Submitted:
05 June 2023
Posted:
06 June 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. LOX
3. Production of Anti-LOX Protein Hydrolysates and Peptides
4. Potency and Modes of Action
5. Future Directions
- a)
- Protein hydrolysates and peptide fractions that have shown anti-LOX activity but whose constituent anti-LOX peptides have not yet been identified can be subjected to peptide identification as a next step. The identification of peptides with LOX inhibitory properties from protein hydrolysates remains scarce. Peptide identification followed by validation of their activities with synthetic peptides would further our understanding of the relationship between peptide structure and anti-LOX activity. In cases where a protein hydrolysate or partially purified peptide fractions exhibit stronger anti-LOX activity than the individual peptides, it will then be possible to test the hypothesis that the anti-LOX peptides act synergistically to account for the activity of the former. Anti-LOX protein hydrolysates and peptide fractions from feather keratins [19], fish scales [20], and insects [40] are promising candidates for the identification of anti-LOX peptides. Meanwhile, the three putative anti-LOX peptides (HYGGPPGGCR, SPKDLALPPGALPPVQ, and TGPSPTAGPPAPGGGTH) identified from chia seed proteins [39] that have not been validated for activity should proceed to synthesis and subsequence activity validation. In the long term, when a large dataset of anti-LOX peptides could be amassed, such information is useful for the development of a machine learning based anti-LOX peptide prediction server.
- b)
- To date, none of the studies discussed above have reported protein hydrolysates and peptides that are more potent than established anti-LOX inhibitors. Whether this is an intrinsic property of the peptides as anti-LOX agents is unclear. Nevertheless, future research may consider exploring different biological sources and proteases for anti-LOX protein hydrolysate and peptide discovery. The diversity of samples from which anti-LOX protein hydrolysates and peptides have been produced (Table 1) suggests that anti-LOX capacities may be part of the protein hydrolysates and peptides of many other protein-rich raw materials, which could be explored more intensively in the future. In particular, the exploration of the anti-LOX properties of protein hydrolysates and peptides prepared from low-value agricultural wastes or by-products may contribute towards an efficient use of resources, a direction in line with Sustainable Development Goals (SDGs), e.g., SDG 12: Responsible Consumption and Production and SDG 3: Good Health and Well-being [59]. Meanwhile, more than 40 proteases of plant, animal and bacterial origins are commercially available [11]. However, as shown in Table 1, fewer than 10 types of proteases have been used for the production of anti-LOX protein hydrolysates and peptides. Therefore, more enzymes should be tested in future. A promising strategy to be attempted in the future would be to systematically optimize the production of anti-LOX protein hydrolysates using the Response Surface Methodology (RSM) approach [60]. RSM can be applied to identify the optimal levels of parameters, such as enzyme type, enzyme: substrate ratio, hydrolysis time, temperature, that maximize the anti-LOX activity of protein hydrolysates. The RSM approach has been used in previous studies to optimize the yield of bioactive protein hydrolysates and peptides [61,62].
- c)
- Future evaluation of the anti-LOX capacity of all protein hydrolysates and peptides should include a well-established LOX inhibitor or an anti-inflammatory drug with anti-LOX capacity for comparison. This would allow for a more objective and convincing interpretation of anti-LOX potency, making it easier to compare between studies the anti-LOX potency of anti-LOX protein hydrolysates and peptides.
- d)
- The in silico or cheminformatics strategy has not been sufficiently utilized to accelerate the discovery of anti-LOX peptides. In particular, molecular docking and molecular dynamics simulations can be more widely used to facilitate anti-LOX peptide discovery [63]. This can overcome the problem of a lack of in silico screening or prediction servers specifically designed for predicting anti-LOX peptides. This may also increase the chance of identifying peptides that inhibit LOX activity by forming a complex with LOX directly. On the other hand, if the sequences of major proteins in a sample targeted for anti-LOX peptide discovery are available in protein databases, e.g., UniProt Knowledgebase (https://www.uniprot.org/)[64], in silico hydrolysis can also be attempted to identify potential protease treatments for the sample. The BIOPEP-UWM server (https://biochemia.uwm.edu.pl/en/biopep-uwm-2/) is a free and user-friendly tool that allows users to perform enzymatic hydrolysis virtually using 33 proteases either singly or up to three proteases simultaneously [65]. The server has been used to conduct in silico GI digestion of proteins in recent cheminformatic studies on bioactive peptides [66,67,68].
- e)
- In view of the potential application of the anti-LOX protein hydrolysates and peptides as functional food ingredients, the effects of heat processing, pH conditions and simulated GI digestion on the stability of such samples can also be investigated in future research [69]. Meanwhile, there are at least 20 peptidases and proteases in the human blood [70]. Thus, the stability of anti-LOX protein hydrolysates and peptides in human blood is also of interest in the context of bioavailability. A simple RP-HPLC-based assay to evaluate the stability of cytotoxic peptides in human blood has been reported previously [18], which could be applied to evaluate the stability of anti-LOX peptides in human blood.
- f)
- The activity of the anti-LOX protein hydrolysates and peptides discussed above has yet to be demonstrated in biological models, both at the cellular and in vivo levels. Nair and Funk [71] developed a 96-well microplate fluorescence assay that can be used to screen samples for intracellular anti-LOX activity using mammalian HEK 293 cells stably expressing 5-LOX, p12-LOX and 15-LOX1 isoforms. Such cell-based anti-LOX assays can be used to confirm the potency of the anti-LOX protein hydrolysates and peptide fractions discussed above for further validation of bioactivity. This can serve as a further screen for promising candidates prior to proceeding to in vivo pharmacological evaluation, which is more costly and requires ethical approval.
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Chai, T.-T.; Ee, K.-Y.; Kumar, D. T.; Manan, F. A.; Wong, F.-C. , Plant bioactive peptides: Current status and prospects towards use on human health. Protein & Peptide Letters 2021, 28, 623–642. [Google Scholar]
- Chai, T.-T.; Law, Y.-C.; Wong, F.-C.; Kim, S.-K. , Enzyme-assisted discovery of antioxidant peptides from edible marine invertebrates: A review. Marine Drugs 2017, 15, 42. [Google Scholar] [CrossRef] [PubMed]
- Wong, F.-C.; Xiao, J.; Wang, S.; Ee, K.-Y.; Chai, T.-T. , Advances on the antioxidant peptides from edible plant sources. Trends in Food Science & Technology 2020, 99, 44–57. [Google Scholar]
- Minkiewicz, P.; Iwaniak, A.; Darewicz, M. , BIOPEP-UWM database of bioactive peptides: Current opportunities. International Journal of Molecular Sciences 2019, 20, 5978. [Google Scholar] [CrossRef] [PubMed]
- Singh, B. P.; Aluko, R. E.; Hati, S.; Solanki, D. , Bioactive peptides in the management of lifestyle-related diseases: Current trends and future perspectives. Critical Reviews in Food Science and Nutrition 2022, 62, 4593–4606. [Google Scholar] [CrossRef]
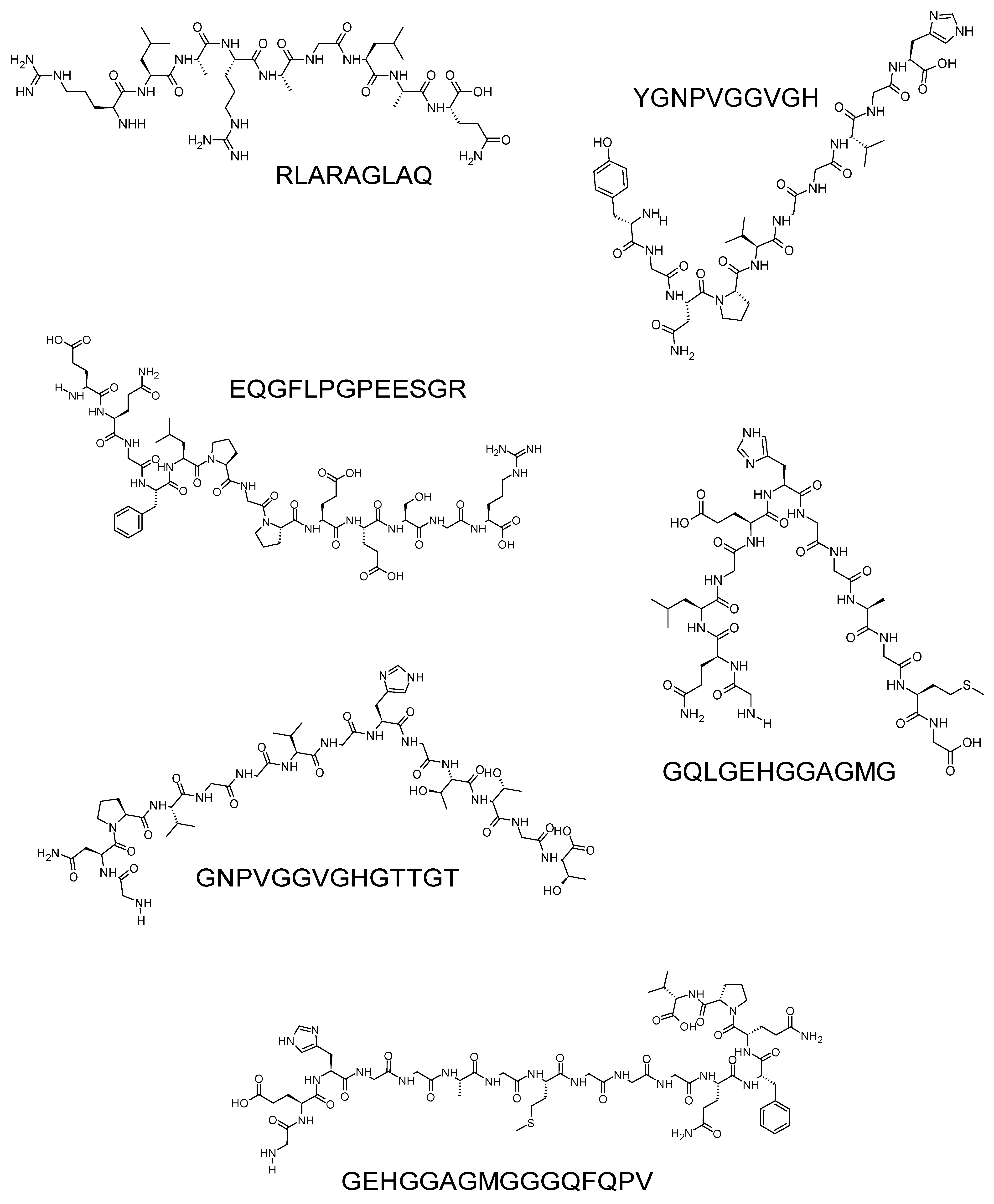
- Złotek, U.; Jakubczyk, A.; Rybczyńska-Tkaczyk, K.; Ćwiek, P.; Baraniak, B.; Lewicki, S. , Characteristics of new peptides GQLGEHGGAGMG, GEHGGAGMGGGQFQPV, EQGFLPGPEESGR, RLARAGLAQ, YGNPVGGVGH, and GNPVGGVGHGTTGT as inhibitors of enzymes involved in metabolic syndrome and antimicrobial potential. Molecules 2020, 25, 2492. [Google Scholar] [CrossRef]
- Tawalbeh, D.; Al-U’datt, M. H.; Wan Ahmad, W. A. N.; Ahmad, F.; Sarbon, N. M. , Recent advances in in vitro and in vivo studies of antioxidant, ace-inhibitory and anti-inflammatory peptides from legume protein hydrolysates. Molecules 2023, 28. [Google Scholar] [CrossRef]
- Thaha, A.; Wang, B.-S.; Chang, Y.-W.; Hsia, S.-M.; Huang, T.-C.; Shiau, C.-Y.; Hwang, D.-F.; Chen, T.-Y. , Food-derived bioactive peptides with antioxidative capacity, xanthine oxidase and tyrosinase inhibitory activity. Processes 2021, 9, 747. [Google Scholar] [CrossRef]
- Wu, J.; Sun, B.; Luo, X.; Zhao, M.; Zheng, F.; Sun, J.; Li, H.; Sun, X.; Huang, M. , Cytoprotective effects of a tripeptide from Chinese Baijiu against AAPH-induced oxidative stress in HepG2 cells: Via Nrf2 signaling. RSC Advances 2018, 8, 10898–10906. [Google Scholar] [CrossRef]
- Wen, C.; Zhang, J.; Zhang, H.; Duan, Y.; Ma, H. , Study on the structure–activity relationship of watermelon seed antioxidant peptides by using molecular simulations. Food Chemistry 2021, 364. [Google Scholar] [CrossRef]
- Cruz-Casas, D. E.; Aguilar, C. N.; Ascacio-Valdés, J. A.; Rodríguez-Herrera, R.; Chávez-González, M. L.; Flores-Gallegos, A. C. , Enzymatic hydrolysis and microbial fermentation: The most favorable biotechnological methods for the release of bioactive peptides. Food Chem (Oxf) 2021, 3, 100047. [Google Scholar] [CrossRef]
- Ye, H.; Tao, X.; Zhang, W.; Chen, Y.; Yu, Q.; Xie, J. , Food-derived bioactive peptides: production, biological activities, opportunities and challenges. Journal of Future Foods 2022, 2, 294–306. [Google Scholar] [CrossRef]
- Wong, F.-C.; Xiao, J.; Ong, M. G. L.; Pang, M.-J.; Wong, S.-J.; Teh, L.-K.; Chai, T.-T. , Identification and characterization of antioxidant peptides from hydrolysate of blue-spotted stingray and their stability against thermal, pH and simulated gastrointestinal digestion treatments. Food Chemistry 2019, 271, 614–622. [Google Scholar] [CrossRef]
- Quah, Y.; Tong, S.-R.; Bojarska, J.; Giller, K.; Tan, S.-A.; Ziora, Z. M.; Esatbeyoglu, T.; Chai, T.-T. , Bioactive peptide discovery from edible insects for potential applications in human health and agriculture. Molecules 2023, 28, 1233. [Google Scholar] [CrossRef]
- Mohana, D. S.; Chai, T.-T.; Wong, F.-C. , Antioxidant and protein protection potentials of fennel seed-derived protein hydrolysates and peptides. Modern Food Science and Technology 2019, 35, 22–29and21. [Google Scholar]
- Chai, T.-T.; Xiao, J.; Mohana Dass, S.; Teoh, J.-Y.; Ee, K.-Y.; Ng, W.-J.; Wong, F.-C. , Identification of antioxidant peptides derived from tropical jackfruit seed and investigation of the stability profiles. Food Chemistry 2021, 340, 127876. [Google Scholar] [CrossRef]
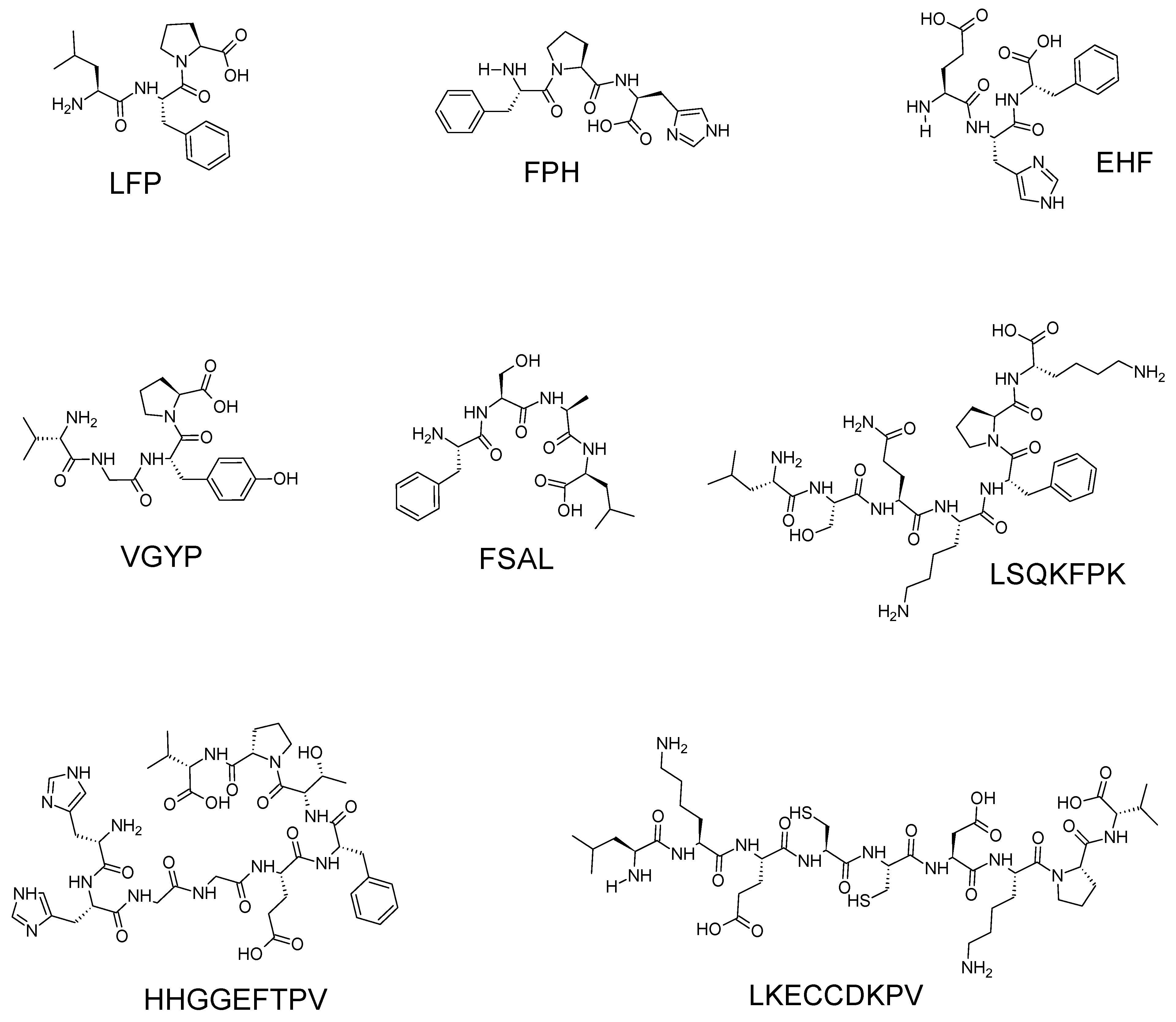
- Ding, C.; Hao, M.; Ma, S.; Zhang, Y.; Yang, J.; Ding, Q.; Sun, S.; Zhang, J.; Zhang, Y.; Liu, W. , Identification of peptides with antioxidant, anti-lipoxygenase, anti-xanthine oxidase and anti-tyrosinase activities from velvet antler blood. LWT 2022, 168. [Google Scholar] [CrossRef]
- Quah, Y.; Mohd Ismail, N. I.; Ooi, J. L. S.; Affendi, Y. A.; Abd Manan, F.; Wong, F.-C.; Chai, T.-T. , Identification of novel cytotoxic peptide KENPVLSLVNGMF from marine sponge Xestospongia testudinaria, with characterization of stability in human serum. International Journal of Peptide Research and Therapeutics 2018, 24, 189–199. [Google Scholar] [CrossRef]
- Kshetri, P.; Singh, P. L.; Chanu, S. B.; Singh, T. S.; Rajiv, C.; Tamreihao, K.; Singh, H. N.; Chongtham, T.; Devi, A. K.; Sharma, S. K.; Chongtham, S.; Singh, M. N.; Devi, Y. P.; Devi, H. S.; Roy, S. S. , Biological activity of peptides isolated from feather keratin waste through microbial and enzymatic hydrolysis. Electronic Journal of Biotechnology 2022, 60, 11–18. [Google Scholar] [CrossRef]
- Chen, Y. P.; Liang, C. H.; Wu, H. T.; Pang, H. Y.; Chen, C.; Wang, G. H.; Chan, L. P. , Antioxidant and anti-inflammatory capacities of collagen peptides from milkfish (Chanos chanos) scales. Journal of Food Science and Technology 2018, 55, 2310–2317. [Google Scholar] [CrossRef]
- Ong, J.-H.; Koh, J.-A.; Cao, H.; Tan, S.-A.; Manan, F. A.; Wong, F.-C.; Chai, T.-T. , Purification, identification and characterization of antioxidant peptides from corn silk tryptic hydrolysate: An integrated in vitro-in silico approach. Antioxidants 2021, 10. [Google Scholar] [CrossRef]
- Peña-Ramos, E. A.; Xiong, Y. L. , Antioxidative activity of whey protein hydrolysates in a liposomal system. Journal of Dairy Science 2001, 84, 2577–2583. [Google Scholar] [CrossRef] [PubMed]
- Aluko, R. E. , Amino acids, peptides, and proteins as antioxidants for food preservation. In Handbook of Antioxidants for Food Preservation, Shahidi, F., Ed. Elsevier Inc: Cambridge, UK, 2015; pp 105-140.
- Dullius, A.; Goettert, M. I.; de Souza, C. F. V. , Whey protein hydrolysates as a source of bioactive peptides for functional foods – Biotechnological facilitation of industrial scale-up. Journal of Functional Foods 2018, 42, 58–74. [Google Scholar] [CrossRef]
- Sierra Lopera, L. M.; Sepúlveda Rincón, C. T.; Vásquez Mazo, P.; Figueroa Moreno, O. A.; Zapata Montoya, J. E. , Byproducts of aquaculture processes: development and prospective uses. Review. Vitae 2018, 25, 128–140. [Google Scholar] [CrossRef]
- Siddik, M. A. B.; Howieson, J.; Fotedar, R.; Partridge, G. J. , Enzymatic fish protein hydrolysates in finfish aquaculture: a review. Reviews in Aquaculture 2021, 13, 406–430. [Google Scholar] [CrossRef]
- Lau, J. L.; Dunn, M. K. , Therapeutic peptides: historical perspectives and current development trends. In Peptide and Peptidomimetic Therapeutics: From Bench to Bedside, 2022; pp 3-33.
- Fetse, J.; Kandel, S.; Mamani, U.-F.; Cheng, K. , Recent advances in the development of therapeutic peptides. Trends in Pharmacological Sciences 2023. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Zhang, Q.; Zhang, B.; Zhao, Y.; Wang, N. , Potential active marine peptides as anti-aging drugs or drug candidates. Marine Drugs 2023, 21, 144. [Google Scholar] [CrossRef]
- Shi, Y.; Mandal, R.; Singh, A.; Pratap Singh, A. , Legume lipoxygenase: Strategies for application in food industry. Legume Science 2020, 2, e44. [Google Scholar] [CrossRef]
- Lončarić, M.; Strelec, I.; Moslavac, T.; Šubarić, D.; Pavić, V.; Molnar, M. , Lipoxygenase inhibition by plant extracts. Biomolecules 2021, 11, 1–17. [Google Scholar] [CrossRef]
- Sinha, S.; Doble, M.; Manju, S. L. , 5-Lipoxygenase as a drug target: A review on trends in inhibitors structural design, SAR and mechanism based approach. Bioorganic & Medicinal Chemistry 2019, 27, 3745–3759. [Google Scholar]
- Mashima, R.; Okuyama, T. , The role of lipoxygenases in pathophysiology; new insights and future perspectives. Redox Biology 2015, 6, 297–310. [Google Scholar] [CrossRef] [PubMed]
- Heinrich, L.; Booijink, R.; Khurana, A.; Weiskirchen, R.; Bansal, R. , Lipoxygenases in chronic liver diseases: current insights and future perspectives. Trends in Pharmacological Sciences 2022, 43, 188–205. [Google Scholar] [CrossRef] [PubMed]
- Ciganović, P.; Jakimiuk, K.; Tomczyk, M.; Zovko Končić, M. , Glycerolic licorice extracts as active cosmeceutical ingredients: Extraction optimization, chemical characterization, and biological activity. Antioxidants 2019, 8, 445. [Google Scholar] [CrossRef] [PubMed]
- Jakupović, L.; Bačić, I.; Jablan, J.; Marguí, E.; Marijan, M.; Inić, S.; Nižić Nodilo, L.; Hafner, A.; Zovko Končić, M. , Hydroxypropyl-β-cyclodextrin-based Helichrysum italicum extracts: Antioxidant and cosmeceutical activity and biocompatibility. Antioxidants 2023, 12. [Google Scholar] [CrossRef] [PubMed]
- Krieg, P.; Fürstenberger, G. , The role of lipoxygenases in epidermis. Biochimica et Biophysica Acta - Molecular and Cell Biology of Lipids 2014, 1841, 390–400. [Google Scholar] [CrossRef] [PubMed]
- Jakubczyk, A.; Szymanowska, U.; Karaś, M.; Złotek, U.; Kowalczyk, D. , Potential anti-inflammatory and lipase inhibitory peptides generated by in vitro gastrointestinal hydrolysis of heat treated millet grains. CyTA - Journal of Food 2019, 17, 324–333. [Google Scholar] [CrossRef]
- Grancieri, M.; Martino, H. S. D.; Gonzalez de Mejia, E. , Digested total protein and protein fractions from chia seed (Salvia hispanica L.) had high scavenging capacity and inhibited 5-LOX, COX-1-2, and iNOS enzymes. Food Chemistry 2019, 289, 204–214. [Google Scholar] [CrossRef] [PubMed]
- Zielińska, E.; Baraniak, B.; Karaś, M. , Antioxidant and anti-inflammatory activities of hydrolysates and peptide fractions obtained by enzymatic hydrolysis of selected heat-treated edible insects. Nutrients 2017, 9, 970. [Google Scholar] [CrossRef]
- Jakubczyk, A.; Karaś, M.; Baraniak, B.; Pietrzak, M. , The impact of fermentation and in vitro digestion on formation angiotensin converting enzyme (ACE) inhibitory peptides from pea proteins. Food Chemistry 2013, 141, 3774–3780. [Google Scholar] [CrossRef]
- Sheng, Z.; Turchini, G. M.; Xu, J.; Fang, Z.; Chen, N.; Xie, R.; Zhang, H.; Li, S. , Functional properties of protein hydrolysates on growth, digestive enzyme activities, protein metabolism, and intestinal health of larval largemouth bass (Micropterus salmoides). Frontiers in Immunology 2022, 13. [Google Scholar] [CrossRef]
- Kshetri, P.; Roy, S. S.; Sharma, S. K.; Singh, T. S.; Ansari, M. A.; Sailo, B.; Singh, S.; Prakash, N. , Feather degrading, phytostimulating, and biocontrol potential of native actinobacteria from North Eastern Indian Himalayan Region. Journal of Basic Microbiology 2018, 58, 730–738. [Google Scholar] [CrossRef] [PubMed]
- Kshetri, P.; Roy, S. S.; Chanu, S. B.; Singh, T. S.; Tamreihao, K.; Sharma, S. K.; Ansari, M. A.; Prakash, N. , Valorization of chicken feather waste into bioactive keratin hydrolysate by a newly purified keratinase from Bacillus sp. RCM-SSR-102. Journal of Environmental Management 2020, 273, 111195. [Google Scholar] [CrossRef] [PubMed]
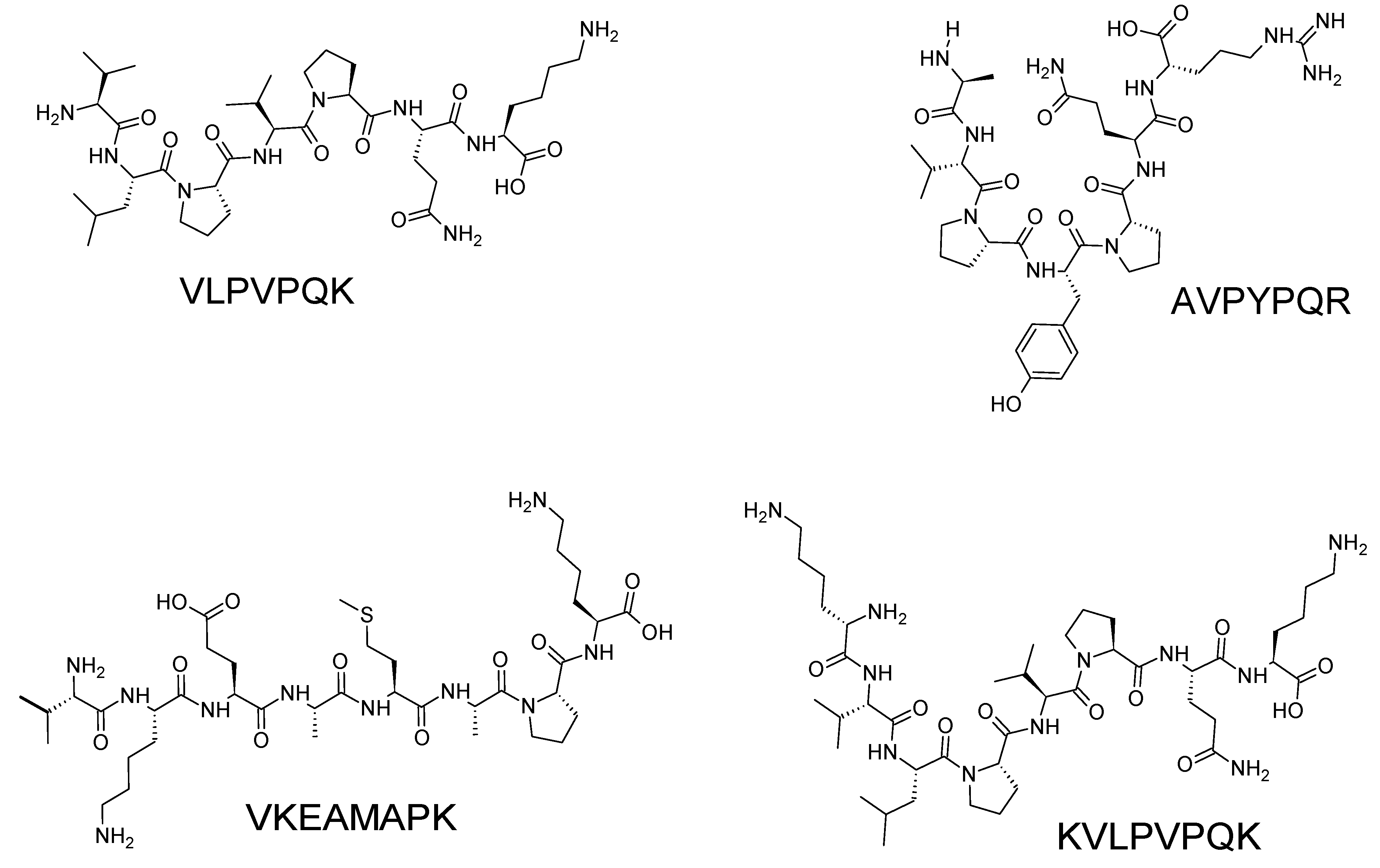
- Rival, S. G.; Fornaroli, S.; Boeriu, C. G.; Wichers, H. J. , Caseins and casein hydrolysates. 1. Lipoxygenase inhibitory properties. Journal of Agricultural and Food Chemistry 2001, 49, 287–294. [Google Scholar] [CrossRef] [PubMed]
- Mooney, C.; Haslam, N. J.; Pollastri, G.; Shields, D. C. , Towards the improved discovery and design of functional peptides: Common features of diverse classes permit generalized prediction of bioactivity. PLOS ONE 2012, 7, e45012. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Sharma, A. K.; Shastri, V.; Madhu, M. K.; Sharma, V. K. , Prediction of anti-inflammatory proteins/peptides: an insilico approach. J Transl Med 2017, 15, 7. [Google Scholar] [CrossRef]
- Purcell, D.; Packer, M. A.; Hayes, M. , Identification of bioactive peptides from a Laminaria digitata protein hydrolysate using in silico and in vitro methods to identify angiotensin-1-converting enzyme (ACE-1) inhibitory peptides. Marine Drugs 2023, 21, 90. [Google Scholar] [CrossRef]
- Garcia-Vaquero, M.; Mora, L.; Hayes, M. , In vitro and in silico approaches to generating and identifying angiotensin-converting enzyme i inhibitory peptides from green macroalga Ulva lactuca. Marine Drugs 2019, 17, 204. [Google Scholar] [CrossRef]
- Xiao, C.; Toldrá, F.; Zhao, M.; Zhou, F.; Luo, D.; Jia, R.; Mora, L. , In vitro and in silico analysis of potential antioxidant peptides obtained from chicken hydrolysate produced using Alcalase. Food Research International 2022, 157, 111253. [Google Scholar] [CrossRef]
- Heissel, S.; Frederiksen, S. J.; Bunkenborg, J.; Højrup, P. , Enhanced trypsin on a budget: Stabilization, purification and high-temperature application of inexpensive commercial trypsin for proteomics applications. PLOS ONE 2019, 14, e0218374. [Google Scholar] [CrossRef]
- Rival, S. G.; Boeriu, C. G.; Wichers, H. J. , Caseins and casein hydrolysates. 2. Antioxidative properties and relevance to lipoxygenase inhibition. Journal of Agricultural and Food Chemistry 2001, 49, 295–302. [Google Scholar] [CrossRef]
- Cengiz Şahin, S.; Cavas, L. , Can soybean lipoxygenases be real models for mammalian lipoxygenases? A bioinformatics approach. Journal of the Turkish Chemical Society, Section A: Chemistry 2020, 8, 79–102. [Google Scholar] [CrossRef]
- Muñoz-Ramírez, A.; Mascayano-Collado, C.; Barriga, A.; Echeverría, J.; Urzúa, A. , Inhibition of soybean 15-lipoxygenase and human 5-lipoxygenase by extracts of leaves, stem bark, phenols and catechols isolated from Lithraea caustica (Anacardiaceae). Frontiers in Pharmacology 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Karaś, M.; Jakubczyk, A.; Szymanowska, U.; Jęderka, K.; Lewicki, S.; Złotek, U. , Different temperature treatments of millet grains affect the biological activity of protein hydrolyzates and peptide fractions. Nutrients 2019, 11. [Google Scholar] [CrossRef] [PubMed]
- Gan, T. J. , Diclofenac: an update on its mechanism of action and safety profile. Current Medical Research and Opinion 2010, 26, 1715–1731. [Google Scholar] [CrossRef] [PubMed]
- Cattaneo, F.; Sayago, J. E.; Alberto, M. R.; Zampini, I. C.; Ordoñez, R. M.; Chamorro, V.; Pazos, A.; Isla, M. I. , Anti-inflammatory and antioxidant activities, functional properties and mutagenicity studies of protein and protein hydrolysate obtained from Prosopis alba seed flour. Food Chemistry 2014, 161, 391–399. [Google Scholar] [CrossRef] [PubMed]
- Framroze, B.; Havaldar, F.; Misal, S. , An in-vitro study on the regulation of oxidative protective genes in human gingival and intestinal epithelial cells after treatment with salmon protein hydrolysate peptides. Functional Foods in Health and Disease 2018, 8, 398–411. [Google Scholar] [CrossRef]
- UN General Assembly, Transforming our world: the 2030 agenda for sustainable development, 2015. Available online: https://sustainabledevelopment.un.org/content/documents/21252030%20Agenda%20for%20Sustainable%20Development%20web.pdf.
- Weremfo, A.; Abassah-Oppong, S.; Adulley, F.; Dabie, K.; Seidu-Larry, S. , Response surface methodology as a tool to optimize the extraction of bioactive compounds from plant sources. Journal of the Science of Food and Agriculture 2023, 103, 26–36. [Google Scholar] [CrossRef] [PubMed]
- Yan, F.; Wang, Q.; Teng, J.; Wu, F.; He, Z. , Preparation process optimization and evaluation of bioactive peptides from Carya cathayensis Sarg meal. Current Research in Food Science 2023, 6, 100408. [Google Scholar] [CrossRef]
- Lee, C. H.; Hamdan, N.; Ling, L. I.; Wong, S. L.; Nyakuma, B. B.; Khoo, S. C.; Ramachandran, H.; Jamaluddin, H.; Wong, K. Y.; Lee, T. H. , Antioxidant and antidiabetic properties of bioactive peptides from soursop (Annona muricata) leaf biomass. Biomass Conversion and Biorefinery 2023. [Google Scholar] [CrossRef]
- Vidal-Limon, A.; Aguilar-Toalá, J. E.; Liceaga, A. M. , Integration of molecular docking analysis and molecular dynamics simulations for studying food proteins and bioactive peptides. Journal of Agricultural and Food Chemistry 2022, 70, 934–943. [Google Scholar] [CrossRef]
- Consortium, T. U. , UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Research 2020, (D1), D480–D489. [Google Scholar]
- Minkiewicz, P.; Iwaniak, A.; Darewicz, M. , BIOPEP-UWM Database of Bioactive Peptides: Current Opportunities. Int J Mol Sci 2019, 20. [Google Scholar] [CrossRef] [PubMed]
- Barati, M.; Javanmardi, F.; Jabbari, M.; Mokari-Yamchi, A.; Farahmand, F.; Eş, I.; Farhadnejad, H.; Davoodi, S. H.; Mousavi Khaneghah, A. , An in silico model to predict and estimate digestion-resistant and bioactive peptide content of dairy products: A primarily study of a time-saving and affordable method for practical research purposes. LWT 2020, 130, 109616. [Google Scholar] [CrossRef]
- Sansi, M. S.; Iram, D.; Zanab, S.; Vij, S.; Puniya, A.; Singh, A.; Ashutosh, A.; Meena, S. , Antimicrobial bioactive peptides from goat Milk proteins: In silico prediction and analysis. Journal of Food Biochemistry 2022, 46. [Google Scholar] [CrossRef] [PubMed]
- Wongngam, W.; Hamzeh, A.; Tian, F.; Roytrakul, S.; Yongsawatdigul, J. , Purification and molecular docking of angiotensin converting enzyme-inhibitory peptides derived from corn gluten meal hydrolysate and from in silico gastrointestinal digestion. Process Biochemistry 2023, 129, 113–120. [Google Scholar] [CrossRef]
- Wu, Y. H. S.; Chen, Y. C. , Trends and applications of food protein-origin hydrolysates and bioactive peptides. Journal of Food and Drug Analysis 2022, 30, 172–184. [Google Scholar] [CrossRef]
- Werle, M.; Bernkop-Schnürch, A. , Strategies to improve plasma half life time of peptide and protein drugs. Amino Acids 2006, 30, 351–367. [Google Scholar] [CrossRef]
- Nair, D. G.; Funk, C. D. , A cell-based assay for screening lipoxygenase inhibitors. Prostaglandins & Other Lipid Mediators 2009, 90, 98–104. [Google Scholar]
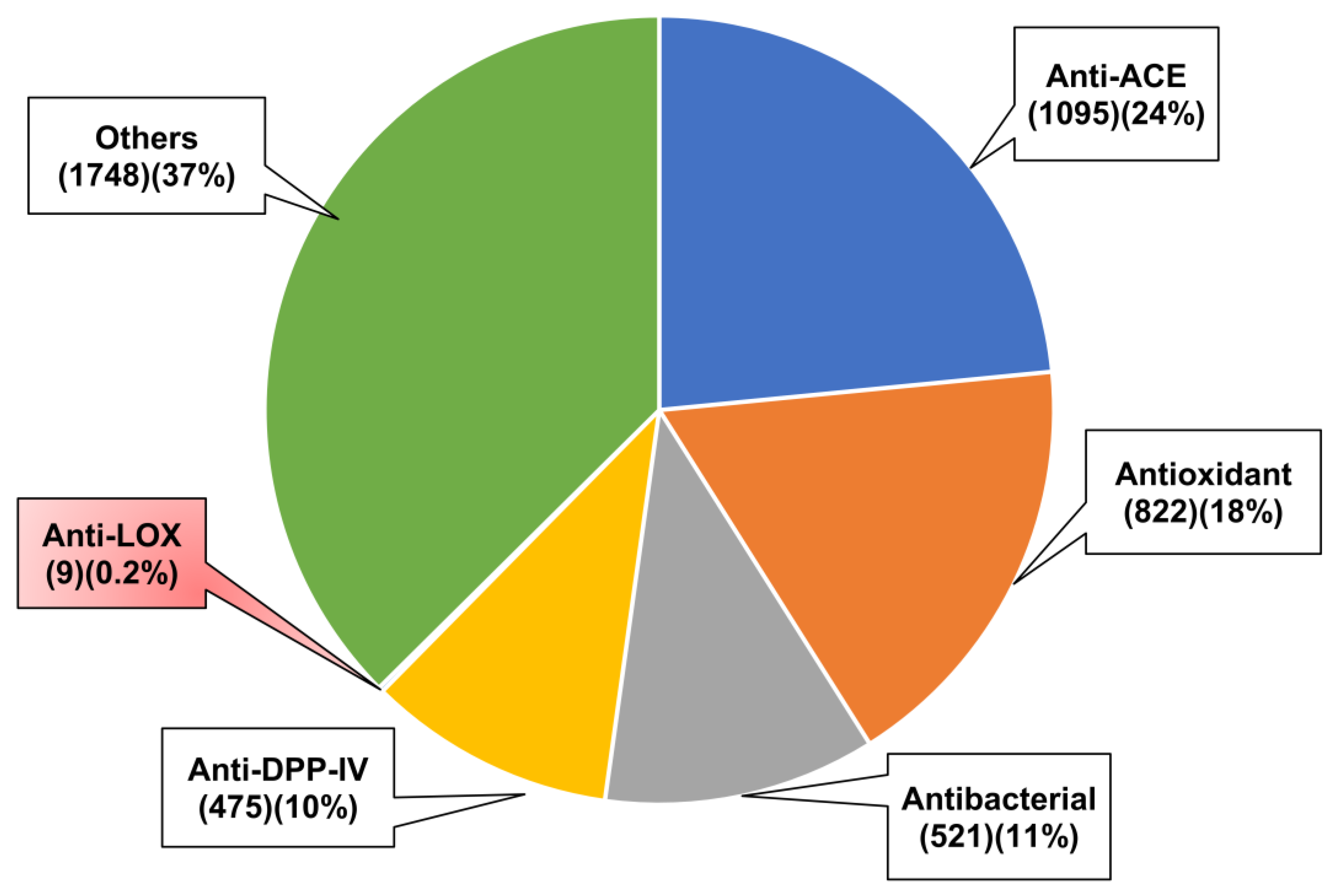
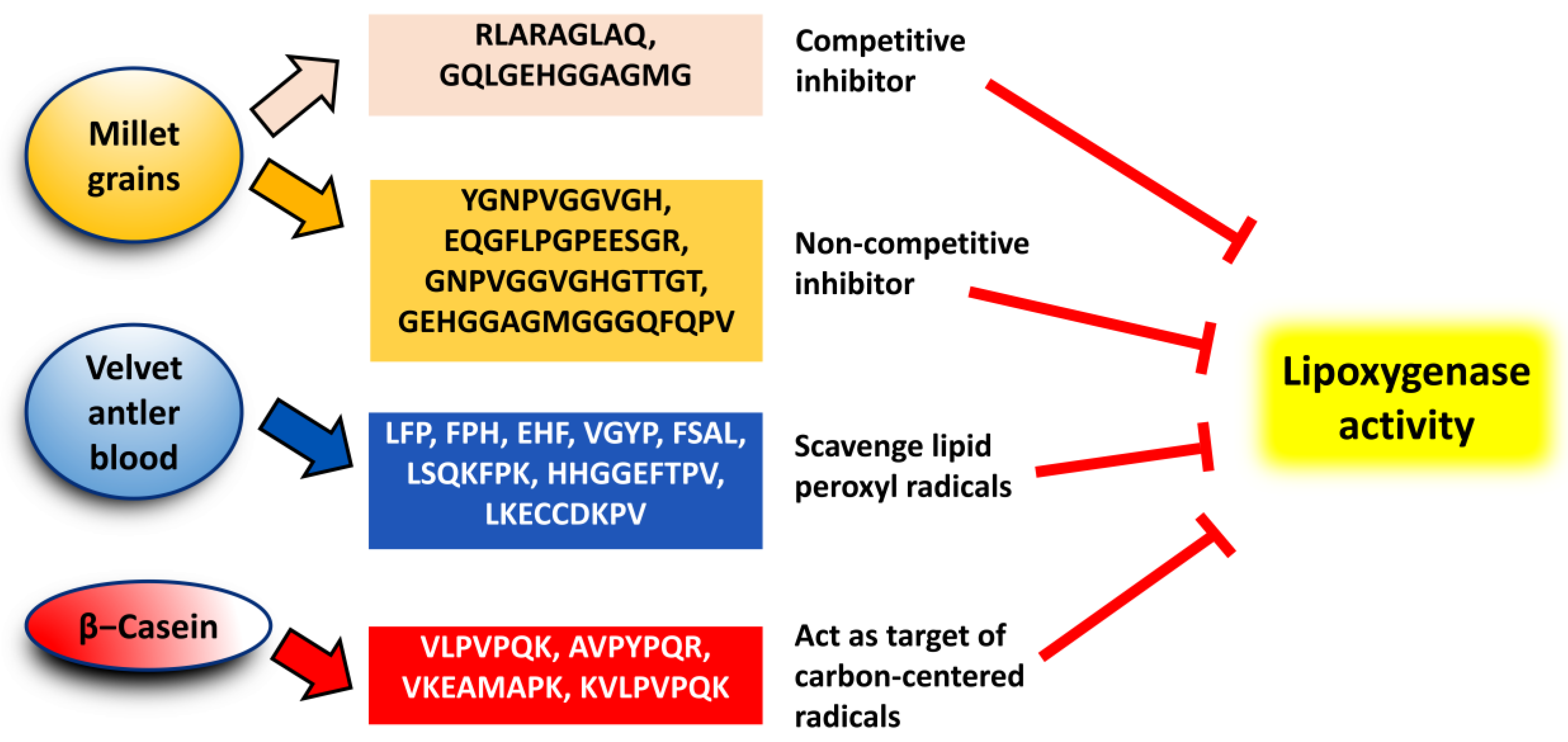
| Strategy | Raw Material | Reference |
|---|---|---|
| Pepsin and pancreatin | Velvet antler blood | [17] |
| Pepsin and pancreatin | Chia seed total protein isolate; chia seed protein fractions (albumin, globulin, prolamin, and glutelin) | [39] |
| α-Amylase, pepsin, pancreatin, and bile extract | Mealworm larvae, locusts, and crickets | [40] |
| α-Amylase, pepsin, pancreatin, and bile extract | Millet protein fractions (albumin, globulin 7S, globulin 11S, prolamin, and glutelin) | [38] |
| Trypsin | β-casein | [45] |
| Neutral protease and keratinase | Fish diet consisting of white fish meal, fermented soybean meal, shrimp meal, and blood meal | [42] |
| Pepsin-soluble collagen extraction method | Scales of the milkfish (Chanos chanos) | [20] |
| Keratinolytic bacteria; purified keratinase enzyme | Poultry feather keratin wastes | [19] |
| Peptide Sequence | Molecular Mass (Da) | Source | Reference |
|---|---|---|---|
| LFP | 375.22# | Velvet antler blood | [17] |
| FPH | 399.19# | ||
| EHF | 431.19# | ||
| VGYP | 434.22# | ||
| FSAL | 436.24# | ||
| LSQKFPK | 846.51# | ||
| HHGGEFTPV | 979.47# | ||
| LKECCDKPV | 1147.55# | ||
| VLPVPQK | 955.12 | β-casein | [45] |
| AVPYPQR | 956.03 | ||
| VKEAMAPK | 1402.48 | ||
| KVLPVPQK | 1070.14 | ||
| RLARAGLAQ | 1210.27 | Proso millet | [6,55] |
| YGNPVGGVGH | 1485.59 | ||
| EQGFLPGPEESGR | 955.12 | ||
| GQLGEHGGAGMG | 956.03 | ||
| GNPVGGVGHGTTGT | 1402.48 | ||
| GEHGGAGMGGGQFQPV | 1070.14 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).





