Submitted:
07 June 2023
Posted:
07 June 2023
You are already at the latest version
Abstract
Keywords:
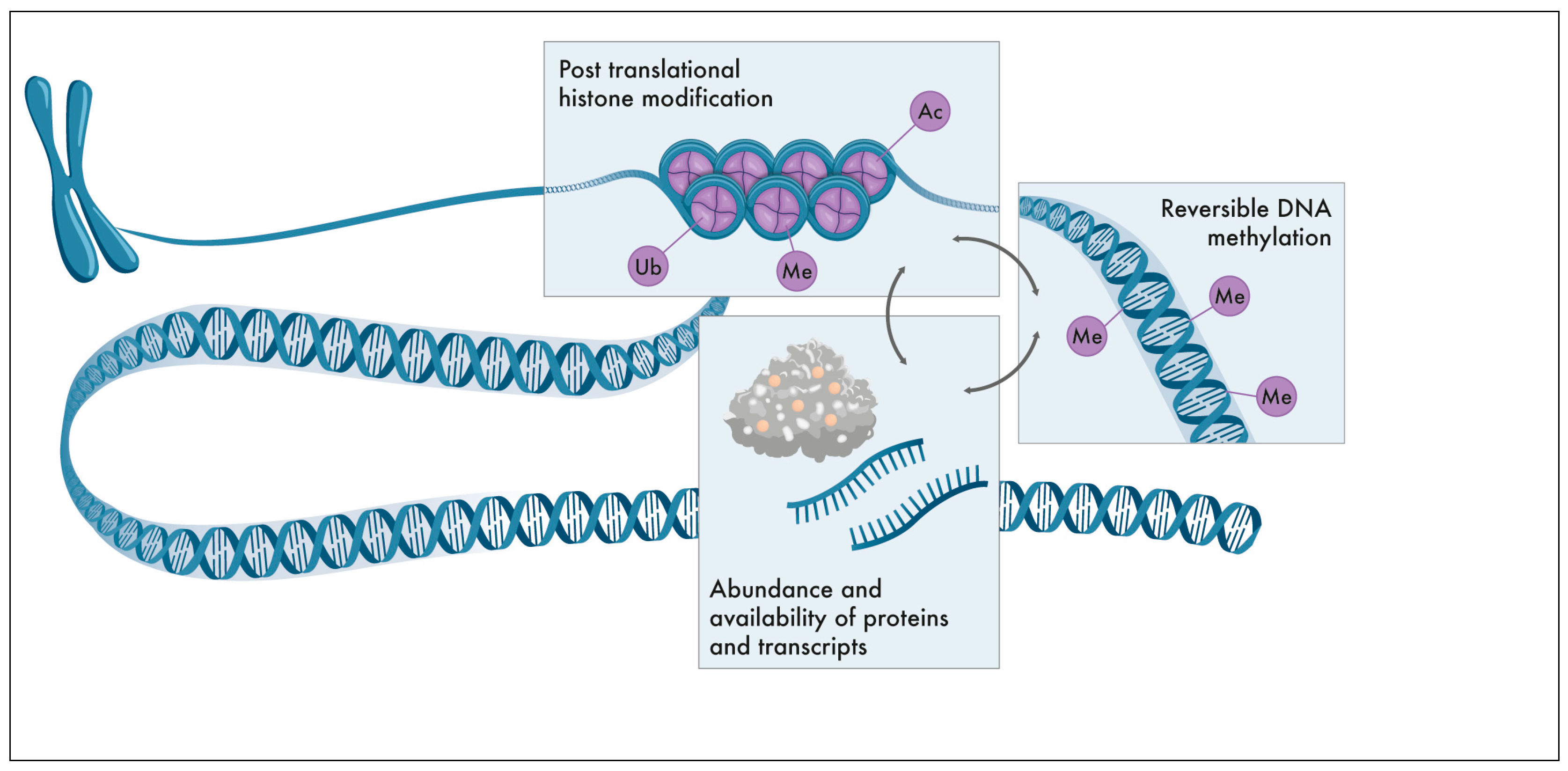
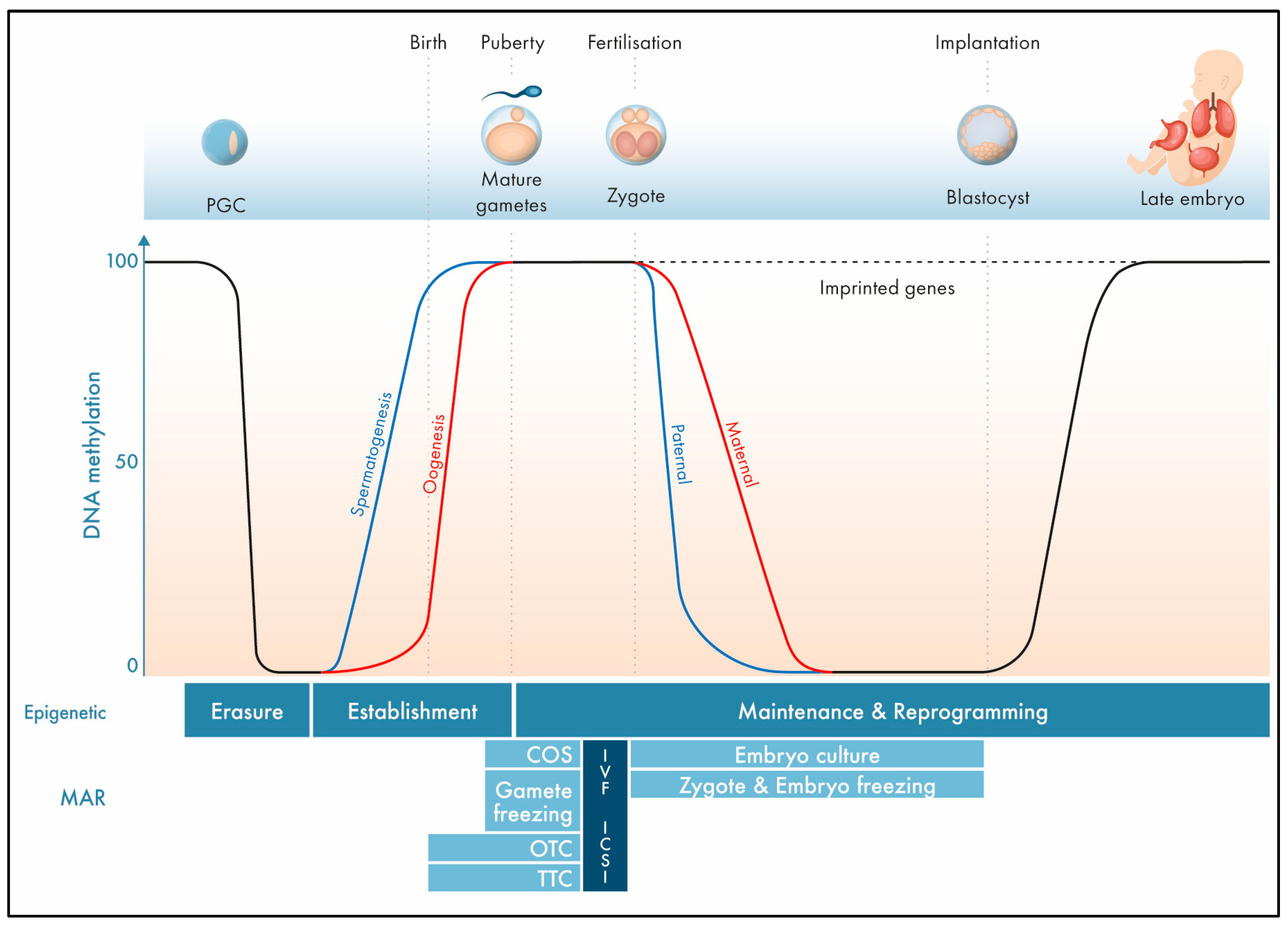
1. The Epigenome
2. Imprinting disorders
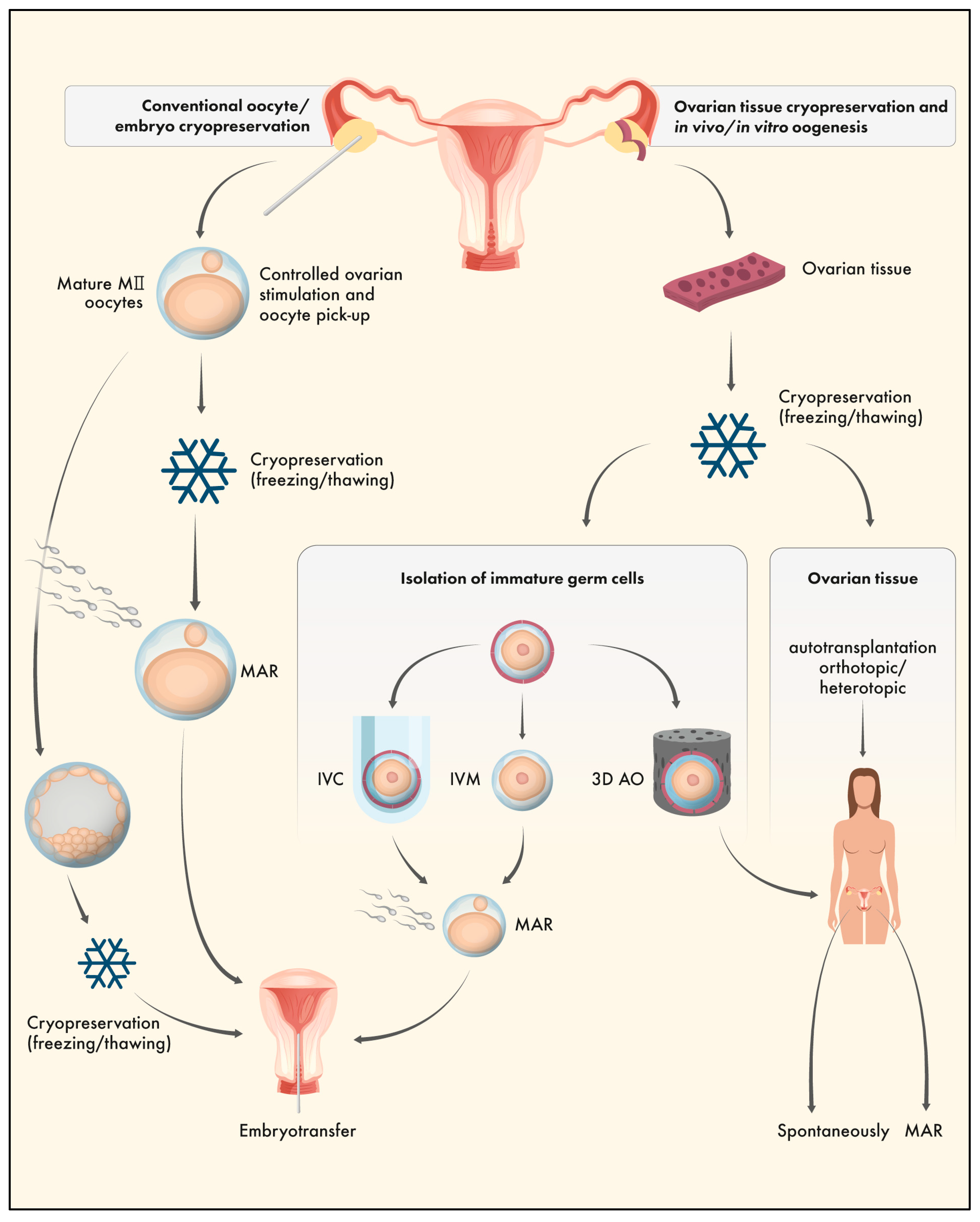
3. Ovarian tissue cryopreservation (OTC)
4. OTC and the Epigenome
Gene expression and protein abundance
DNA methylation patterns
Post-translational histone modifications
| Reference | Type/Species | Analysis | Main Outcome |
|---|---|---|---|
| Shirazi et al. (2016) [42] | Ovine GV oocytes, IVM | Epigenetically-relevant mRNA abundance in GV/embryos | Alteration of DNMT3B and HDAC1 |
| Demant et al. (2012) [44] | Murine pre-antral follicles, IVC | Proteome analysis in GV/MII | No differences between vitrified and non-vitrified GV/MII |
| He at al. (2018) [41] | Murine OTC | mRNA expression and protein abundance | Decreased mRNA/ protein levels for Dnmt1 |
| Yan et al. (2020) [43] | Murine OTC and orthotopic transplantation | Epigenetically relevant mRNA abundance in offspring | mRNA differences in H19, Igf2r and PLAGL1 but normal Snrpn expression |
| Yodrug et al. (2021) [49] | Bovine GV oocytes, IVM | Global DNA methylation in MII and embryos | Normal gDNA pattern in MII but altered in blastocysts |
| Al-Khtib et al. (2011) [48] | Human GV oocytes, IVM | Imprinted genes in MII | Normal pattern for H19 and KCNQ1OT1 |
| Yan et al. (2014) [47] | Murine GV oocytes, IVM | Global DNA methylation in MII | Normal gDNA pattern |
| Trapphoff et al. (2010) [50] | Murine pre-antral follicles, IVC | Imprinted genes in GV | Normal establishment of H19 and Igf2r imprinting but some single CpG errors in Snrpn |
| Yan et al. (2020) [43] | Murine OTC and orthotopic transplantation | Imprinted genes in offspring | Significant variations in H19, Igf2r, and PLAGL1 but normal Snrpn methylation |
| He et al. (2018) [41] | Murine OTC | Methylation pattern | Hypermethylation of the Grb10 promoter region |
| Wang et al. (2013) [46] | Murine OTC | Methylation pattern in GV after vitrification/warming | Normal Snrpn methylation |
| Sauvat et al. (2008) [52] | Murine OTC and grafting | Imprinted genes in offspring | Normal H19 and Lit1 methylation |
| Sauvat et al. (2013) [53] | OTC and grafting in ewes | Imprinted gene in offspring | Normal Igf2r methylation |
| Damavandi et al. (2021) [51] | Murine pre-antral follicles, IVC | CpG methylation in granulosa cells | Altered Inhba/Inhbb methylation |
| Yodrug et al. (2021) [49] | Bovine GV oocytes, IVM | Histone modifications | Normal H3K9me pattern in MII/blastocysts |
| Tian et al. (2022) [57] | Murine pre-antral follicles, IVC | Histone modifications in embryos | Normal histone pattern (H3K9me3, H3K4me3, H3K27ac) |
| Lee and Comizzoli (2019) [58] | Domestic cat GV | Histone modifications after vitrification | Normal H3K9me3 but altered H3K4me3 |
5. Conclusion - OTC and the Epigenome
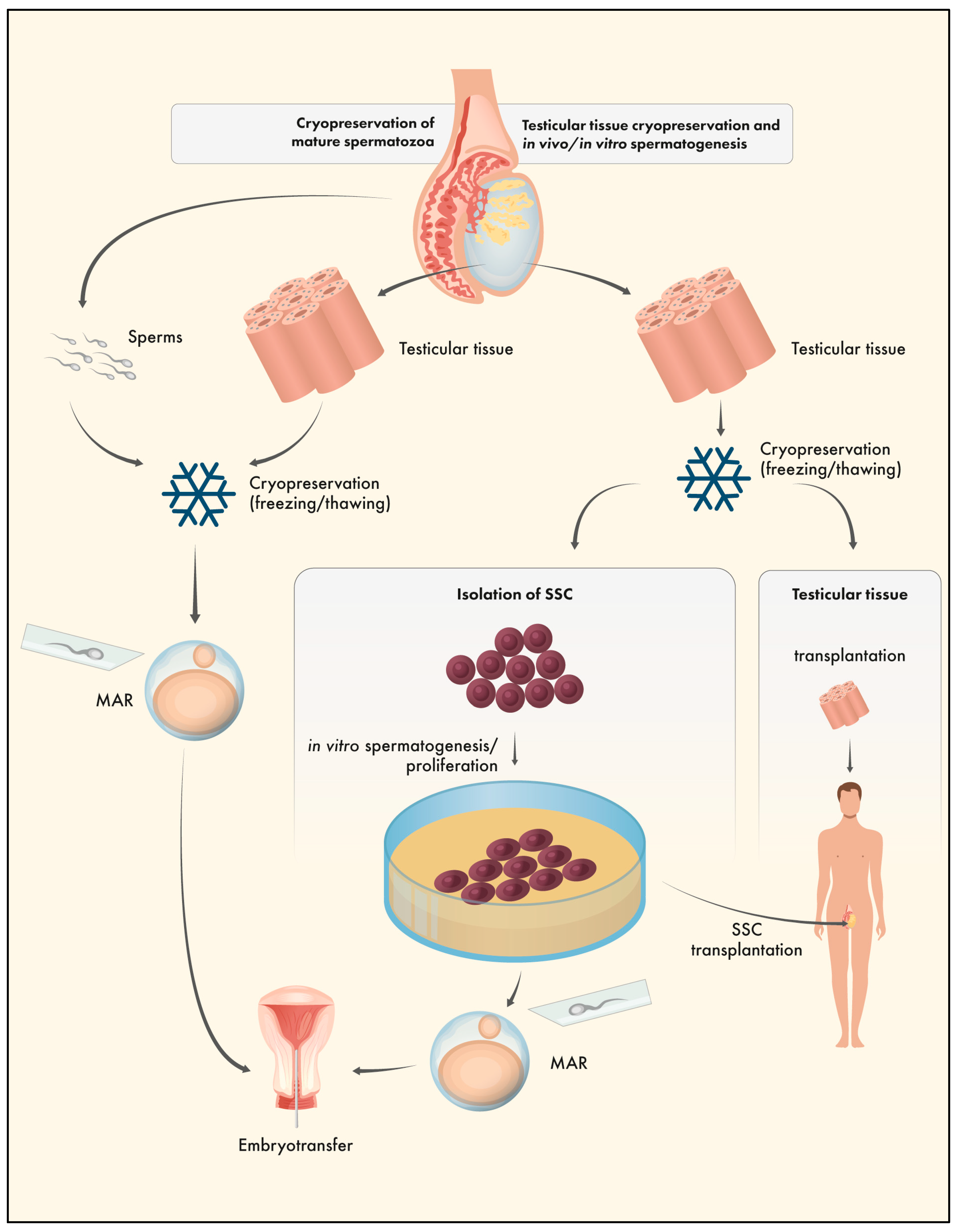
6. Testicular tissue cryopreservation (TTC)
7. TTC and the Epigenome
| Reference | Type/Species | Analysis | Main Outcome |
|---|---|---|---|
| Oblette et al. (2019) [77] | Murine in vitro culture of TT | Testicular tissue after IVC | Normal expression of epigenetic modification enzymes and gDNA methylation, but differences in histone modification |
| Oblette et al. (2021) [76] | Murine in vitro culture of TT | Pre-implantation embryo | Normal post-translational histone modifications and altered gDNA methylation |
8. Conclusion - TTC and the Epigenome
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Tucci, V.; Isles, A.R.; Kelsey, G.; Ferguson-Smith, A.C. Genomic Imprinting and Physiological Processes in Mammals. Cell 2019, 176, 952–965. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Gao, Z.; Qin, D.; Li, L. A Maternal Functional Module in the Mammalian Oocyte-To-Embryo Transition. Trends Mol. Med. 2017, 23, 1014–1023. [Google Scholar] [CrossRef] [PubMed]
- Swales, A.K.E.; Spears, N. Genomic Imprinting and Reproduction. Reproduction 2005, 130, 389–399. [Google Scholar] [CrossRef]
- Anvar, Z.; Chakchouk, I.; Demond, H.; Sharif, M.; Kelsey, G.; Van den Veyver, I.B. DNA Methylation Dynamics in the Female Germline and Maternal-Effect Mutations That Disrupt Genomic Imprinting. Genes (Basel). 2021, 12, 1214. [Google Scholar] [CrossRef] [PubMed]
- Demond, H.; Kelsey, G. The Enigma of DNA Methylation in the Mammalian Oocyte. F1000Research 2020, 9, F1000. [Google Scholar] [CrossRef] [PubMed]
- Weaver, J.R.; Bartolomei, M.S. Chromatin Regulators of Genomic Imprinting. Biochim. Biophys. Acta 2014, 1839, 169–177. [Google Scholar] [CrossRef]
- Nicholls, R.D.; Saitoh, S.; Horsthemke, B. Imprinting in Prader-Willi and Angelman Syndromes. Trends Genet. 1998, 14, 194–200. [Google Scholar] [CrossRef] [PubMed]
- Poole, R.L.; Leith, D.J.; Docherty, L.E.; Shmela, M.E.; Gicquel, C.; Splitt, M.; Temple, I.K.; Mackay, D.J.G. Beckwith-Wiedemann Syndrome Caused by Maternally Inherited Mutation of an OCT-Binding Motif in the IGF2/H19-Imprinting Control Region, ICR1. Eur. J. Hum. Genet. 2012, 20, 240–243. [Google Scholar] [CrossRef]
- Ishida, M. New Developments in Silver-Russell Syndrome and Implications for Clinical Practice. Epigenomics 2016, 8, 563–580. [Google Scholar] [CrossRef]
- Begemann, M.; Rezwan, F.I.; Beygo, J.; Docherty, L.E.; Kolarova, J.; Schroeder, C.; Buiting, K.; Chokkalingam, K.; Degenhardt, F.; Wakeling, E.L.; et al. Maternal Variants in NLRP and Other Maternal Effect Proteins Are Associated with Multilocus Imprinting Disturbance in Offspring. J. Med. Genet. 2018, 55, 497–504. [Google Scholar] [CrossRef]
- Boonen, S.E.; Mackay, D.J.G.; Hahnemann, J.M.D.; Docherty, L.; Grønskov, K.; Lehmann, A.; Larsen, L.G.; Haemers, A.P.; Kockaerts, Y.; Dooms, L.; et al. Transient Neonatal Diabetes, ZFP57, and Hypomethylation of Multiple Imprinted Loci: A Detailed Follow-Up. Diabetes Care 2013, 36, 505–512. [Google Scholar] [CrossRef] [PubMed]
- Kou, Y.C.; Shao, L.; Peng, H.H.; Rosetta, R.; del Gaudio, D.; Wagner, A.F.; Al-Hussaini, T.K.; Van den Veyver, I.B. A Recurrent Intragenic Genomic Duplication, Other Novel Mutations in NLRP7 and Imprinting Defects in Recurrent Biparental Hydatidiform Moles. Mol. Hum. Reprod. 2008, 14, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Mackay, D.J.G.; Callaway, J.L.A.; Marks, S.M.; White, H.E.; Acerini, C.L.; Boonen, S.E.; Dayanikli, P.; Firth, H.V.; Goodship, J.A.; Haemers, A.P.; et al. Hypomethylation of Multiple Imprinted Loci in Individuals with Transient Neonatal Diabetes Is Associated with Mutations in ZFP57. Nat. Genet. 2008, 40, 949–951. [Google Scholar] [CrossRef] [PubMed]
- Barberet, J.; Binquet, C.; Guilleman, M.; Doukani, A.; Choux, C.; Bruno, C.; Bourredjem, A.; Chapusot, C.; Bourc’his, D.; Duffourd, Y.; Fauque, P. Do Assisted Reproductive Technologies and in Vitro Embryo Culture Influence the Epigenetic Control of Imprinted Genes and Transposable Elements in Children? Hum. Reprod. 2021, 36, 479–492. [Google Scholar] [CrossRef] [PubMed]
- Barberet, J.; Romain, G.; Binquet, C.; Guilleman, M.; Bruno, C.; Ginod, P.; Chapusot, C.; Choux, C.; Fauque, P. Do Frozen Embryo Transfers Modify the Epigenetic Control of Imprinted Genes and Transposable Elements in Newborns Compared with Fresh Embryo Transfers and Natural Conceptions? Fertil. Steril. 2021, 116, 1468–1480. [Google Scholar] [CrossRef]
- Hattori, H.; Hiura, H.; Kitamura, A.; Miyauchi, N.; Kobayashi, N.; Takahashi, S.; Okae, H.; Kyono, K.; Kagami, M.; Ogata, T.; Arima, T. Association of Four Imprinting Disorders and ART. Clin. Epigenetics 2019, 11, 21. [Google Scholar] [CrossRef]
- Yao, J.-F.; Huang, Y.-F.; Huang, R.-F.; Lin, S.-X.; Guo, C.-Q.; Hua, C.-Z.; Wu, P.-Y.; Hu, J.-F.; Li, Y.-Z. Effects of Vitrification on the Imprinted Gene Snrpn in Neonatal Placental Tissue. Reprod. Dev. Med. 2020, 4, 25–31. [Google Scholar] [CrossRef]
- Ma, Y.; Ma, Y.; Wen, L.; Lei, H.; Chen, S.; Wang, X. Changes in DNA Methylation and Imprinting Disorders in E9.5 Mouse Fetuses and Placentas Derived from Vitrified Eight-Cell Embryos. Mol. Reprod. Dev. 2019, 86, 404–415. [Google Scholar] [CrossRef]
- Wang, Z.; Xu, L.; He, F. Embryo Vitrification Affects the Methylation of the H19/Igf2 Differentially Methylated Domain and the Expression of H19 and Igf2. Fertil. Steril. 2010, 93, 2729–2733. [Google Scholar] [CrossRef]
- Henningsen, A.A.; Gissler, M.; Rasmussen, S.; Opdahl, S.; Wennerholm, U.B.; Spangsmose, A.L.; Tiitinen, A.; Bergh, C.; Romundstad, L.B.; Laivuori, H.; et al. Imprinting Disorders in Children Born after ART: A Nordic Study from the CoNARTaS Group. Hum. Reprod. 2020, 35, 1178–1184. [Google Scholar] [CrossRef]
- Lazaraviciute, G.; Kauser, M.; Bhattacharya, S.; Haggarty, P.; Bhattacharya, S. A Systematic Review and Meta-Analysis of DNA Methylation Levels and Imprinting Disorders in Children Conceived by IVF/ICSI Compared with Children Conceived Spontaneously. Hum. Reprod. Update 2014, 20, 840–852. [Google Scholar] [CrossRef]
- Deanesly, R. Immature Rat Ovaries Grafted after Freezing and Thawing. J. Endocrinol. 1954, 11, 197–200. [Google Scholar] [CrossRef] [PubMed]
- Donnez, J.; Dolmans, M.M.; Demylle, D.; Jadoul, P.; Pirard, C.; Squifflet, J.; Martinez-Madrid, B.; van Langendonckt, A. Livebirth after Orthotopic Transplantation of Cryopreserved Ovarian Tissue. Lancet 2004, 364, 1405–1410. [Google Scholar] [CrossRef] [PubMed]
- Yding Andersen, C.; Mamsen, L.S.; Kristensen, S.G. Fertility preservation: Freezing of Ovarian Tissue and Clinical Opportunities. Reproduction 2019, 158, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Kawamura, K.; Cheng, Y.; Suzuki, N.; Deguchi, M.; Sato, Y.; Takae, S.; Ho, C.; Kawamura, N.; Tamura, M.; Hashimoto, S.; et al. Hippo Signaling Disruption and Akt Stimulation of Ovarian Follicles for Infertility Treatment. Proc. Natl. Acad. Sci. U. S. A. 2013, 110, 17474–17479. [Google Scholar] [CrossRef]
- Suzuki, N.; Yoshioka, N.; Takae, S.; Sugishita, Y.; Tamura, M.; Hashimoto, S.; Morimoto, Y.; Kawamura, K. Successful Fertility Preservation Following Ovarian Tissue Vitrification in Patients with Primary Ovarian Insufficiency. Hum. Reprod. 2015, 30, 608–615. [Google Scholar] [CrossRef]
- Rivas Leonel, E.C.; Lucci, C.M.; Amorim, C.A. Cryopreservation of Human Ovarian Tissue: A Review. Transfus. Med. hemotherapy 2019, 46, 173–181. [Google Scholar] [CrossRef]
- Cheng, H.; Han, Y.; Zhang, J.; Zhang, S.; Zhai, Y.; An, X.; Li, Q.; Duan, J.; Zhang, X.; Li, Z.; Tang, B.; Shen, H. Effects of Dimethyl Sulfoxide (DMSO) on DNA Methylation and Histone Modification in Parthenogenetically Activated Porcine Embryos. Reprod. Fertil. Dev. 2022, 34, 598–607. [Google Scholar] [CrossRef]
- Iwatani, M.; Ikegami, K.; Kremenska, Y.; Hattori, N.; Tanaka, S.; Yagi, S.; Shiota, K. Dimethyl Sulfoxide Has an Impact on Epigenetic Profile in Mouse Embryoid Body. Stem Cells 2006, 24, 2549–2556. [Google Scholar] [CrossRef]
- Verheijen, M.; Lienhard, M.; Schrooders, Y.; Clayton, O.; Nudischer, R.; Boerno, S.; Timmermann, B.; Selevsek, N.; Schlapbach, R.; Gmuender, H.; et al. DMSO Induces Drastic Changes in Human Cellular Processes and Epigenetic Landscape in Vitro. Sci. Rep. 2019, 9, 4641. [Google Scholar] [CrossRef]
- Nakamura, Y.; Obata, R.; Okuyama, N.; Aono, N.; Hashimoto, T.; Kyono, K. Residual Ethylene Glycol and Dimethyl Sulphoxide Concentration in Human Ovarian Tissue during Warming/Thawing Steps Following Cryopreservation. Reprod. Biomed. Online 2017, 35, 311–313. [Google Scholar] [CrossRef]
- Dolmans, M.-M.; Donnez, J.; Cacciottola, L. Fertility Preservation: The Challenge of Freezing and Transplanting Ovarian Tissue. Trends Mol. Med. 2021, 27, 777–791. [Google Scholar] [CrossRef]
- O’Brien, M.J.; Pendola, J.K.; Eppig, J.J. A Revised Protocol for in Vitro Development of Mouse Oocytes from Primordial Follicles Dramatically Improves Their Developmental Competence. Biol. Reprod. 2003, 68, 1682–1686. [Google Scholar] [CrossRef]
- Telfer, E.E.; Andersen, C.Y. In Vitro Growth and Maturation of Primordial Follicles and Immature Oocytes. Fertil. Steril. 2021, 115, 1116–1125. [Google Scholar] [CrossRef]
- McLaughlin, M.; Albertini, D.F.; Wallace, W.H.B.; Anderson, R.A.; Telfer, E.E. Metaphase II Oocytes from Human Unilaminar Follicles Grown in a Multi-Step Culture System. Mol. Hum. Reprod. 2018, 24, 135–142. [Google Scholar] [CrossRef]
- Chen, H.; Zhang, L.; Wang, Z.; Chang, H.; Xie, X.; Fu, L.; Zhang, Y.; Quan, F. Resveratrol Improved the Developmental Potential of Oocytes after Vitrification by Modifying the Epigenetics. Mol. Reprod. Dev. 2019, 86, 862–870. [Google Scholar] [CrossRef]
- Cheng, K.-R.; Fu, X.-W.; Zhang, R.-N.; Jia, G.-X.; Hou, Y.-P.; Zhu, S.-E. Effect of Oocyte Vitrification on Deoxyribonucleic Acid Methylation of H19, Peg3, and Snrpn Differentially Methylated Regions in Mouse Blastocysts. Fertil. Steril. 2014, 102, 1183–1190. [Google Scholar] [CrossRef]
- Ma, Y.; Long, C.; Liu, G.; Bai, H.; Ma, L.; Bai, T.; Zuo, Y.; Li, S. WGBS Combined with RNA-Seq Analysis Revealed That Dnmt1 Affects the Methylation Modification and Gene Expression Changes during Mouse Oocyte Vitrification. Theriogenology 2022, 177, 11–21. [Google Scholar] [CrossRef]
- Gu, R.; Ge, N.; Huang, B.; Fu, J.; Zhang, Y.; Wang, N.; Xu, Y.; Li, L.; Peng, X.; Zou, Y.; Sun, Y.; Sun, X. Impacts of Vitrification on the Transcriptome of Human Ovarian Tissue in Patients with Gynecological Cancer. Front. Genet. 2023, 14, 1114650. [Google Scholar] [CrossRef]
- Zhou, Y.; Wang, W.; Todorov, P.; Pei, C.; Isachenko, E.; Rahimi, G.; Mallmann, P.; Nawroth, F.; Isachenko, V. RNA Transcripts in Human Ovarian Cells: Two-Time Cryopreservation Does Not Affect Developmental Potential. Int. J. Mol. Sci. 2023, 24, 6880. [Google Scholar] [CrossRef]
- He, Z.-Y.; Wang, H.-Y.; Zhou, X.; Liang, X.-Y.; Yan, B.; Wang, R.; Ma, L.-H.; Wang, Y.-L. Evaluation of Vitrification Protocol of Mouse Ovarian Tissue by Effect of DNA Methyltransferase-1 and Paternal Imprinted Growth Factor Receptor-Binding Protein 10 on Signaling Pathways. Cryobiology 2018, 80, 89–95. [Google Scholar] [CrossRef]
- Shirazi, A.; Naderi, M.M.; Hassanpour, H.; Heidari, M.; Borjian, S.; Sarvari, A.; Akhondi, M.M. The Effect of Ovine Oocyte Vitrification on Expression of Subset of Genes Involved in Epigenetic Modifications during Oocyte Maturation and Early Embryo Development. Theriogenology 2016, 86, 2136–2146. [Google Scholar] [CrossRef]
- Yan, Z.; Li, Q.; Zhang, L.; Kang, B.; Fan, W.; Deng, T.; Zhu, J.; Wang, Y. The Growth and Development Conditions in Mouse Offspring Derived from Ovarian Tissue Cryopreservation and Orthotopic Transplantation. J. Assist. Reprod. Genet. 2020, 37, 923–932. [Google Scholar] [CrossRef]
- Demant, M.; Trapphoff, T.; Fröhlich, T.; Arnold, G.J.; Eichenlaub-Ritter, U. Vitrification at the Pre-Antral Stage Transiently Alters Inner Mitochondrial Membrane Potential but Proteome of in Vitro Grown and Matured Mouse Oocytes Appears Unaffected. Hum. Reprod. 2012, 27, 1096–1111. [Google Scholar] [CrossRef]
- Pfeiffer, M.J.; Siatkowski, M.; Paudel, Y.; Balbach, S.T.; Baeumer, N.; Crosetto, N.; Drexler, H.C.A.; Fuellen, G.; Boiani, M. Proteomic Analysis of Mouse Oocytes Reveals 28 Candidate Factors of the “Reprogrammome”. J. Proteome Res. 2011, 10, 2140–2153. [Google Scholar] [CrossRef]
- Wang, H.-Y.; Li, Y.-H.; Sun, L.; Gao, X.; You, L.; Wang, Y.; Ma, J.-L.; Chen, Z.-J. Allotransplantation of Cryopreserved Prepubertal Mouse Ovaries Restored Puberty and Fertility without Affecting Methylation Profile of Snrpn-DMR. Fertil. Steril. 2013, 99, 241–247. [Google Scholar] [CrossRef]
- Yan, J.; Zhang, L.; Wang, T.; Li, R.; Liu, P.; Yan, L.; Qiao, J. Effect of Vitrification at the Germinal Vesicle Stage on the Global Methylation Status in Mouse Oocytes Subsequently Matured in Vitro. Chin. Med. J. (Engl). 2014, 127, 4019–4024. [Google Scholar] [CrossRef]
- Al-Khtib, M.; Perret, A.; Khoueiry, R.; Ibala-Romdhane, S.; Blachère, T.; Greze, C.; Lornage, J.; Lefèvre, A. Vitrification at the Germinal Vesicle Stage Does Not Affect the Methylation Profile of H19 and KCNQ1OT1 Imprinting Centers in Human Oocytes Subsequently Matured in Vitro. Fertil. Steril. 2011, 95, 1955–1960. [Google Scholar] [CrossRef]
- Yodrug, T.; Parnpai, R.; Hirao, Y.; Somfai, T. Effect of Vitrification at Different Meiotic Stages on Epigenetic Characteristics of Bovine Oocytes and Subsequently Developing Embryos. Anim. Sci. J. 2021, 92, e13596. [Google Scholar] [CrossRef]
- Trapphoff, T.; El Hajj, N.; Zechner, U.; Haaf, T.; Eichenlaub-Ritter, U. DNA Integrity, Growth Pattern, Spindle Formation, Chromosomal Constitution and Imprinting Patterns of Mouse Oocytes from Vitrified Pre-Antral Follicles. Hum. Reprod. 2010, 25, 3025–3042. [Google Scholar] [CrossRef]
- Damavandi, M.; Farrokh, P.; Zavareh, S. Effect of Mouse Ovarian Vitrification on Promoter Methylation of Inhba and Inhbb in Granulosa Cells of Follicles. Cryo Letters 2021, 42, 67–72. [Google Scholar]
- Sauvat, F.; Capito, C.; Sarnacki, S.; Poirot, C.; Bachelot, A.; Meduri, G.; Dandolo, L.; Binart, N. Immature Cryopreserved Ovary Restores Puberty and Fertility in Mice without Alteration of Epigenetic Marks. PLoS One 2008, 3, e1972. [Google Scholar] [CrossRef]
- Sauvat, F.; Bouilly, J.; Capito, C.; Lefèvre, A.; Blachère, T.; Borenstein, N.; Sarnacki, S.; Dandolo, L.; Binart, N. Ovarian Function Is Restored after Grafting of Cryopreserved Immature Ovary in Ewes. FASEB J. 2013, 27, 1511–1518. [Google Scholar] [CrossRef]
- Bonnet-Garnier, A.; Feuerstein, P.; Chebrout, M.; Fleurot, R.; Jan, H.-U.; Debey, P.; Beaujean, N. Genome Organization and Epigenetic Marks in Mouse Germinal Vesicle Oocytes. Int. J. Dev. Biol. 2012, 56, (10–12). [Google Scholar] [CrossRef]
- Kageyama, S.; Liu, H.; Kaneko, N.; Ooga, M.; Nagata, M.; Aoki, F. Alterations in Epigenetic Modifications during Oocyte Growth in Mice. Reproduction 2007, 133, 85–94. [Google Scholar] [CrossRef]
- Stewart, K.R.; Veselovska, L.; Kim, J.; Huang, J.; Saadeh, H.; Tomizawa, S.; Smallwood, S.A.; Chen, T.; Kelsey, G. Dynamic Changes in Histone Modifications Precede de Novo DNA Methylation in Oocytes. Genes Dev. 2015, 29, 2449–2462. [Google Scholar] [CrossRef]
- Tian, C.; Shen, L.; Gong, C.; Cao, Y.; Shi, Q.; Zhao, G. Microencapsulation and Nanowarming Enables Vitrification Cryopreservation of Mouse Preantral Follicles. Nat. Commun. 2022, 13, 7515. [Google Scholar] [CrossRef]
- Lee, P.-C.; Comizzoli, P. Desiccation and Supra-Zero Temperature Storage of Cat Germinal Vesicles Lead to Less Structural Damage and Similar Epigenetic Alterations Compared to Cryopreservation. Mol. Reprod. Dev. 2019, 86, 1822–1831. [Google Scholar] [CrossRef]
- Tamburrino, L.; Traini, G.; Marcellini, A.; Vignozzi, L.; Baldi, E.; Marchiani, S. Cryopreservation of Human Spermatozoa: Functional, Molecular and Clinical Aspects. Int. J. Mol. Sci. 2023, 24, 4656. [Google Scholar] [CrossRef]
- Picton, H.M.; Wyns, C.; Anderson, R.A.; Goossens, E.; Jahnukainen, K.; Kliesch, S.; Mitchell, R.T.; Pennings, G.; Rives, N.; Tournaye, H.; et al. A European Perspective on Testicular Tissue Cryopreservation for Fertility Preservation in Prepubertal and Adolescent Boys. Hum. Reprod. 2015, 30, 2463–2475. [Google Scholar] [CrossRef]
- Delgouffe, E.; Braye, A.; Goossens, E. Testicular Tissue Banking for Fertility Preservation in Young Boys: Which Patients Should Be Included? Front. Endocrinol. (Lausanne). 2022, 13, 854186. [Google Scholar] [CrossRef]
- Sanou, I.; van Maaren, J.; Eliveld, J.; Lei, Q.; Meißner, A.; de Melker, A.A.; Hamer, G.; van Pelt, A.M.M.; Mulder, C.L. Spermatogonial Stem Cell-Based Therapies: Taking Preclinical Research to the Next Level. Front. Endocrinol. (Lausanne). 2022, 13, 850219. [Google Scholar] [CrossRef]
- Wyns, C.; Kanbar, M.; Giudice, M.G.; Poels, J. Fertility Preservation for Prepubertal Boys: Lessons Learned from the Past and Update on Remaining Challenges towards Clinical Translation. Hum. Reprod. Update 2021, 27, 433–459. [Google Scholar] [CrossRef]
- Onofre, J.; Baert, Y.; Faes, K.; Goossens, E. Cryopreservation of Testicular Tissue or Testicular Cell Suspensions: A Pivotal Step in Fertility Preservation. Hum. Reprod. Update 2016, 22, 744–761. [Google Scholar] [CrossRef]
- Onofre, J.; Kadam, P.; Baert, Y.; Goossens, E. Testicular Tissue Cryopreservation Is the Preferred Method to Preserve Spermatogonial Stem Cells Prior to Transplantation. Reprod. Biomed. Online 2020, 40, 261–269. [Google Scholar] [CrossRef]
- Fayomi, A.P.; Peters, K.; Sukhwani, M.; Valli-Pulaski, H.; Shetty, G.; Meistrich, M.L.; Houser, L.; Robertson, N.; Roberts, V.; Ramsey, C.; et al. Autologous Grafting of Cryopreserved Prepubertal Rhesus Testis Produces Sperm and Offspring. Science 2019, 363, 1314–1319. [Google Scholar] [CrossRef]
- Goossens, E.; De Rycke, M.; Haentjens, P.; Tournaye, H. DNA Methylation Patterns of Spermatozoa and Two Generations of Offspring Obtained after Murine Spermatogonial Stem Cell Transplantation. Hum. Reprod. 2009, 24, 2255–2263. [Google Scholar] [CrossRef]
- Serrano, J.B.; van Eekelen, R.; de Winter-Korver, C.M.; van Daalen, S.K.M.; Tabeling, N.C.; Catsburg, L.A.E.; Gijbels, M.J.J.; Mulder, C.L.; van Pelt, A.M.M. Impact of Restoring Male Fertility with Transplantation of in Vitro Propagated Spermatogonial Stem Cells on the Health of Their Offspring throughout Life. Clin. Transl. Med. 2021, 11, e531. [Google Scholar] [CrossRef]
- Nickkholgh, B.; Mizrak, S.C.; van Daalen, S.K.M.; Korver, C.M.; Sadri-Ardekani, H.; Repping, S.; van Pelt, A.M.M. Genetic and Epigenetic Stability of Human Spermatogonial Stem Cells during Long-Term Culture. Fertil. Steril. 2014, 102, 1700–1707. [Google Scholar] [CrossRef] [PubMed]
- Kanatsu-Shinohara, M.; Ogonuki, N.; Iwano, T.; Lee, J.; Kazuki, Y.; Inoue, K.; Miki, H.; Takehashi, M.; Toyokuni, S.; Shinkai, Y.; et al. Genetic and Epigenetic Properties of Mouse Male Germline Stem Cells during Long-Term Culture. Development 2005, 132, 4155–4163. [Google Scholar] [CrossRef]
- Langenstroth-Röwer, D.; Gromoll, J.; Wistuba, J.; Tröndle, I.; Laurentino, S.; Schlatt, S.; Neuhaus, N. De Novo Methylation in Male Germ Cells of the Common Marmoset Monkey Occurs during Postnatal Development and Is Maintained in Vitro. Epigenetics 2017, 12, 527–539. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; Katagiri, K.; Gohbara, A.; Inoue, K.; Ogonuki, N.; Ogura, A.; Kubota, Y.; Ogawa, T. In Vitro Production of Functional Sperm in Cultured Neonatal Mouse Testes. Nature 2011, 471, 504–507. [Google Scholar] [CrossRef] [PubMed]
- Yokonishi, T.; Sato, T.; Komeya, M.; Katagiri, K.; Kubota, Y.; Nakabayashi, K.; Hata, K.; Inoue, K.; Ogonuki, N.; Ogura, A.; Ogawa, T. Offspring Production with Sperm Grown in Vitro from Cryopreserved Testis Tissues. Nat. Commun. 2014, 5, 4320. [Google Scholar] [CrossRef]
- Wu, X.; Goodyear, S.M.; Abramowitz, L.K.; Bartolomei, M.S.; Tobias, J.W.; Avarbock, M.R.; Brinster, R.L. Fertile Offspring Derived from Mouse Spermatogonial Stem Cells Cryopreserved for More than 14 Years. Hum. Reprod. 2012, 27, 1249–1259. [Google Scholar] [CrossRef] [PubMed]
- Serrano, J.B.; Tabeling, N.C.; de Winter-Korver, C.M.; van Daalen, S.K.M.; van Pelt, A.M.M.; Mulder, C.L. Sperm DNA Methylation Is Predominantly Stable in Mice Offspring Born after Transplantation of Long-Term Cultured Spermatogonial Stem Cells. Clin. Epigenetics 2023, 15, 58. [Google Scholar] [CrossRef] [PubMed]
- Oblette, A.; Rives-Feraille, A.; Dumont, L.; Delessard, M.; Saulnier, J.; Rives, N.; Rondanino, C. Dynamics of Epigenetic Modifications in ICSI Embryos from in Vitro-Produced Spermatozoa. Andrology 2021, 9, 640–656. [Google Scholar] [CrossRef] [PubMed]
- Oblette, A.; Rondeaux, J.; Dumont, L.; Delessard, M.; Saulnier, J.; Rives, A.; Rives, N.; Rondanino, C. DNA Methylation and Histone Post-Translational Modifications in the Mouse Germline Following in-Vitro Maturation of Fresh or Cryopreserved Prepubertal Testicular Tissue. Reprod. Biomed. Online 2019, 39, 383–401. [Google Scholar] [CrossRef]
- Kawai, K.; Li, Y.-S.; Song, M.-F.; Kasai, H. DNA Methylation by Dimethyl Sulfoxide and Methionine Sulfoxide Triggered by Hydroxyl Radical and Implications for Epigenetic Modifications. Bioorg. Med. Chem. Lett. 2010, 20, 260–265. [Google Scholar] [CrossRef]
- Zeng, C.; Peng, W.; Ding, L.; He, L.; Zhang, Y.; Fang, D.; Tang, K. A Preliminary Study on Epigenetic Changes during Boar Spermatozoa Cryopreservation. Cryobiology 2014, 69, 119–127. [Google Scholar] [CrossRef]
- Khosravizadeh, Z.; Hassanzadeh, G.; Tavakkoly Bazzaz, J.; Alizadeh, F.; Totonchi, M.; Salehi, E.; Khodamoradi, K.; Khanehzad, M.; Hosseini, S.R.; Abolhassani, F. The Effect of Cryopreservation on DNA Methylation Patterns of the Chromosome 15q11-Q13 Region in Human Spermatozoa. Cell Tissue Bank. 2020, 21, 433–445. [Google Scholar] [CrossRef]
- Kläver, R.; Bleiziffer, A.; Redmann, K.; Mallidis, C.; Kliesch, S.; Gromoll, J. Routine Cryopreservation of Spermatozoa Is Safe-Evidence from the DNA Methylation Pattern of Nine Spermatozoa Genes. J. Assist. Reprod. Genet. 2012, 29, 943–950. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




