, 88 (C2-3), 233-238.
- Jiao, J., Li, Z.-G., Gai, Q.-Y., Li, X.-J., Wei, F.-Y., Fu, Y.-J., & Ma, W. (2014). Microwave-assisted aqueous enzymatic extraction of oil from pumpkin seeds and evaluation of its physicochemical properties, fatty acid compositions and antioxidant activities. Food Chemistry, 147, 17-24.
- Khor, V. K., Shen, W.-J., & Kraemer, F. B. (2013). Lipid droplet metabolism. Current Opinion in Clinical Nutrition and Metabolic Care, 16 (6), 632-637.
- Lamsal, B. P., & Johnson, L. A. (2007). Separating oil from aqueous extraction fractions of soybean. Journal of the American Oil Chemists Society, 84 (8), 785-792.
- Latif, S., Diosady, L. L., & Anwar, F. (2008). Enzyme-assisted aqueous extraction of oil and protein from canola (Brassica napus L.) seeds. European Journal of Lipid Science and Technology, 110 (10), 887-892.
- Li, J., Zu, Y.-G., Luo, M., Gu, C.-B., Zhao, C.-J., Efferth, T., & Fu, Y.-J. (2013). Aqueous enzymatic process assisted by microwave extraction of oil from yellow horn (Xanthoceras sorbifolia Bunge.) seed kernels and its quality evaluation. Food Chemistry, 138 (4), 2152-2158.
- Li, P. F. , Zhang, W. B., Han, X., Liu, J. J., Liu, Y. Y., Gasmalla, M. A. A., & Yang, R. J. (2017). Demulsification of oil-rich emulsion and characterization of protein hydrolysates from peanut cream emulsion of aqueous extraction processing. Journal of Food Engineering, 204, 64-72.
- Li, X.-J., Li, Z.-G., Wang, X., Han, J.-Y., Zhang, B., Fu, Y.-J., & Zhao, C.-J. (2016). Application of cavitation system to accelerate aqueous enzymatic extraction of seed oil from Cucurbita pepo L. and evaluation of hypoglycemic effect. Food Chemistry, 212, 403-410.
- Li, Z. , Geng, H., Wang, X., Jing, B., Liu, Y., & Tan, Y. (2018). Noval tannic acid-based polyether as an effective demulsifier for water-in-aging crude oil emulsions. Chemical Engineering Journal, 354, 1110-1119.
- Lin, C., He, G., Li, X., Peng, L., Dong, C., Gu, S., & Xiao, G. (2007). Freeze/thaw induced demulsification of water-in-oil emulsions with loosely packed droplets. Separation and Purification Technology, 56 (2), 175-183.
- Liu, B., Du, J., Zeng, J., Chen, C., & Niu, S. (2009). Characterization and antioxidant activity of dihydromyricetin-lecithin complex. European Food Research and Technology, 230 (2), 325-331.
- Liu, C., Chen, F.-s., & Xia, Y.-m. (2022). Composition and structural characterization of peanut crude oil bodies extracted by aqueous enzymatic method. Journal of Food Composition and Analysis, 105.
- Liu, C., Hao, L.-h., Chen, F.-s., & Zhu, T.-w. (2020). The Mechanism of Extraction of Peanut Protein and Oil Bodies by Enzymatic Hydrolysis of the Cell Wall. Journal of Oleo Science, 69 (11), 1467-1479.
- Liu, Q., Li, P., Chen, J., Li, C., Jiang, L., Luo, M., & Sun, A. (2019). Optimization of Aqueous Enzymatic Extraction of Castor (Ricinus communis) Seeds Oil Using Response Surface Methodology. Journal of Biobased Materials and Bioenergy, 13 (1), 114-122.
- Longzheng, Z., Fusheng, C., Kunlun, L., Tingwei, Z., & Lianzhou, J. (2020). Combination of alcalase 2.4 L and CaCl2 for aqueous extraction of peanut oil. Journal of Food Science, 85 (6), 1772-1780.
- Mahatma, M. K., Thawait, L. K., Bishi, S. K., Khatediya, N., Rathnakumar, A. L., Lalwani, H. B., & Misra, J. B. (2016). Nutritional composition and antioxidant activity of Spanish and Virginia groundnuts (Arachis hypogaea L.): a comparative study. Journal of Food Science and Technology-Mysore, 53 (5), 2279-2286.
- Nagao, S., Takahashi, T., Shono, A., & Otake, K. (2010). Effects of high-pressure carbon dioxide on the demulsification of O/W emulsion. Desalination and Water Treatment, 17 (1-3), 80-89.
- Passos, C. P., Yilmaz, S., Silva, C. M., & Coimbra, M. A. (2009). Enhancement of grape seed oil extraction using a cell wall degrading enzyme cocktail. Food Chemistry, 115 (1), 48-53.
- Pengfei, L., Wenbin, Z., Xin, H., Junjun, L., Yuanyuan, L., Gasmalla, M. A. A., & Ruijin, Y. (2017). Demulsification of oil-rich emulsion and characterization of protein hydrolysates from peanut cream emulsion of aqueous extraction processing. Journal of Food Engineering, 204, 64-72.
- Penno, A., Hackenbroich, G., & Thiele, C. (2013). Phospholipids and lipid droplets. Biochimica Et Biophysica Acta-Molecular and Cell Biology of Lipids, 1831 (3), 589-594.
- Ramin, G., & Karamatollah, R. (2017). Optimization of an aqueous extraction process for pomegranate seed oil. Journal of the American Oil Chemists' Society, 94 (12), 1491-1501.
- Su, C.-H., Hoang Chinh, N., Thi Loan, B., & Huang, D.-L. (2019). Enzyme-assisted extraction of insect fat for biodiesel production. Journal of Cleaner Production, 223, 436-444.
- Tabtabaei, S., & Diosady, L. L. (2013). Aqueous and enzymatic extraction processes for the production of food-grade proteins and industrial oil from dehulled yellow mustard flour. Food Research International, 52 (1), 547-556.
- Tuntiwiwattanapun, N., Tongcumpou, C., Haagenson, D., & Wiesenborn, D. (2013). Development and Scale-up of Aqueous Surfactant-Assisted Extraction of Canola Oil for Use as Biodiesel Feedstock. Journal of the American Oil Chemists Society, 90 (7), 1089-1099.
- Tzen, J. T., Peng, C. C., Cheng, D. J., Chen, E. C., & Chiu, J. M. (1997). A new method for seed oil body purification and examination of oil body integrity following germination. Journal of Biochemistry, 121 (4), 762-768.
- Tzen, J. T. C., Cao, Y. Z., Laurent, P., Ratnayake, C., & Huang, A. H. C. (1993). LIPIDS, PROTEINS, AND STRUCTURE OF SEED OIL BODIES FROM DIVERSE SPECIES. Plant Physiology, 101 (1), 267-276.
- Wang, Q., Gao, C., Yang, N., & Nishinari, K. (2021). Effect of simulated saliva components on the in vitro digestion of peanut oil body emulsion. Rsc Advances, 11 (49), 30520-30531.
- Wang, W., Cui, C., Wang, Q., Sun, C., Jiang, L., & Hou, J. (2019). Effect of pH on physicochemical properties of oil bodies from different oil crops. Journal of Food Science and Technology-Mysore, 56 (1), 49-58.
- Xu, D. , Gao, Q., Ma, N., Hao, J., Yuan, Y., Zhang, M., Cao, Y., & Ho, C.-T. (2021). Structures and physicochemical characterization of enzyme extracted oil bodies from rice bran. Lwt-Food Science and Technology, 135.
- Yan, Z., Zhao, L., Kong, X., Hua, Y., & Chen, Y. (2016). Behaviors of particle size and bound proteins of oil bodies in soymilk processing. Food Chemistry, 194, 881-890.
- Yang, M., Cao, J., Cao, F., Lu, C., & Su, E. (2018). Efficient Extraction of Bioactive Flavonoids from Ginkgo biloba Leaves Using Deep Eutectic Solvent/Water Mixture as Green Media. Chemical and Biochemical Engineering Quarterly, 32 (3), 315-324.
- Yusoff, M. M., Gordon, M. H., Ezeh, O., & Niranjan, K. (2016). Aqueous enzymatic extraction of Moringa oleifera oil. Food Chemistry, 211, 400-408.
- Zhou, L., Chen, F., Liu, K., Zhu, T., & Jiang, L. (2020). Combination of Alcalase 2.4 L and CaCl2 for aqueous extraction of peanut oil. Journal of Food Science, 85 (6), 1772-1780.


| Variables | Factor | Coded levels | ||
| -1 | 0 | 1 | ||
| X1 | Concentration (%) | 0.10 | 0.20 | 0.30 |
| X2 | Materials-to-liquid ratio (w/v) | 1:4 | 1:5 | 1:6 |
| X3 | Time (min) | 40 | 60 | 80 |
| X4 | Temperature (ºC) | 60 | 70 | 80 |
| Run | Variable | Response, Y | |||
| X1 | X2 | X3 | X4 | ||
| 1 | -1 | 0 | 0 | -1 | 89.00±0.21 |
| 2 | -1 | 1 | 0 | 0 | 91.44±0.18 |
| 3 | -1 | -1 | 0 | 0 | 92.67±0.33 |
| 4 | -1 | 0 | 1 | 0 | 92.51±0.18 |
| 5 | -1 | 0 | 0 | 1 | 93.89±0.20 |
| 6 | -1 | 0 | -1 | 0 | 87.08±0.13 |
| 7 | 0 | -1 | 0 | -1 | 91.23±0.37 |
| 8 | 0 | -1 | 1 | 0 | 94.56±0.25 |
| 9 | 0 | 0 | 0 | 0 | 97.15±0.43 |
| 10 | 0 | 0 | 0 | 0 | 97.00±0.37 |
| 11 | 0 | 0 | 1 | 1 | 97.83±0.55 |
| 12 | 0 | 0 | 0 | 0 | 96.81±0.28 |
| 13 | 0 | -1 | 0 | 1 | 96.11±0.34 |
| 14 | 0 | 1 | 0 | -1 | 92.18±0.22 |
| 15 | 0 | 1 | -1 | 0 | 93.00±0.17 |
| 16 | 0 | -1 | -1 | 0 | 90.34±0.31 |
| 17 | 0 | 0 | -1 | -1 | 89.58±0.10 |
| 18 | 0 | 1 | 0 | 1 | 95.83±0.50 |
| 19 | 0 | 1 | 1 | 0 | 94.66±0.34 |
| 20 | 0 | 0 | -1 | 1 | 96.12±0.15 |
| 21 | 0 | 0 | 0 | 0 | 96.59±0.26 |
| 22 | 0 | 0 | 0 | 0 | 95.99±0.34 |
| 23 | 0 | 0 | 1 | -1 | 93.89±0.42 |
| 24 | 1 | 0 | -1 | 0 | 92.58±0.19 |
| 25 | 1 | 0 | 0 | 1 | 95.37±0.29 |
| 26 | 1 | 0 | 1 | 0 | 95.42±0.31 |
| 27 | 1 | 0 | 0 | -1 | 92.21±0.20 |
| 28 | 1 | -1 | 0 | 0 | 93.60±0.22 |
| 29 | 1 | 1 | 0 | 0 | 94.76±0.26 |
| Source | Sum of squares | Degree of freedom | Mean squares | F value | p value | Significance |
| Model | 191.71 | 14 | 13.69 | 18.78 | < 0.0001 | ** |
| X1 | 25.09 | 1 | 25.09 | 34.40 | < 0.0001 | ** |
| X2 | 0.94 | 1 | 0.94 | 1.29 | 0.2751 | |
| X3 | 33.90 | 1 | 33.90 | 46.49 | < 0.0001 | ** |
| X4 | 61.02 | 1 | 61.02 | 83.49 | < 0.0001 | ** |
| X1X2 | 1.43 | 1 | 1.43 | 1.96 | 0.1835 | |
| X1X3 | 1.68 | 1 | 1.68 | 2.30 | 0.1517 | |
| X1X4 | 0.75 | 1 | 0.75 | 1.03 | 0.3283 | |
| X2X3 | 1.64 | 1 | 1.64 | 2.25 | 0.1561 | |
| X2X4 | 0.38 | 1 | 0.38 | 0.52 | 0.4833 | |
| X3X4 | 1.69 | 1 | 1.69 | 2.32 | 0.1502 | |
| X12 | 47.24 | 1 | 47.24 | 64.77 | < 0.0001 | ** |
| X22 | 13.97 | 1 | 13.97 | 19.15 | 0.0006 | * |
| X32 | 21.45 | 1 | 21.45 | 29.41 | < 0.0001 | ** |
| X42 | 7.99 | 1 | 7.99 | 10.95 | 0.0052 | ** |
| Residual (error) | 10.21 | 14 | 0.73 | |||
| Lack of fit | 9.39 | 10 | 0.94 | 4.58 | 0.0779 | No significance |
| Pure error | 0.82 | 4 | 0.21 | |||
| Total | 201.92 | 28 |
| Index | Pressed oil | Soxhlet-extracted oil | Caproic acid demulsified oil |
| Acid value (mg KOH/g) |
0.18±0.03b | 0.38±0.02b | 0.62±0.06a |
| Peroxide value (g/100g) | 0.07±0.00b | 0.08±0.00b | 0.23±0.00a |
| Fatty acid | Pressed oil | Soxhlet-extracted oil | Caproic acid demulsified oil |
| Palmitic acid (C16:0) | 11.89±0.01c | 12.43±0.03b | 12.69±0.02a |
| Stearic acid (C18:0) | 3.65±0.01c | 3.82±0.02b | 3.94±0.01a |
| Oleic acid (C18:1) | 38.04±0.10a | 37.75±0.01b | 38.19±0.04a |
| Linoleic acid (C18:2) | 40.67±0.03a | 40.25±0.013b | 38.90±0.03c |
| Arachidic acid (C20:0) | 1.47±0.03b | 1.44±0.00b | 1.49±0.01a |
| Arachidonic acid (C20:1) | 0.77±0.01a | 0.82±0.02a | 0.71±0.12a |
| Behenic acid (C22:0) | 2.29±0.03a | 2.28±0.01a | 2.33±0.09a |
| Erucic acid (C22:1) | 0.00±0.00 | 0.00±0.00 | 0.25±0.02 |
| Lignoceric acid (C24:0) | 1.23±0.04b | 1.22±0.04b | 1.50±0.03a |
| MUFA | 38.81±0.12b | 38.57±0.03c | 39.16±0.10a |
| PUFA | 40.67±0.03a | 40.25±0.01b | 38.90±0.03c |
| UFA | 79.41±0.09a | 78.82±0.05b | 78.05±0.07c |
| SFA | 20.53±0.09c | 21.18±0.05b | 21.95±.07a |
| UFA/SFA | 3.87±0.02a | 3.72±0.01b | 3.56±0.01c |
| O/L | 0.94±0.00b | 0.94±0.68b | 0.98±0.00a |
| Pressed oil | Soxhlet-extracted oil | Caproic acid demulsified oil | |
| α-tocopherol (µg/g) | 5.61±0.44a | 5.37±0.03b | 5.62±0.01a |
| α-tocotrienol (µg/g) | 0.13±0.01a | 0.10±0.01b | 0.08±0.00b |
| β- tocopherol (µg/g) | 0.41±0.00a | 0.44±0.02a | 0.32±0.00b |
| γ- tocopherol (µg/g) | 4.04±0.25a | 3.45±0.01b | 3.98±0.01a |
| γ- tocotrienol (µg/g) | 0.81±0.06a | 0.78±0.02a | 0.78±0.01a |
| δ- tocopherol (µg/g) | 0.46±0.00a | 0.46±0.00a | 0.31±0.00b |
| Total content (µg/g) | 11.46±0.08a | 10.60±0.05c | 11.09±0.02b |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





