1. Introduction
Deformable Image Registration (DIR) is a technique used in medical imaging to align and correlate two or more images of the same anatomical region. It aims to generate a deformation field that aligns a “moving image” with a “fixed image” by optimizing a similarity metric. This metric measures the correspondence between corresponding image reigions. By maximizing the similarity metric. DIR establishes a coordinate transformation the aligns the images, resulting in congruent anatomical features, boundaries, and instensity information.
In practical terms, when applying DIR to medical images, the moving image is spatially transformed to align with the coordinate system of the fixed image. This alignment ensures that corresponding anatomical structures, boundaries, and intensity values in the moving image occupy the same positions (i.e., coordinates) as their counterparts in the fixed image. By achieving this spatial correspondence, DIR facilitates a comprehensive and accurate comparison of images obtained at different time points or using different modalities, enabling healthcare professionals to monitor disease progression, assess treatment efficacy, and plan interventions with improved precision.
DIR algorithms employ various mathematical models and optimization techniques to estimate the deformation field. Conventional DIR algorithms typically employ intensity-based or feature-based methods to establish spatial alignment between two or more images by mapping them to a common coordinate system. Recently, advancements in deep learning have led to the emergence of learning-based registration methods that have shown promising results. These approaches either employ deep learning to model the transformation function through supervised training with synthesized deformations [
1,
2,
3,
13,
14,
17] or through unsupervised training with a spatial transformer network [
5], as demonstrated in works presented in [
6,
7].
There are several commercial DIR software packages that are commonly used in radiation oncology, including Velocity AI (Velocity Medical Solutions LLC, Atlanta, GA, USA), Mirada RTx (Mirada Medical Ltd, Oxford, UK), MIM software (MIM Software Inc., Cleveland, OH, USA), Raystation (RaySearch Laboratories AB, Stockholm, Sweden), and Eclipse (Varian Medical Systems, Palo Alto, CA, USA). These software packages are widely used in radiation oncology for a variety of applications, including treatment planning, dose accumulation, and monitoring of changes in the patient's anatomy during treatment. Each software package has its strengths and weaknesses, and the choice of software depends on the specific needs and requirements of the radiation oncology team.
However, the wide adoption of DIR in routine clinical operations is limited due to concerns about the accuracy and uncertainty of many DIR algorithms and the challenges of efficiently integrating them into the complex workflows of Radiation Oncology clinics. This includes difficulties in accurately registering a series of multimodal images in an adaptive radiotherapy workflow or using assistive devices. These limitations can make it difficult for healthcare providers to trust and effectively use DIR in their daily operations [
4].
This study aims to evaluate the clinical applicability and usefulness of current state-of-the-art algorithms and traditional algorithms by applying them to real clinical patients. By conducting comprehensive evaluations on diverse patient cases, we seek to assess the potential and efficacy of these algorithms in clinical settings.
2. Materials and Methods
2.1. Patient dataset
In our study, we utilized a publicly available head and neck squamous cell carcinoma dataset from The Cancer Imaging Archive (TCIA) [
8]. The dataset consists of CT images from 31 HNSCC patients who underwent VMAT (volumetric modulated arc therapy) at Miami Hospital between 2011 and 2017 [
9]. VMAT was chosen due to its superior dose conformity and tissue sparing capabilities. Each patient had three CT scans acquired throughout the duration of the radiation treatment course. Pre-treatment CT, Mid-treatment CT, Post Treatment CT. The Pre-treatment CT scans were performed with a median of 13 days(range:2 ~ 27) before treatment, the Mid-treatment CT was performed at fraction 17(range: 8 ~ 26), and the Post-treatment CT around fraction 30(range: 26th fraction - 21 weeks Post-treatment. The patients received RT treatment to a total dose of 58-70 Gy, delivered in daily fractions of 2.0-2.20 Gy over 30-35 fractions. The fan beam CT images were acquired using a Siemens 16-slice CT scanner head protocol with 120 kV and a current of 400 mAs. A helical scan with 1 rotation per second was used, with a slice thickness of 2 mm and a table increment of 1.2 mm. In addition to the imaging data, RT structure Dicom, demographic, and outcome measurements were provided. An experienced radiation oncologist delineated the RT structures at each treatment. To verify the deformable image registration algorithms used in our study, we focused on the patient's organs at risk (OAR). For quantitative analysis, the datasets of patients' OAR were limited to the spinal cord, optic chiasm, brain stem, parotid gland, cochlear, and PTV
2.2. Deformable image registration models and hyperparameter optimization
We conducted a comparative analysis among various state-of-the-art registration methods to assess their performance. Initially, we compared two non-deep learning approaches, namely ANTs [
1] SyN and NiftyReg [
10], along with three deep learning methods: Voxelmorph [
6], Cyclemorph [
11], and Transmorph [
12].
ANTs SyN, developed by Avants et al., is a non-linear diffeomorphic registration method within the Advanced Normalization Tools (ANTs) framework. Like LDDMM, ANTs SyN utilizes Euler-Lagrange equations to solve the stationary velocity field, representing the transformation model of the moving image. This technique employs cross-correlation as a similarity metric, ensuring diffeomorphic and symmetric normalization. The computation of cross-correlation Euler-Lagrange equations is made symmetrical through a Jacobian operator, significantly reducing computation time. Inclusion of invertibility constraints during optimization guarantees diffeomorphic transformations.
NiftyReg offers both global and deformable image registration capabilities. Global registration encompasses rigid and affine registration, while deformable registration utilizes Free-Form Deformation (FFD) by defining control points. The computational consumption of deformable registration can be adjusted by manipulating the number of control points.
Voxelmorph is a convolutional neural network (CNN)-based non-linear diffeomorphic registration algorithm developed by Dalca et al. It employs a stationary velocity field as the transformation model. The CNN learns the stationary velocity field, and spatial transformation layers perform diffeomorphic integration to obtain the deformation field.
Cyclemorph consists of two registration networks that switch inputs and enforce cycle consistency. This cycle consistency in the image domain helps avoid discretization errors that may arise from the real-world implementation of inverse deformation fields.
Transmorph is a hybrid Transformer-CNN method. It utilizes the Swin-Transformer as the encoder to capture spatial correspondence, while a CNN decoder processes the information from the transformer encoder into a dense displacement field. Long skip connections are incorporated to maintain the flow of localization information between the encoder and decoder stages. By evaluating these different registration methods, we aimed to assess their effectiveness and compare their performance in our study.
The hyperparameters of these methods, unless otherwise specified, were empirically set to balance the trade-off between registration accuracy and running time. The methods and their hyperparameter settings are described below.
SyN: We used cross-correlation (CC) as the objective function, Gaussian smoothing of 5, and three scales with 160, 100, and 40 iterations.
NiftyReg: We used the sum of squared differences as the objective function and bending energy as a regularizer.
Next, we compared with several existing deep learning-based methods. For fair comparison, unless otherwise indicated, the loss function that consists of bending energy and Dice loss was used. Recall that the hyperparameters λ and γ define, respectively, the weight for deformation field regularization and Dice loss. The detailed parameter settings used for each method were as follows:
- 3.
Voxelmorph: We used regularization hyperparameters λ and γ equal to 1, where these values were reported as the optimal values in the Voxelmorph authors' paper.
- 4.
Cyclemorph: In Cyclemorph, the hyperparameters alpha, beta, and lambda correspond to the weights for cycle loss, identity loss, and deformation field regularization. We modified Cyclemorph by adding a Dice loss with a weighting of 1 to incorporate organ segmentation during training. We set alpha = 0.1 and beta = 1, which were recommended in the Cyclemorph authors' paper.
- 5.
Transmorph: We used regularization hyperparameters λ and γ equal to 1.
We conducted an experiment to compare the performance of five different registration methods: SyN, NiftyReg, Voxelmorph, Cyclemorph, and Transmorph. We evaluated the registration performance of each model based on the volume overlap between anatomical structures and organ deformation, which was quantified using the Dice score and difference of deformed images and fixed images. We averaged the Dice coefficient scores (DSC) of five anatomical/organ structures for all patients. The mean and standard deviation of the averaged scores were compared across various registration methods. The DSC is a metric that is sensitive to internal image alignment and is commonly used to quantify registration accuracy.
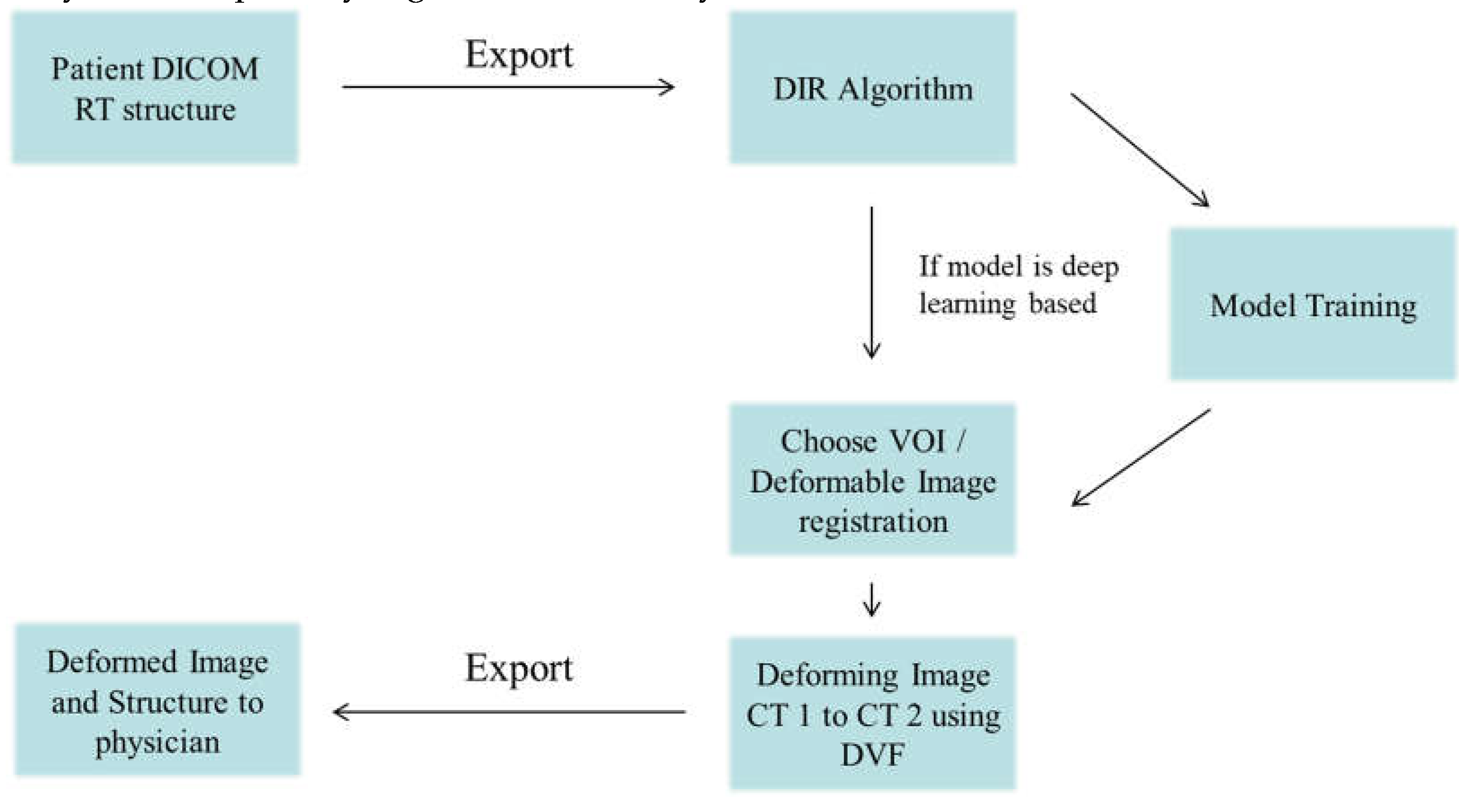
Figure 1.
The framework of Deformable Image Registration workflow.
Figure 1.
The framework of Deformable Image Registration workflow.
2.3. Evaluation metrics.
The DSC is defined as follows:
Here, F and M represent the fixed and moving images, respectively. A DSC of 1 indicates perfect anatomical alignment by the registration algorithm. We also evaluated the DSC of regions of interest (ROIs) according to the American Association of Physicists in Medicine (AAPM) publication Task Group No. 132 (TG-132) [
18]. We did not apply any data augmentation to the CT dataset during training. To train the deep learning DIR algorithms, we split the dataset into training, validation, and test sets in a ratio of 7:1:2. We evaluated the performance of the models using a workstation equipped with an Intel Xeon Silver processor @ 2.10GHz, 128GB RAM, and an Nvidia RTX 6000 graphics card. The virtual environment was configured in Python 3.8.0 + PyTorch 1.12.1 [
15]. All models were trained for 400 epochs using the Adam optimizer, with a learning rate of 1*10^ (-4) and a batch size of 1.
2.4. Evaluation of Deformation in moving image to fixed image.
According to TG132, when dealing with small volumes such as the head and neck, the Dice coefficient score may be evaluated as low during deformation. Additionally, in order to perform quantitative evaluations, the number of organs at risk (OAR) was limited. Therefore, in clinical evaluations after applying patient treatment, there may be regions where the Dice score is not measurable since they were not included as input to the deep learning model. Consequently, subjective considerations may be involved in these cases.
The subjective nature of these evaluations arises from the considerations outlined in TG132 and the limitations imposed by small volume structures and the restricted input to the deep learning model. To facilitate accurate evaluations within these constraints, it is possible to incorporate expert opinions and subjective evaluation criteria. By leveraging the expertise and domain knowledge of clinical professionals, a comprehensive assessment of the deep learning model's performance and clinical applicability can be achieved. It is important to interpret the results considering the subjective nature of these considerations and provide a detailed description of the clinical evaluation outcomes.
3. Results
Assessment of Deformed Position for OARs
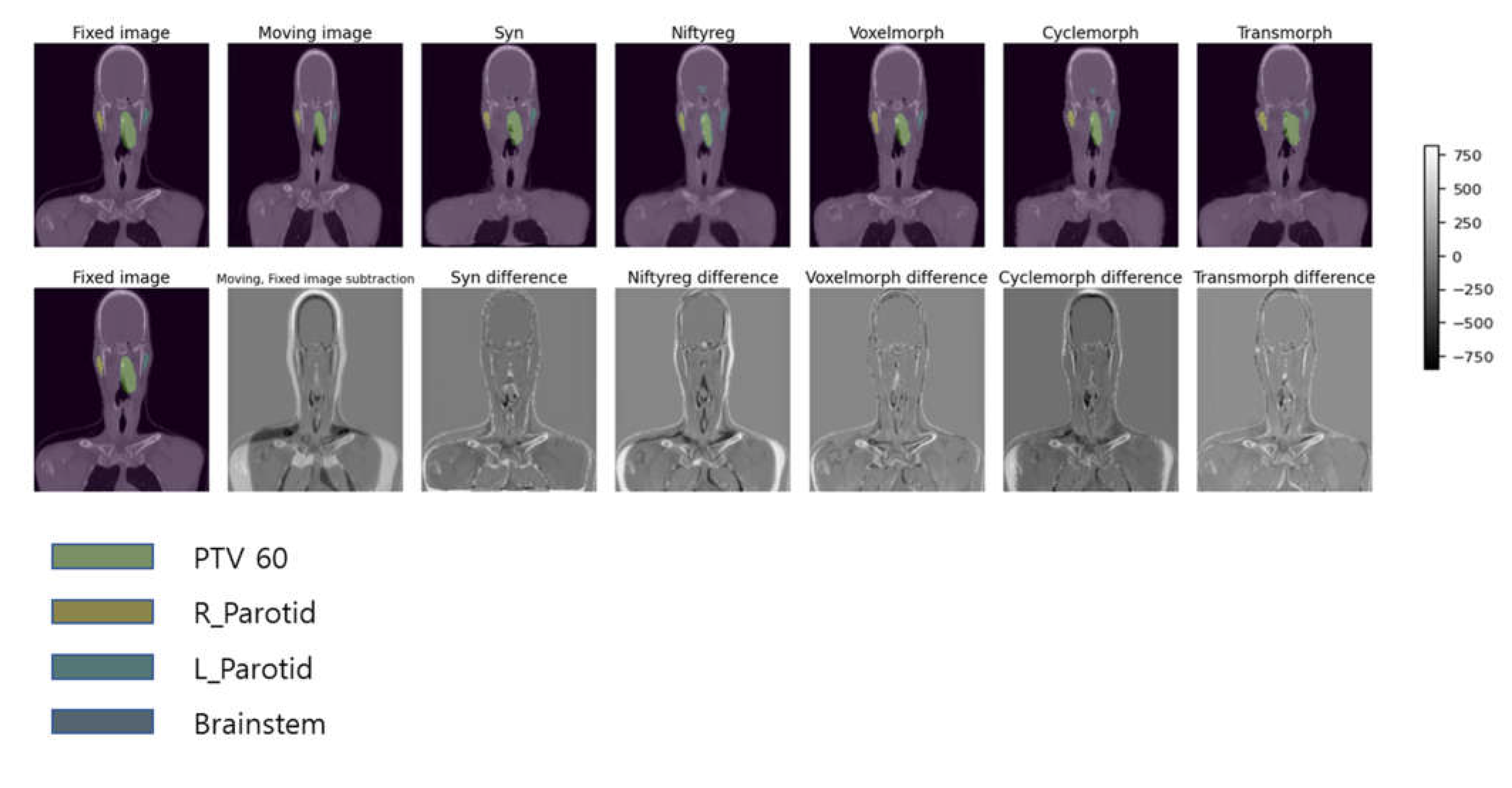
In
Figure 3, we present examples of deformed images obtained using five different DIR algorithms along with their difference maps as compared to the original fixed image. Based on qualitative analysis, it can be concluded that the SyN and Transmorph algorithms produced reasonably accurate deformations, while the other algorithms showed relatively greater discrepancies from the moving image. Among these models, the best-deformed model was SyN (patient mean dice score: 0.76), which was well-deformed in the modified patient’s contour, airway, lung, and bone. The deformed images of SyN and Transmorph accurately denoted the heart between the moving image and the fixed image.
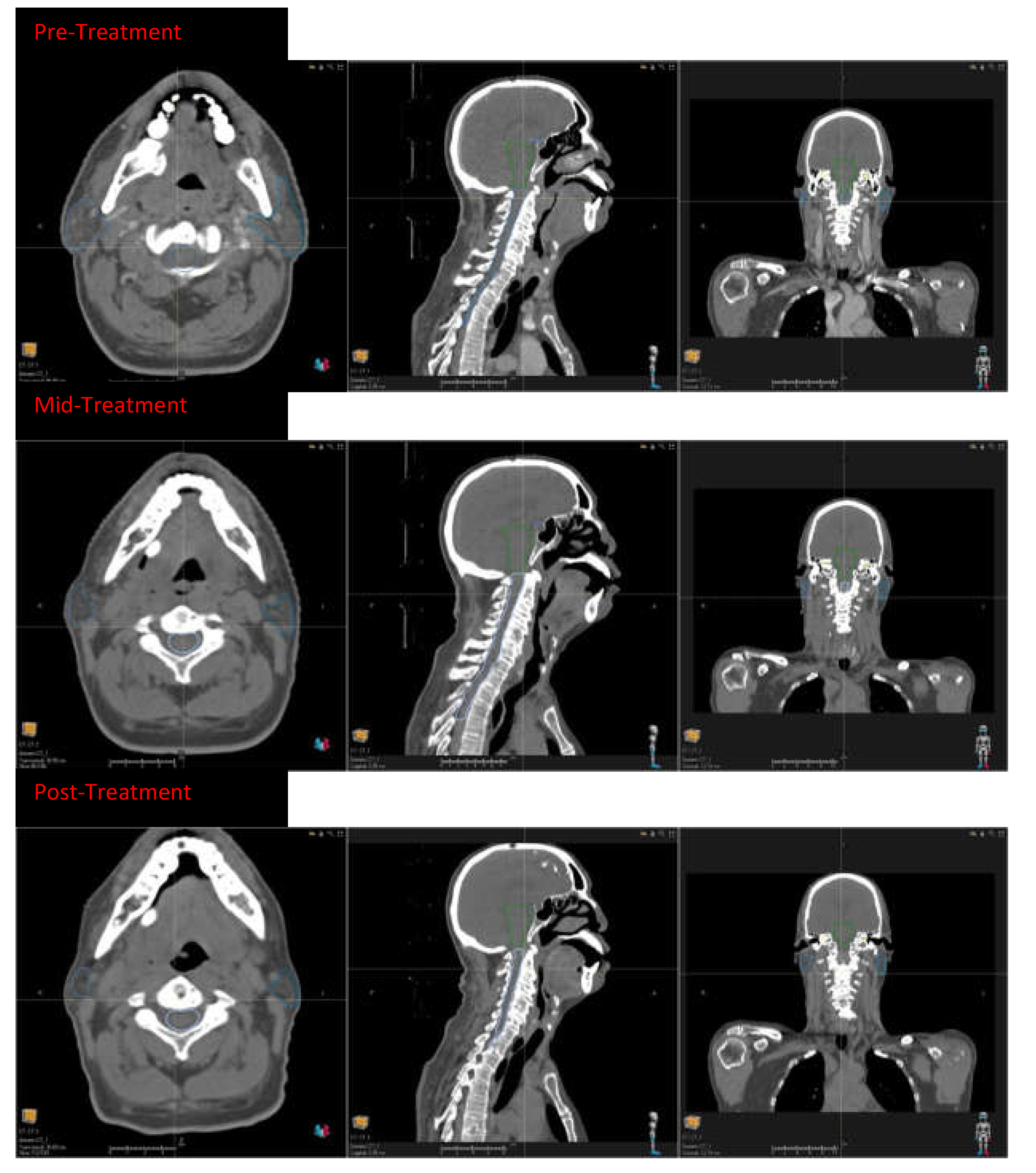
Figure 2.
Head and neck squamous cell carcinoma patients with CT taken during pre-treatment, mid treatment, and the post treatment.
Figure 2.
Head and neck squamous cell carcinoma patients with CT taken during pre-treatment, mid treatment, and the post treatment.
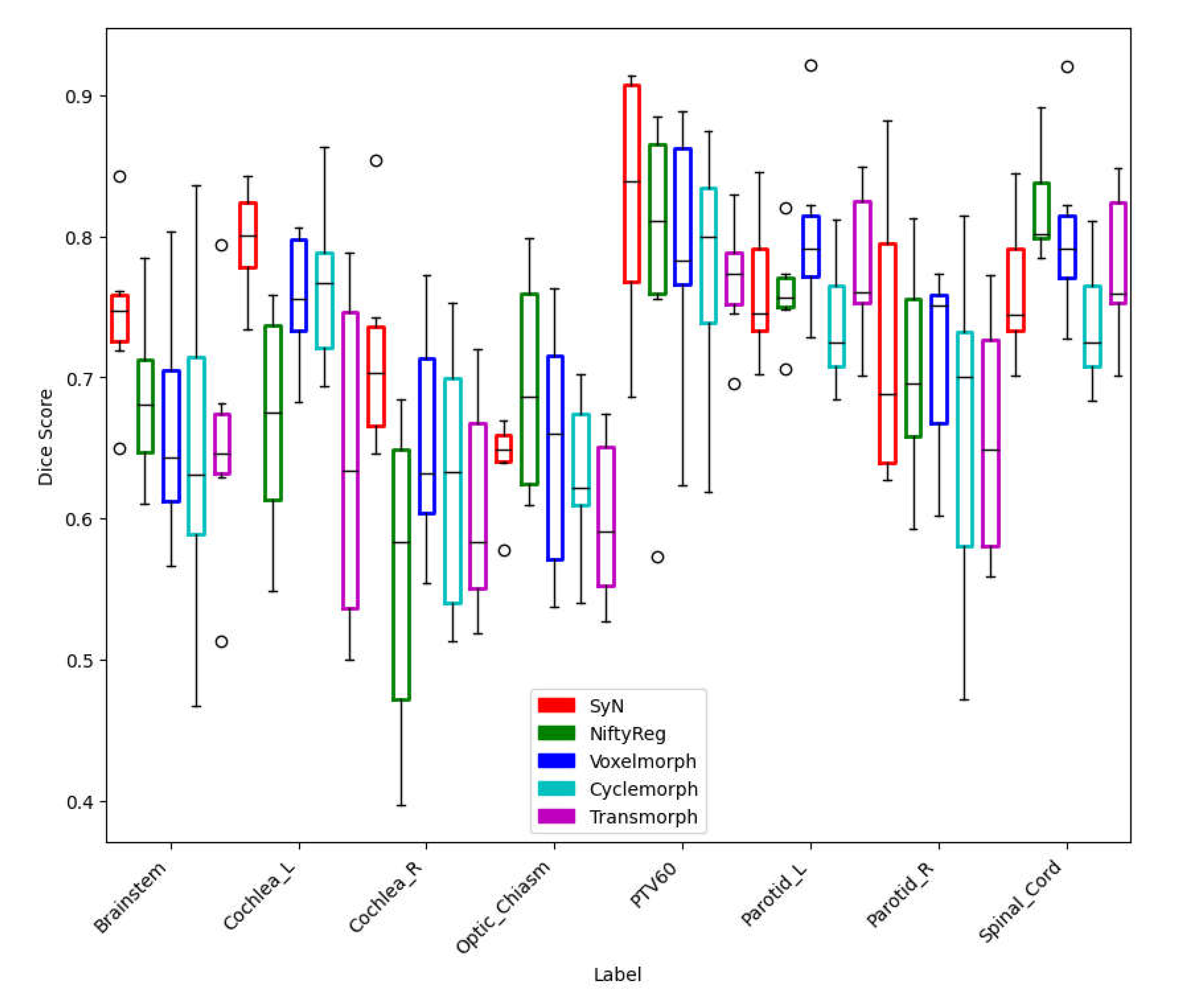
Figure 4 shows the registration results of 10 test datasets (5 patients deformed from pre to mid and 5 patients deformed from mid to post) for each DIR algorithm, along with the Dice coefficient score for each OAR and PTV. Among the classical iterative models, SyN (0.74±0.06) and NiftyReg (0.70±0.12) consistently achieved similar scores. However, for the supervised and unsupervised models, the dice scores were highest for Voxelmorph (0.72±0.08), followed by Cyclemorph (0.68±0.11) and Transmorph. (0.69±0.13).
Table 1 compares the mean Dice coefficient, training time in minutes per epoch, and inference time in seconds per image among the methods used in this paper. Note that the SyN and NiftyReg packages are CPU-based methods, while the Voxelmorph, Cyclemorph, and Transmorph methods are GPU-based deep learning methods. The speed was calculated using an input image size of 160*192*224. The most time-consuming methods to train were Cyclemorph and Transmorph, which required approximately 2~3 days each. Cyclemorph was time-consuming because the cycle-consistent training virtually trains four networks simultaneously in a single epoch.
4. Discussion
The deformed images obtained by the DIR algorithms exhibited the same trends in test patient’s images, and our results show that the deformed Images and contours obtained using SyN showed the best performance. Nevertheless, a few outliers were detected in real patient cases.
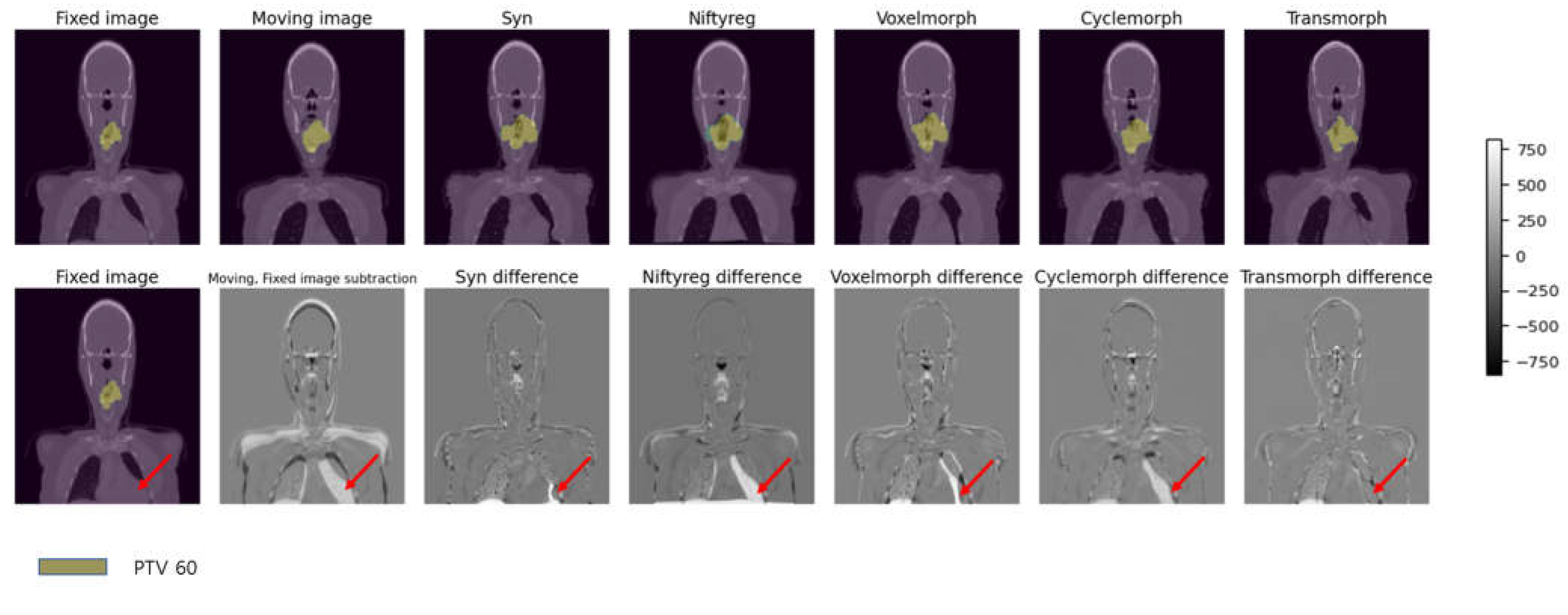
Figure 5 presents the results of five deformable image registration algorithms applied to fixed image CT and moving image CT, namely Syn, Niftyreg, VoxelMorph, CycleMorph, and Transmorph models. Among them, the Syn and Transmorph models effectively capture the size variations in the heart between the fixed and moving images. The Syn model accurately aligns the heart region, while the Transmorph model nearly achieves alignment by reflecting the observed size changes in the images. However, the Niftyreg, VoxelMorph, and CycleMorph models demonstrate less accuracy in representing the size changes compared to the Syn and Transmorph models. The Voxelmorph model partially matches the size of the heart region but fails to accurately reproduce overall size variations. The VoxelMorph model achieves smooth deformations but falls short in adequately reflecting the observed size changes. Lastly, the CycleMorph model disregards size changes, resulting in minimal variation. These findings indicate that the Syn and Transmorph models excel in capturing size variations between the fixed and moving images, achieving detailed alignment compared to the other models.
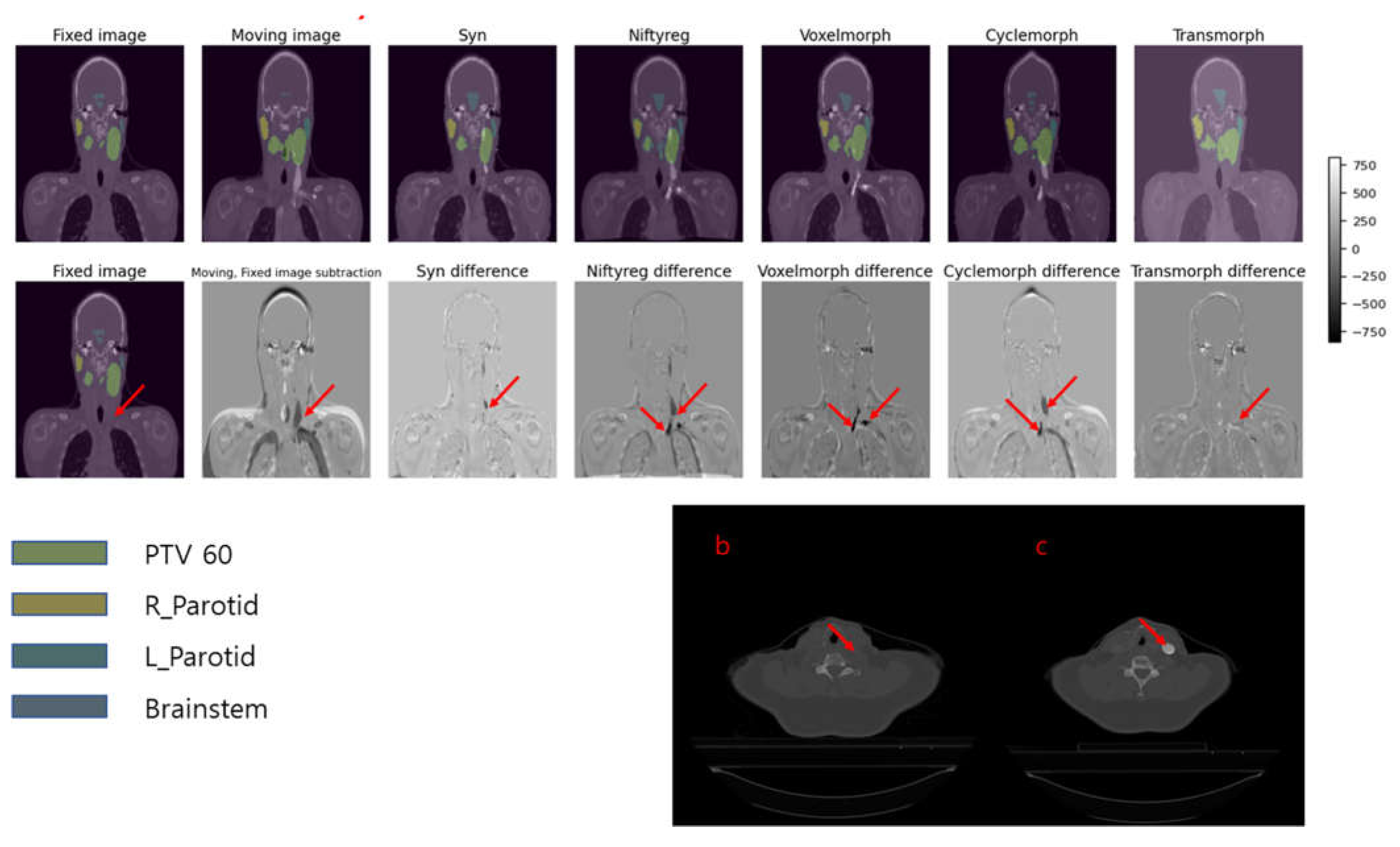
Figure 6 shows a situation where a white artifact is present in the dynamic CT (moving image), but not in the fixed image.
Figure 6a showcases the deformed images and contours obtained by applying five different deformable image registration (DIR) algorithms, incrementally, to the fixed and dynamic images. Among the five models, only Transmorph successfully removes the white artifact from the deformed image.
Figure 6c points out the white contrast medium present in the dynamic image, while
Figure 6b shows the same slice in which the contrast medium is not visible.
These findings demonstrate the effectiveness of using a Transmorph, vision transformer-based model compared to traditional algorithms like Syn and Niftyreg, as well as CNN-based algorithms like VoxelMorph and CycleMorph, in situations involving contrast medium.
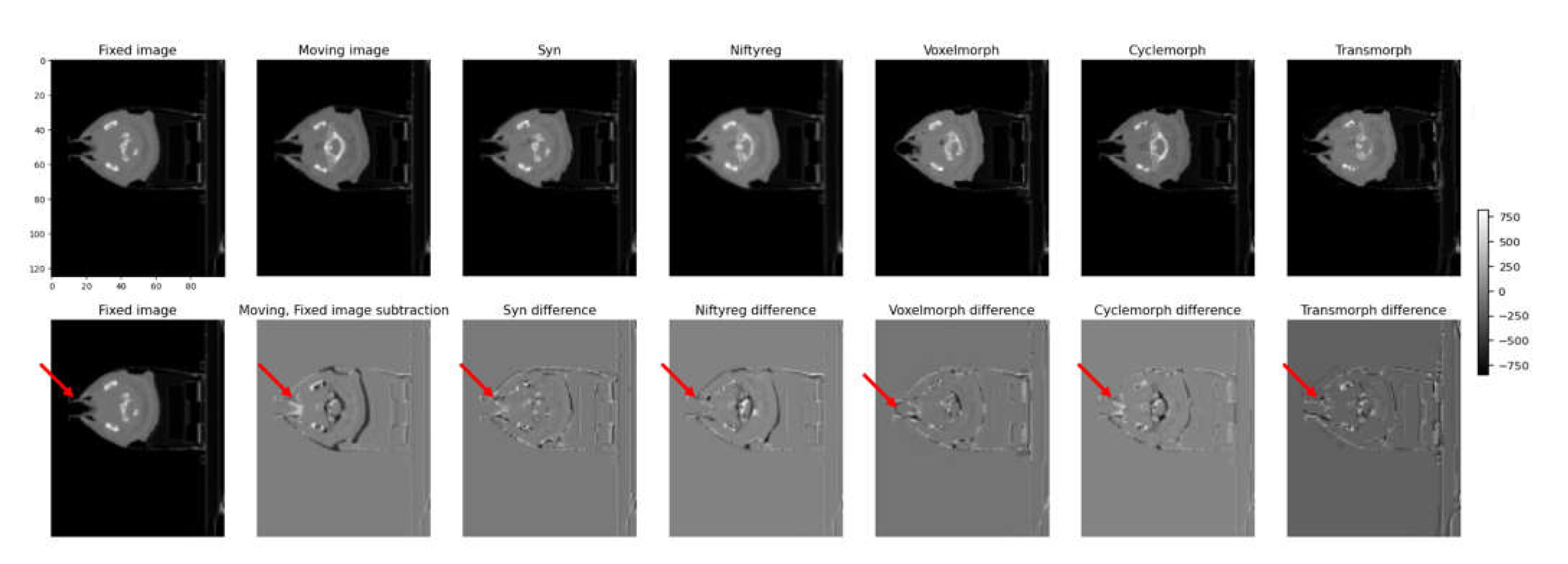
Figure 7 shows the fixed and moving image sequences of a patient wearing an assistive device due to lip surgery. The first row presents the transformed results for each model, while the second row shows the subtraction images between the transformed image and the fixed image. In the first row, each model's transformed results are displayed, indicating how well they align the moving image with the fixed image. The transformed images show the effects of the deformable image registration (DIR) algorithms on the patient's anatomy. The performance of each model can be assessed by comparing the alignment and accuracy of the transformed structures. In the second row, the subtraction images demonstrate the differences between the transformed image and the fixed image. These subtraction images allow for a visual comparison of the discrepancies and deviations between the transformed and fixed images, highlighting areas of misalignment or inaccurate deformation.
By examining both the transformed results and the subtraction images in
Figure 6 it is possible to evaluate the effectiveness of each model in accurately aligning and deforming the moving image to match the fixed image in the context of a patient with an assistive device.
In this study, we identified two main limitations. Firstly, the experiment did not involve generating treatment plans based on the deformed CT images and contours. Consequently, direct comparisons between the deformed images/contours and treatment plans, such as DVH evaluations, were not performed. Although the dataset included information for treatment planning, treatment plans were not generated using the deformed images and contours in this experiment. This limitation hinders a comprehensive analysis of the impact of deformations on treatment outcomes. Future studies should incorporate treatment planning based on deformed images and contours to provide a more comprehensive assessment of different deformation algorithms and their effects on radiation dose delivery and treatment plan quality.
Secondly, the inclusion of datasets encompassing surgical CT images and significant anatomical alterations would greatly enhance the evaluation of deformable image registration (DIR) models in clinical scenarios. By incorporating pre- and post-operative CT images, we can simulate and evaluate the performance of DIR models under conditions involving substantial anatomical changes caused by surgical interventions. The availability of surgical CT images would allow us to assess the effectiveness of DIR models in accurately registering and aligning images before and after surgery. This analysis would provide valuable insights into the capability of DIR models to account for deformations resulting from surgery, facilitating precise treatment planning and evaluation. Furthermore, incorporating datasets with significant anatomical alterations would enable a comprehensive evaluation of the limitations of DIR models. By assessing registration accuracy in the presence of such alterations, we can identify scenarios where DIR models may struggle or exhibit limitations, aiding clinicians in making informed decisions regarding treatment planning and assessment.
5. Conclusions
In conclusion, our preliminary study aimed to assess the clinical applicability of deformable image registration (DIR) algorithms for head and neck patients. We evaluated two traditional DIR algorithms (Syn and NiftyReg) and three deep learning based algorithms (VoxelMorph, CycleMorph and TransMorph).
To evaluate the clinical performance of these algorithms, we referred to the TG 132 guidelines, which recommend an average dice coefficient score of 0.8-0.9 for accurate DIR application. However, upon clinical evaluation, we observed lower dice scores for smaller volumes across various DIR models.
These findings highlight the challenges in achieving high accuracy and precision in deformable image registration for head and neck patients. Specifically, accurately capturing deformations in small volumes proved to be a limitation for the evaluated DIR models. This limitation warrants further investigation and improvement.
For future research, we suggest focusing on refining the algorithms to enhance their performance, especially in accurately registering and aligning smaller volumes. Additionally, we plan to investigate and compare commercial DIR algorithms with DIR algorithms utilizing deep learning. This comparative analysis will enable us to evaluate and identify potential improvements and performance differences between existing deep learning algorithms and commercial solutions [
16].
In summary, our study provides valuable insights into the clinical application of DIR algorithms for head and neck patients, highlighting the need for further advancements in this field to overcome the challenges and improve the accuracy of deformable image registration.
Author Contributions
For research articles with several authors, a short paragraph specifying their individual contributions must be provided. The following statements should be used “Conceptualization, methodology, software, validation, investigation, resources, visualization, writing—original draft preparation, Kang Houn Lee.; writing—review and editing, Kang Houn Lee, Young Nam Kang.; supervision, Young Nam Kang, funding acquisition, Young Nam Kang. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Research Foundation of Korea Grant funded by the Korean Government (NRF-2022R1F1A1072382).
Informed Consent Statement
Patient consent was waived due to open dataset in TCIA.
Data Availability Statement
Acknowledgments
The authors have no Acknowledgements.
Conflicts of Interest
The authors have no conflict to disclose.
References
- Avants, B.B. , Epstein, C.L., Grossman, M., Gee, J.C., 2008. Symmetric dif- feomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Medical image analysis 12, 26–41.
- H. Sokooti, B. de Vos, F. Berendsen, M. Ghafoorian, S. Yousefi, B. P. Lelieveldt, I. Isgum, and M. Staring. “3D convolutional neural networks image registration based on efficient supervised learning from artificial deformations”. In: arXiv preprint arXiv:1908.10235 (2019). arXiv:1908.10235 (2019).
- M.-M. Rohé, M. Datar, T. Heimann, M. Sermesant, and X. Pennec. “SVF-Net: Learn- ing deformable image registration using shape matching”. In: International Confer- ence on Medical Image Computing and Computer-Assisted Intervention. Springer. 2017, pages 266–274.
- Shen D, Wu G, Suk HI (2017) Deep learning in medical image analysis. Annu Rev Biomed Eng 19:221–248.
- M. Jaderberg, K. Simonyan, A. Zisserman, and K. Kavukcuoglu. “Spatial transformer networks”. In: arXiv preprint arXiv:1506.02025 (2015). arXiv:1506.02025 (2015).
- G. Balakrishnan, A. Zhao, M. R. Sabuncu, J. Guttag, and A. V. Dalca. “VoxelMorph: a learning framework for deformable medical image registration”. In: IEEE Transactions on Medical Imaging 38.8 (2019), pages 1788–1800. [CrossRef]
- B. D. de Vos, F. F. Berendsen, M. A. Viergever, H. Sokooti, M. Staring, and I. Išgum. “A deep learning framework for unsupervised affine and deformable image registration”. In: Medical Image Analysis 52 (2019), pages 128–143. [CrossRef]
- Clark K, Vendt B, Smith K, Freymann J, Kirby J, Koppel P, Moore S, Phillips S, Maffitt D, Pringle M, Tarbox L, Prior F. The Cancer Imaging Archive (TCIA): Maintaining and Operating a Public Information Repository, Journal of Digital Imaging, Volume 26, Number 6, December, 2013, pp 1045-1057.
- Bejarano T, De Ornelas-Couto M, Mihaylov IB. (2018). Head-and-neck squamous cell carcinoma patients with CT taken during pre-treatment, mid-treatment, and post-treatment Dataset. The Cancer Imaging Archive.
- , Ridgway, G.R., Taylor, Z.A., Lehmann, M., Barnes, J., Hawkes, D.J., Fox, N.C., Ourselin, S., 2010. Fast free-form deformation using graph- ics processing units. Computer methods and programs in biomedicine 98, 278–284.
- Kim, B., Kim, D.H., Park, S.H., Kim, J., Lee, J.G., Ye, J.C., 2021. Cyclemorph: Cycle consistent unsupervised deformable image registration. Medical Im- age Analysis 71, 102036. [CrossRef]
- Chen, Junyu, et al. "Transmorph: Transformer for unsupervised medical image registration." Medical image analysis 82 (2022): 102615. [CrossRef]
- Yang, Xiao, et al. "Quicksilver: Fast predictive image registration–a deep learning approach." NeuroImage 158 (2017): 378-396.
- Sokooti, Hessam, et al. "Nonrigid image registration using multi-scale 3D convolutional neural networks." Medical Image Computing and Computer Assisted Intervention− MICCAI 2017: 20th International Conference, Quebec City, QC, Canada, -13, 2017, Proceedings, Part I 20. Springer International Publishing, 2017. 11 September.
- Paszke, Adam, et al. "Pytorch: An imperative style, high-performance deep learning library." Advances in neural information processing systems 32 (2019).
- Pukala J, Johnson PB, Shah AP, Langen KM, Bova FJ, Staton RJ, Mañon RR, Kelly P, Meeks SL. Benchmarking of five commercial deformable image registration algorithms for head and neck patients. J Appl Clin Med Phys. 2016 ;17(3):25-40. 8 May.
- Varadhan R, Karangelis G, Krishnan K, Hui S. A framework for deformable image registration validation in radiotherapy clinical applications. J Appl Clin Med Phys. 2013;14(1):192–213. [CrossRef]
- Brock, Kristy K., et al. "Use of image registration and fusion algorithms and techniques in radiotherapy: Report of the AAPM Radiation Therapy Committee Task Group No. 132." Medical physics 44.7 (2017): e43-e76. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).