2. Review of Related works
A study (Idriss et al, 2021) revealed that the proliferation of innovative technologies has led to a significant increase in digital livestock disease data in past decay. In the past few years, it observed that the pace of digital disruption has been spectacular, transforming every sector of the economy, including animal production, health, and welfare The potential benefits of incorporating new digital technologies in animal health are convincing and can likely reveal new models that make national veterinary services more competent for meeting the required standards for animal welfare and health practice (Idriss et al, 2021). However, gaining the full potential advantages and anticipated results of the digital revolution is challenging in all sectors in general veterinary services in particular
In another research study (Bernard, 2012) the capability of easily capturing massive volumes of diagnostic data and health information has created the opportunity to identify patterns and risk factors not only for individual animals but across herds, regions, and species. These insights have renovated the field of diagnostics into a tool of prevention, supporting animal health professionals to take immediate action with confidence. Because the early notification of disease events or outbreaks determines the effective containment and prediction of epidemic diseases
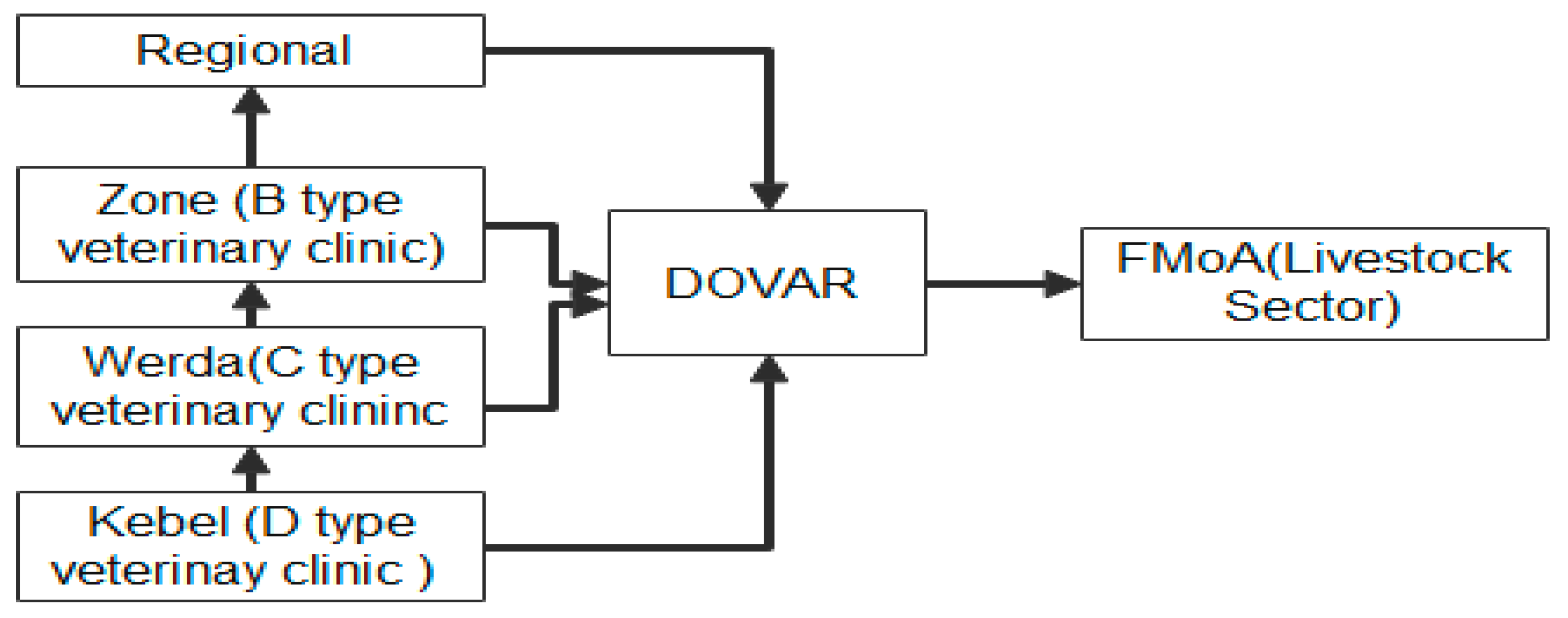
Some of the important reports reported that the Livestock disease data sources can have different types of data that we collect for the aim of analysis. These include routine surveillance such as Disease Outbreak and Vaccination Activity Report (DOVAR), Animal Disease Notification and Investigation System (ADNIS), and the World Animal Health Information System (WAHIS). WAHIS is a web application platform for World Organization for Animal Health (OIE) Member to fulfill their obligation by supplying information on any relevant domestic animal or wildlife disease, including zoonosis, identified or detected within their territory (World Organization For Animal Health, 2014). Though these data are found in the form of electronic data as aggregate data.
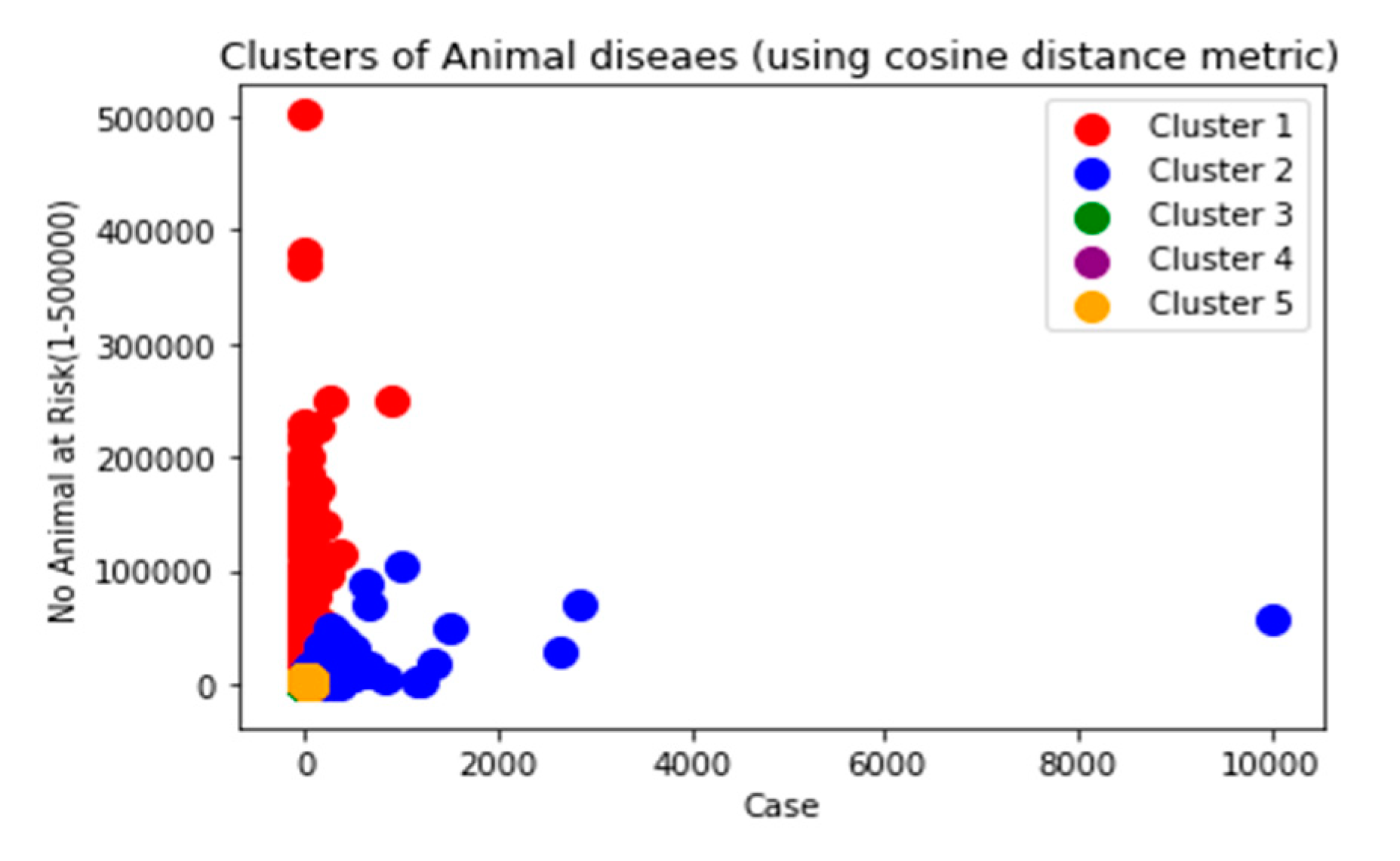
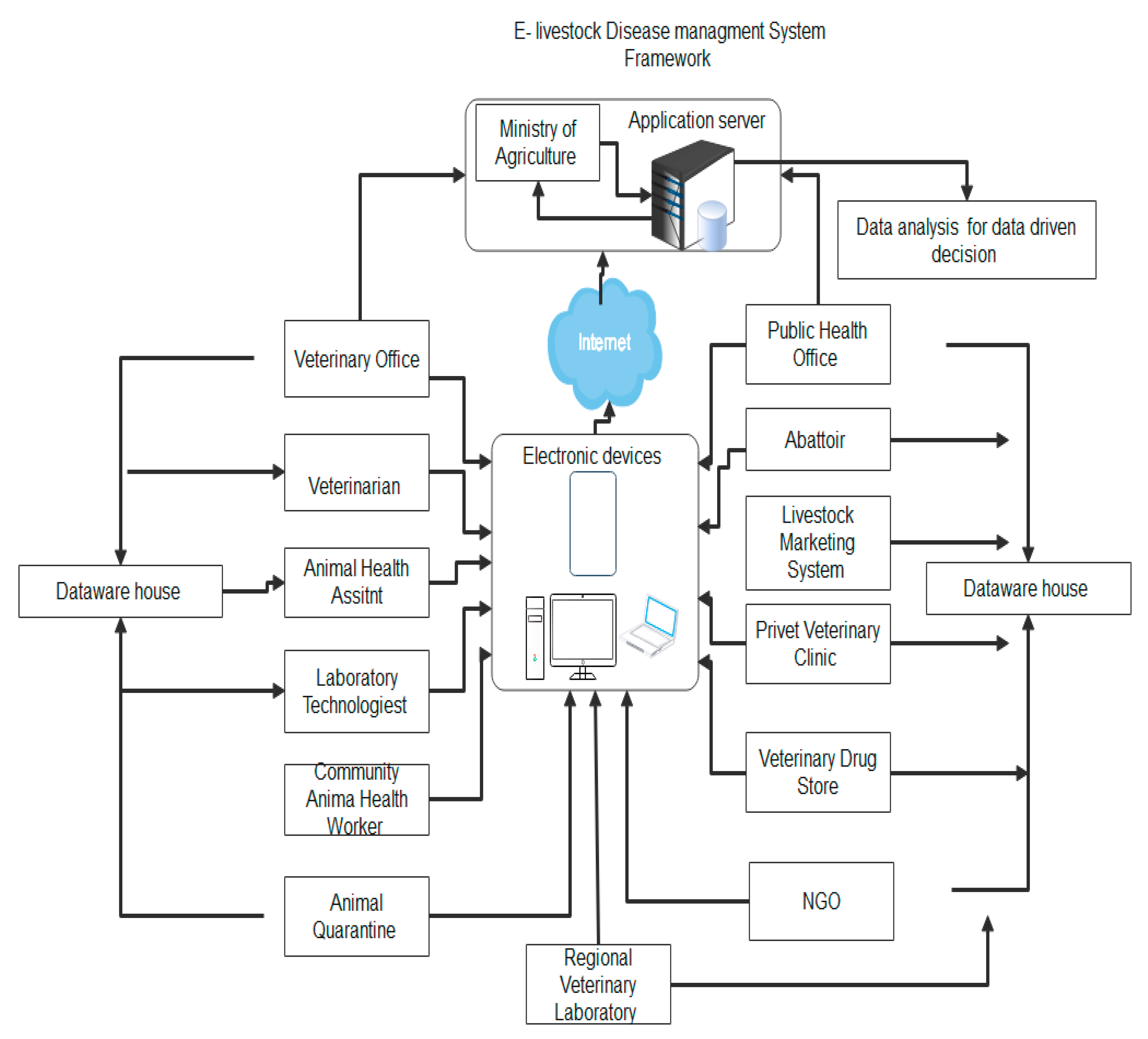
As depicted in
Figure 2 and presented by the Ministry of Agriculture (MoA, Ethiopia, 2021), the future livestock information system will facilitate the storage of data in the Ministry of Agriculture data center or it will be integrated with the National Datacenter as well for the study, analysis, and research purposes. This facilitates a centralized storage capacity that supports storage and gives space to data sourcing from various livestock sectors. Hence, the responsible bodies can access the data to make data-driven decisions.
Despite various advantages allied with the adoption of the electronic livestock disease recording (ELDR) system, there are different concerns raised by Animal Health Professionals (AHPs) during this study and survey on research findings while we engaged in investigating the current status and challenges of the veterinary service provision system in Ethiopia. The survey result revealed that many of the AHPs have no awareness of electronic-based disease diagnosing and treatment of animals, however, few of them think that ELDR systems take more time than paper-based. This survey revealed that Lack of understanding, insufficient training, insufficient funding, lack of awareness, lack of commitment, weak ICT infrastructure, and failure of sustainability of Electronic livestock Health Recording implementation are the major hindrances in adopting the ELDR systems. In enhancing Ethiopia’s agricultural sectors, livestock health has a significant role (MoA, Ethiopia, 2021). Modern livestock healthcare generates and stores vast amounts of detailed livestock disease-associated data. Very few real-world livestock disease data have been utilized to enhance the sector.
One of the bottlenecks revealed by a study (Naemi A et al, 2021) for the utilization of these data is unreachability to researchers. However, making these resources easier to access as well as integrating the data enable more researchers to react to problems of clinical care (Naemi A et al, 2021).
As a benefit of Electronic livestock health Records, a study (Mwanga et al, 2020) argued that the availability of quality and sufficiency of data determine the worthiness of your decision. The ELHR improves evidence-based decisions and policymakers and researchers in the area can easily access the data. A study (Global animal health association, 2020) also explained that it facilitates the data analytics process and building of veterinary intelligence systems to predict a particular disease before the onset of the disease. Similarly (Laura, Falzon C; et al., 2021) noted that ELHR is a means to facilitate the extraction of useful information in a timely fashion.
In contrast to the traditional recording system(paper-based) while reporting the aggregated data from daily to monthly reports there is a chance of losing resolution while also suspending detection of any notable events and, therefore, delaying response time furthermore it makes the reporting labor-intensive and prone to error. Data is an asset for organizations to make fact-based decisions. Hence, salient stakeholders can access comprehensive information to make decisions on disease, control measures, and their consequences.
Likewise, ELHR support generating models or trends for monitoring disease occurrence and control programs (World Organization for Animal Health, 2017). Useful and important results from information systems are obtained if we implement a good surveillance system and comprehensive, accurate data is collected and integrated into an animal health information system.
Digital technologies create the most exhilarating chances in analytics not only using data to understand past performance but also using information to generate insight into what is likely to happen in the future. “Data Analytics refers to the set of quantitative and qualitative approaches for deriving valuable insights from data. It involves many processes that include extracting data and categorizing it in data science, to derive various patterns, relations, connections, and other valuable insights from it” (Mantas, 2022). Today data analytics has become an engineering tool in predictive analysis.
Despite the rapid expansion and application of digital technologies in healthcare are increasing at an astonishing rate. However, in general, the livestock sector has not deployed this technology to the level of data management and analysis necessary to make use of the data in developing countries like Ethiopia. As a result, getting organized and appropriate livestock health records is a bottleneck for researchers and policymakers to execute fact-based decisions.
In Ethiopia, one of the strategic objectives of the livestock Information system roadmap is to improve access to livestock health information by using technological innovative tools. For the achievement of this initiatives and deliverables are designed. From these initiatives enhancing livestock and public healthcare information systems with the currently existing and new information technology is the one which includes the proper use of ELHR.
A report (MoA, Ethiopia, 2021) reveals that ensuring access to consistent reliable, timely, and useful information by salient stakeholders through Web or mobile-based livestock health-related information systems is anticipated to be a deliverable of this initiative. In this way, it is possible to enhance the use of technology and innovation (United Nations, 2021)
A study (Laio et al, 2016) states that Cluster analysis is part of unsupervised machine learning that deals with the practice of segregating a set of data objects (or instances) into meaningful subclasses. Each subclass is a cluster, such that objects in a cluster are similar to one another, yet dissimilar to objects in other clusters. In the process of data clustering they have given unlabeled data and grouped similar samples in one category, called a cluster, and the dissimilar sample in another category.
Clustering is useful in several machine learning and data mining tasks, in medicine to identify different disease categories, for biology Clustering used to find genetic information (Zhao et al, 2014), pattern recognition (Fu-Ru Lin et al, 2017), (Rajib S et al, 2019). Similarly, scholars (Hloušková & Lekešová, 2020) applied clustering to identify the farm outcome in the compound. Cluster analysis was also used by (Ishikawa et al, 2020) to evaluate disease risk in periparturient dairy cattle. Researchers (Zhao et al, 2014) also applied cluster analysis to group breast cancer and Alzheimer's lung cancer from a large biological and medical dataset.
Types of cluster analysis: Though there are various categories of cluster analysis method existed, the selection of the type for analytics purpose depend on the nature of the dataset, the computational complexity, and the specific need of the user are some of the criteria to select cluster analysis (Simon & Suresh, 2022). For instance (Koh, Ahmad, & Lee, 2022) applied hierarchical clustering to detect clusters of highly pathogenic avian influenza, (Komaru, Teruhiko, Yoshifumi, & Nangaku, 2020) to predict 1-Year Mortality After Starting Hemodialysis. Similarly (Cao K, 2022) identified and validated subtypes of Parkinson’s disease based on multimodal MRI data, (Rios, Tatiane, & Mello, 2022) to predict next COVID-19 waves. However, recent trends indicate that hierarchical clustering draws the attention of scholars to apply for grouping, predicting, and validating different disease categories.
Hierarchical cluster analysis (HCA) is an investigative tool intended to reveal natural grouping within a data set that would otherwise not be apparent. The detail of cluster categorization check-in was done by (Teng, Amin, & ElSayed, 2022), and (Praveen P et al, 2020). It is the most suitable unsupervised machine learning algorithm to cluster objects from small to large datasets. Clustering is performed either through Agglomerative or Divisive ways. Agglomerative starts from the dissimilarities between the objects and then step-by-step grouping. As it treats each entity as a cluster 1 (Praveen, Ranjith, Mohammed, R, & R, 2020). (Mahmoud & Zulaiha, 2016), applied agglomerative hierarchical clustering to cluster ground-level ozone in Malaysia. Similarly (Ana, Junshi, Milanović, Nina, & Riccardo, 2020) were used for Clustering Time Series Data. In other way Divisive hierarchical clustering starts from the whole document as a single cluster step by step the algorithm is going to split until it reaches its own cluster. This is exactly the inverse of agglomerative hierarchical clustering.
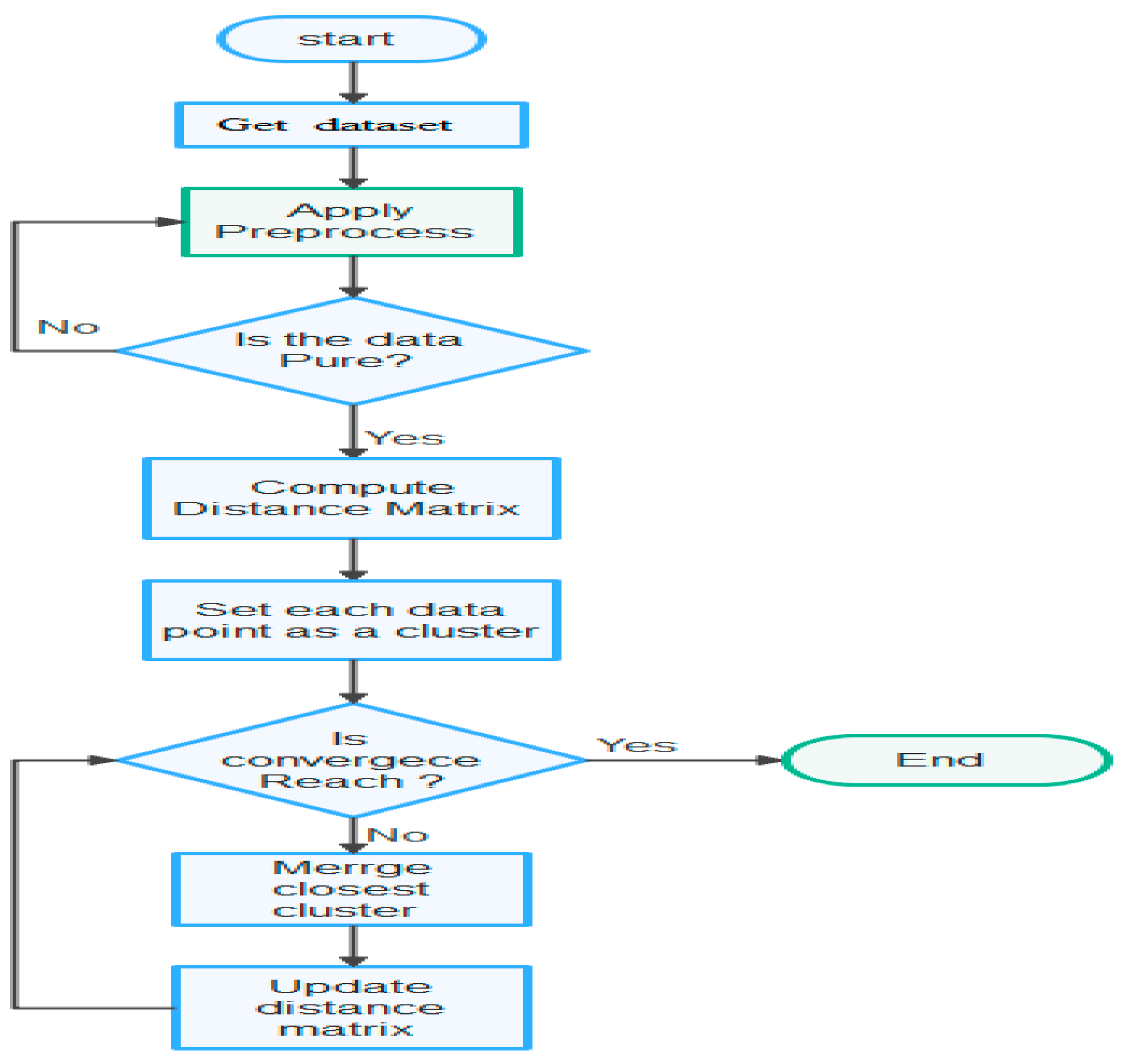
The basic principle of Agglomerative Hierarchical clustering: 1) Get dataset, 2) Apply to preprocess, 3) Check the purity of the dataset, 4) Compute the proximity matrix, 5) Consider each data point to be a cluster, 6) Repeat: Combine the two neighboring clusters and update the proximity matrix, 7) Do the process until it remains with a single cluster.
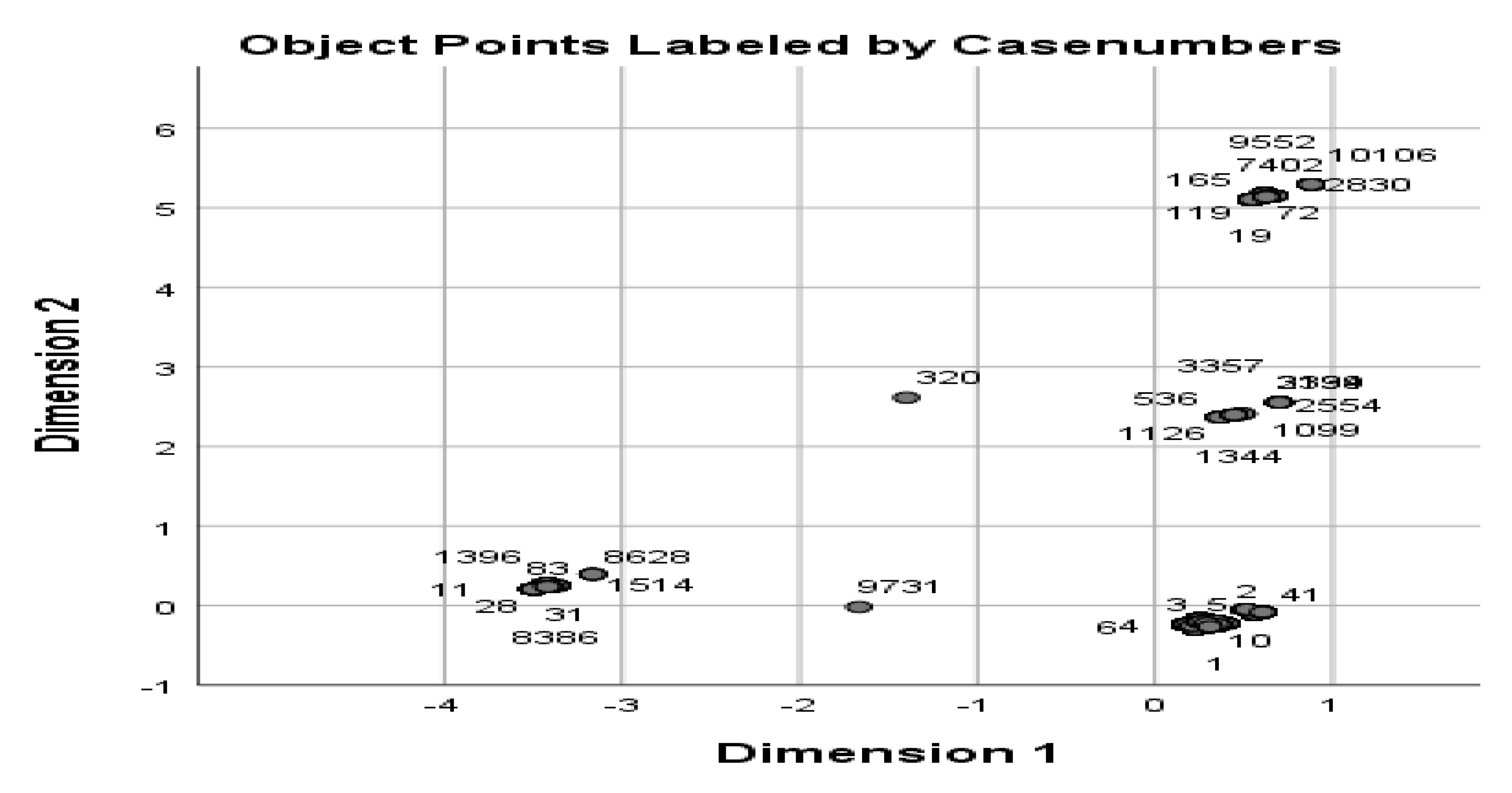
After this, it is possible to represent a dendrogram-like structure by defining distance similarity and merging approach as indicated in
Figure 3. To get useful output (information) out of the clustering, the distance matrix we use should be realistic.
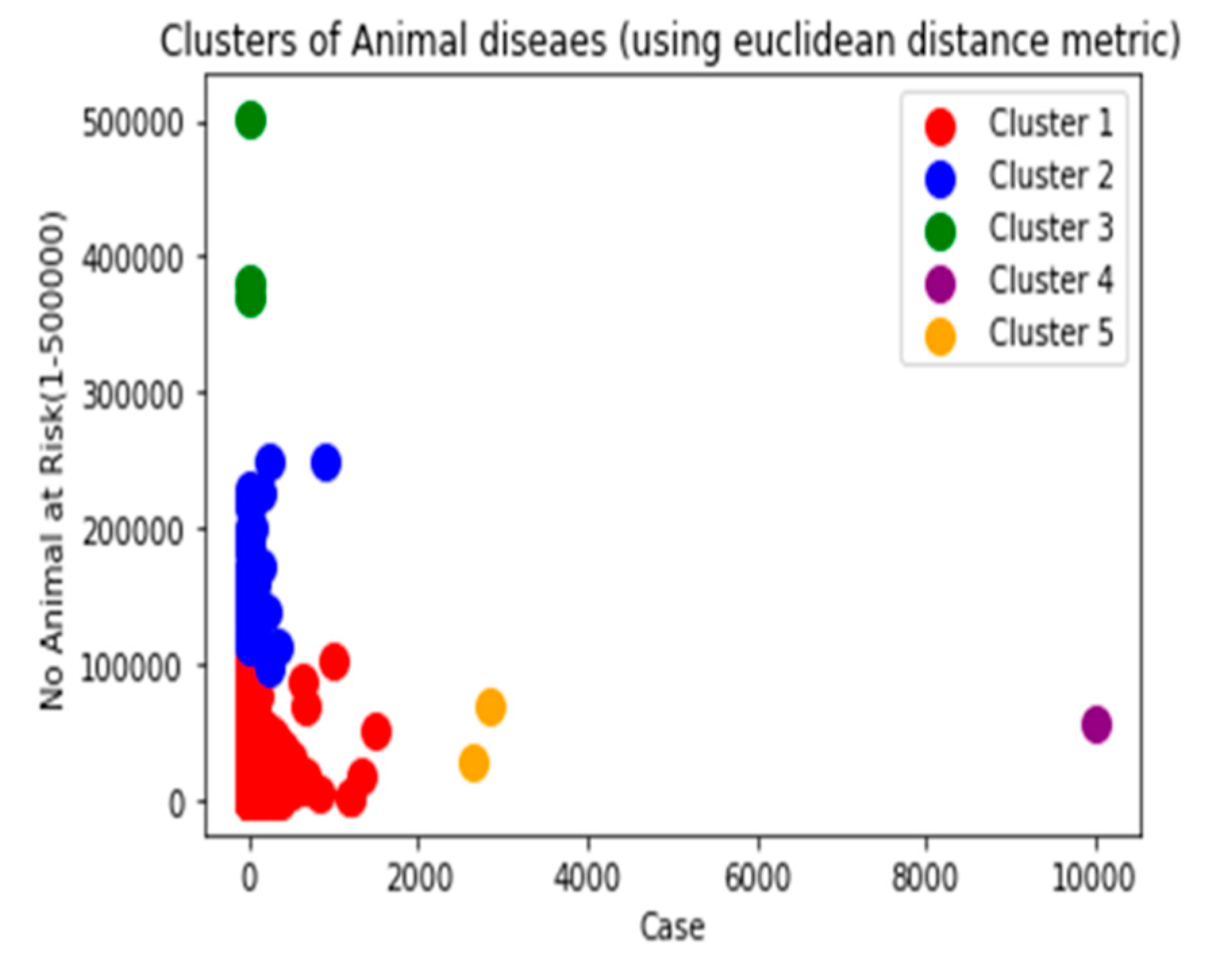
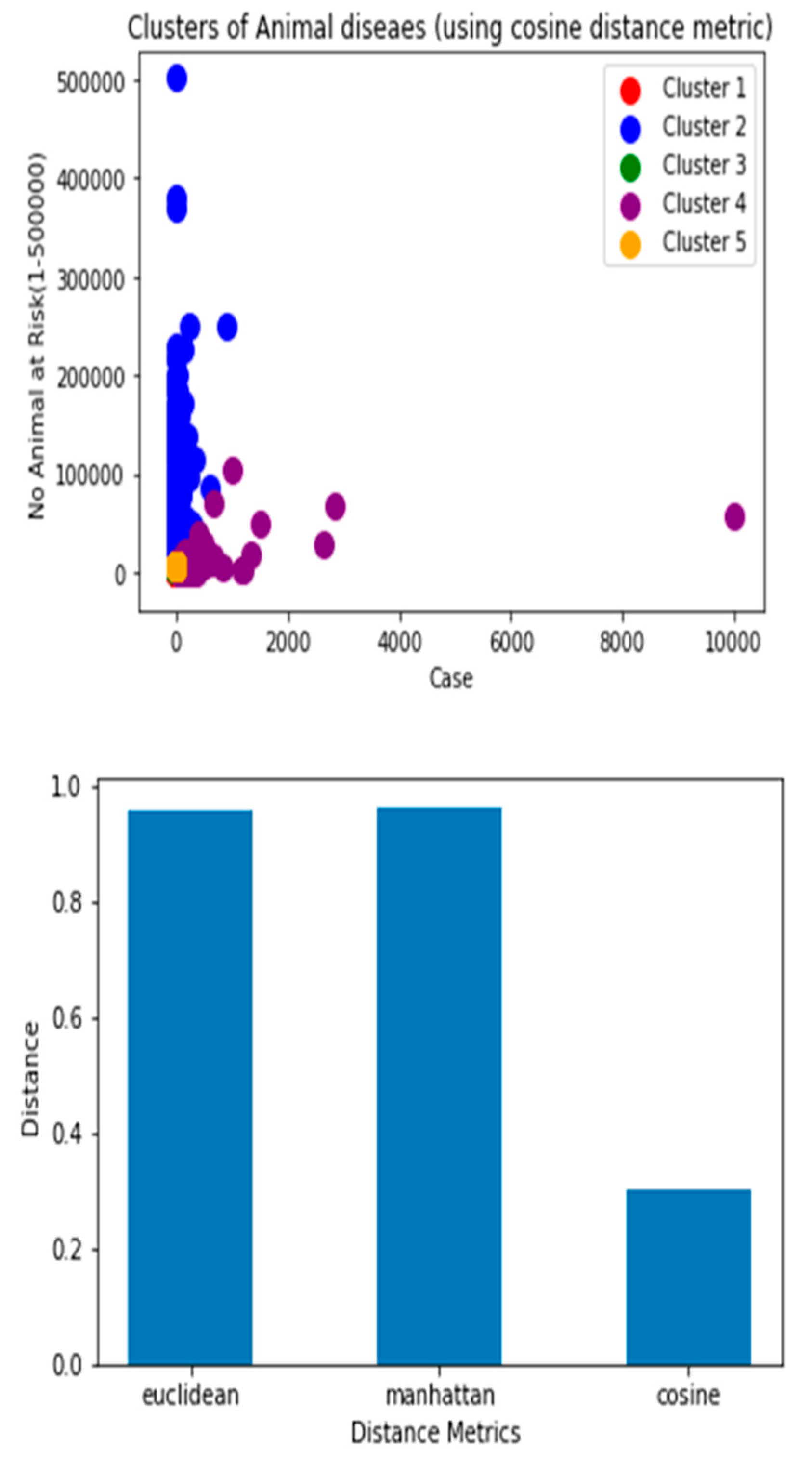
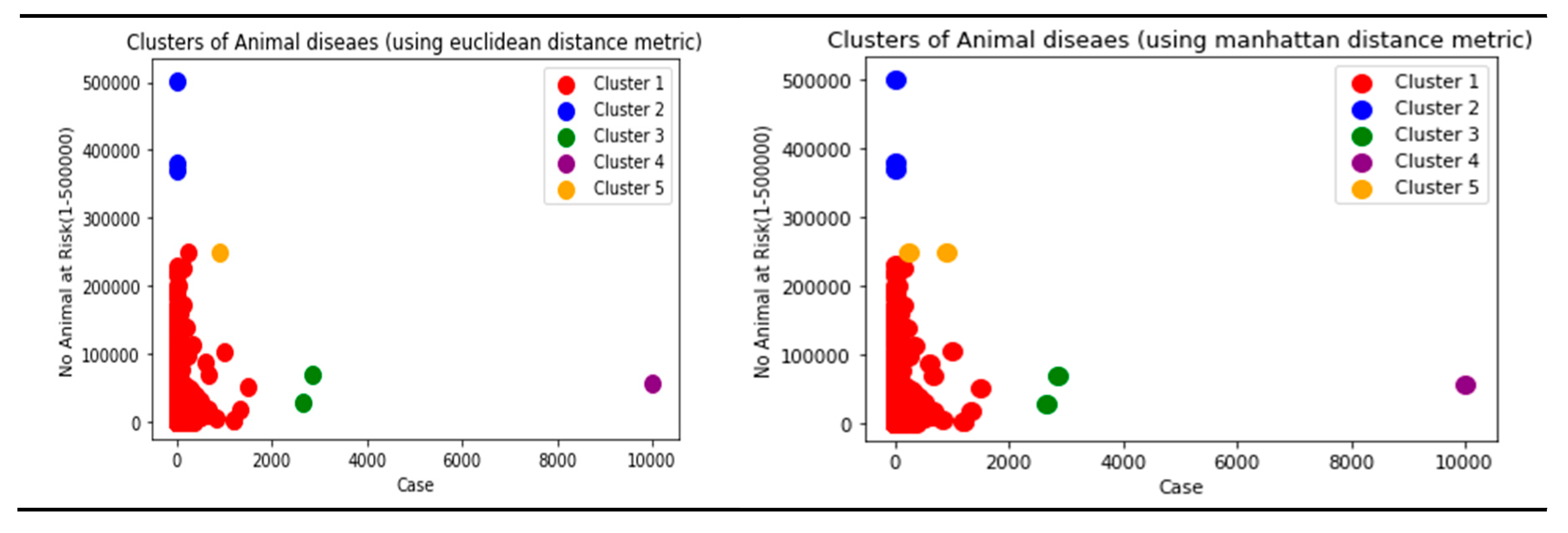
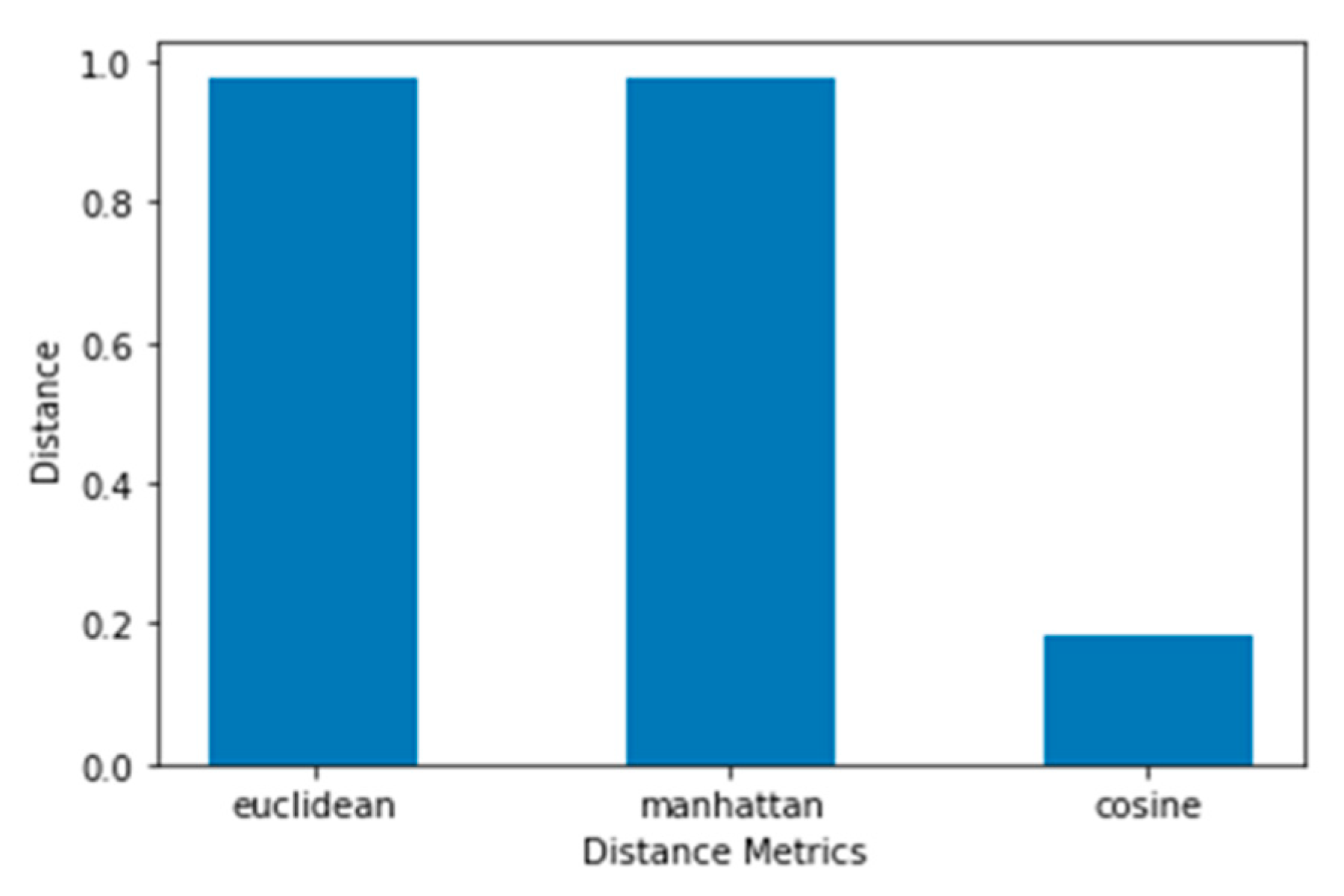
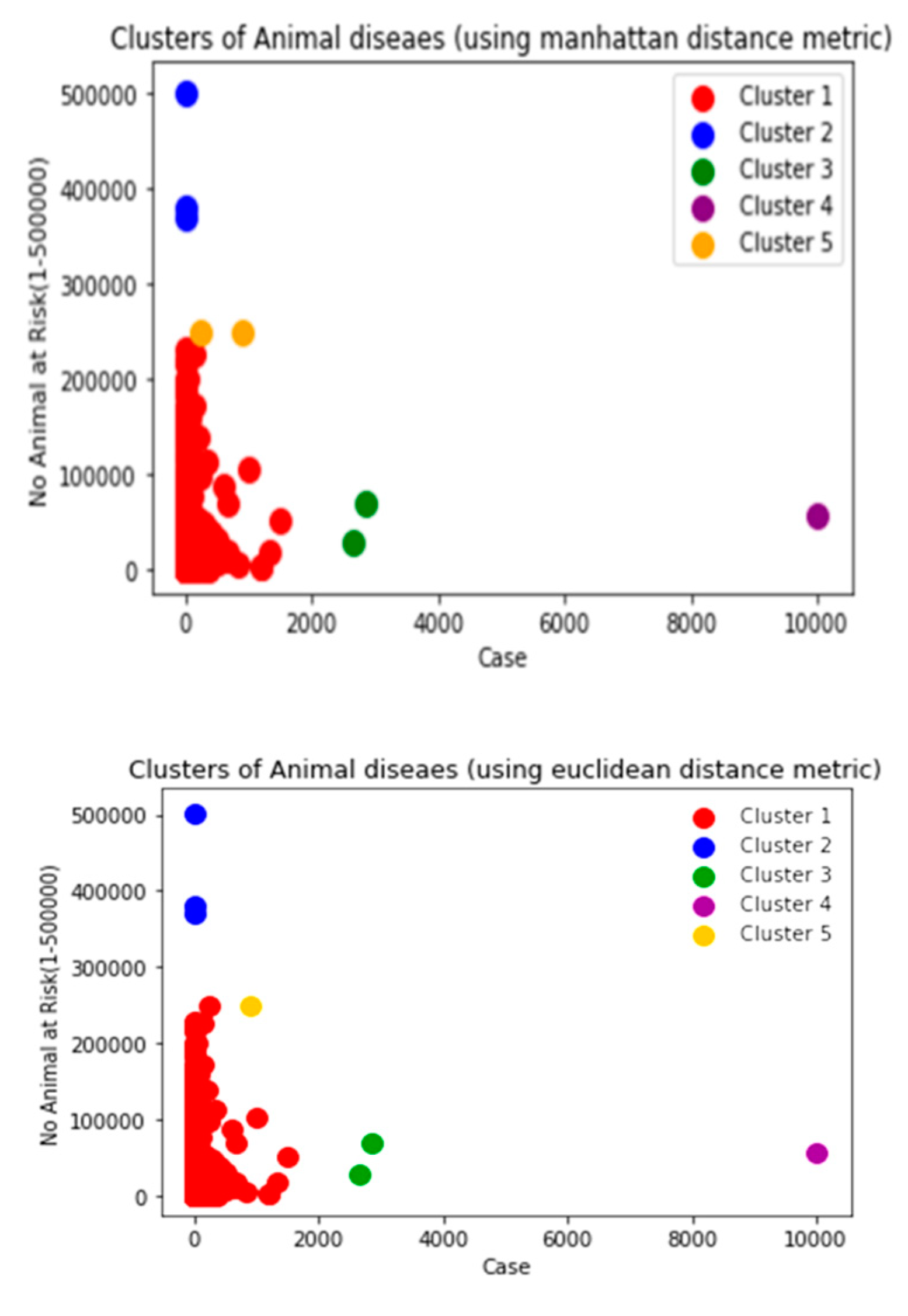
Though there are several commonly used metrics for characterizing distance or its inverse, similarity. In this study, Euclidean distance(ED) was selected. As noted by (Alfred & Jörn, 2022), as it corresponds to the everyday perception of distances, the Euclidean distance is the most intuitive distance metric. The Euclidean distance d of two data cases (x1, x2) is defined as the square root of the sum of squared differences. It is a continuous metric that can be thought of in geometric terms as the'' straight –line” distance between two points.
In general, the formula for Euclidean distance between points X and Y in dataset D is calculated as follows,
The following are points on the X and Y coordinate,
Hence, Euclidean Distance
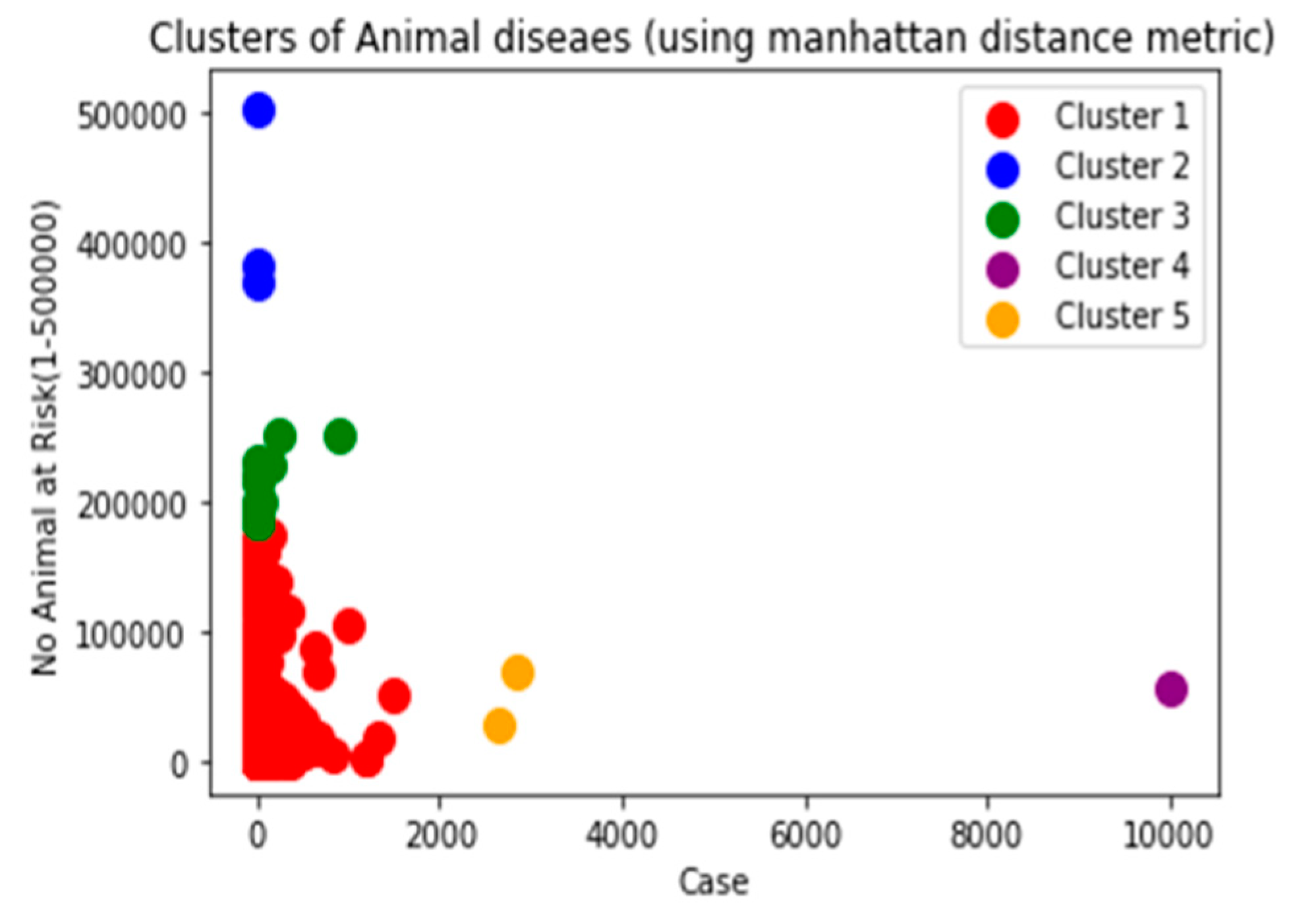
Manhattan and Cosine distance metrics can also be used.
Manhattan distance: is important to compute the absolute differences between coordinates of a pair of objects. Manhattan distance is relatively efficient and easily understandable straightforward forward and gives the best results when the data set has a high dimension.
This equation helps to generate the proximity matrix describing the closeness among objects to be clustered. Moreover, the mathematical notations taken from (Thomas, Jan, & Christoph, 2021) are also important to perform Hierarchical cluster analysis (HCA). Assume n objects to be clustered represented by the set of O where
is the
ith object
A partition T of X divides X into subsets
provided that
j, where Ø is the empty set
As indicated above, all the total n objects resulted from the union of all clusters (Thomas, Jan, & Christoph, 2021). Consequently, a sequence of partitions in which each partition is nested into the next partition in the sequence can be performed through a hierarchical clustering algorithm. The process can continue until a single cluster having all n objects remains.
K-means clustering is the most commonly used unsupervised machine learning algorithm for partitioning a data set into k groups without a predefined target class. It defines the total within-cluster variation as the sum of squared distances between and the corresponding centroid. (Silitonga, 2017) mathematically computed as
Where:
xi is a data point belonging to the cluster
μk is the mean value of the points assigned to the cluster
Each observation (x
i) is assigned to the given cluster such that the sum of squares (SS) distance of the observation to their assigned cluster centres
is a minimum. We can define the total within-cluster variation as follow : The total within a cluster is represented by
The total within the cluster sum of squares measures the compactness (i.e. goodness) of the clustering and it should be as small as possible.
Method of cluster optimization: The optimal number of clusters is one way or another, it is subjective and depends on the method for measuring similarities and the parameters used for grouping objects. For instance (Dimitris, Agni, & Holly, 2021) applied a pairwise distance matrix of the observation to determine the quality of a given cluster assignment in a mixed variable (categorical and numerical variable.
Similarly, (Quan, Fei, Zhongheng, & Nie, 2021) proposed the coordinate descent method to enhance the clustering performance in complex data. According to (Alboukadel, 2017), the direct method, consists of optimizing a criterion, such as the within-cluster sums of squares or average silhouette. The corresponding methods are named elbow and silhouette methods respectively. Since the K-means cluster algorithm depends on the initial selection of centroid thus, this weakness can be overcome by the know cluster optimization techniques called elbow and silhouette (Edy, Jatmiko Endro, & Vincensius, 2020).