1. Introduction
Neuroligin-3 (NLGN3) is an adhesion molecule expressed at neuronal synapses in the central nervous system (CNS) and is well established as a regulator of brain function [
1,
2,
3,
4,
5]. Although some studies suggest NLGN3 is important for proper GI function [
6,
7], its cellular expression profile in the enteric nervous system has not been characterised. In cultured rat hippocampal neurons, NLGN3 is expressed at the postsynaptic membrane of both excitatory and inhibitory neuronal synapses [
8]. NLGN3 is also expressed in non-neuronal cells. For example, Venkatesh and colleagues showed that neurally-secreted NLGN3 invades the microenvironment of tumours and induces NLGN3 expression in glioma cells to promote tumour growth [
9]. In addition, NLGN3 is expressed in many types of glia during rodent development, including olfactory ensheathing glia, retinal astrocytes, Schwann cells and spinal cord astrocytes [
10]. These expression patterns suggest that NLGN3 could also be expressed in enteric glia.
Mutations in the
NLGN3 gene including the missense R451C point mutation as well as
NLGN3 deletion are implicated in autism [
11,
12,
13]. Patients expressing the
NLGN3 R451C mutation show GI dysfunction including diarrhea, faecal incontinence, post-meal regurgitation, oesophageal inflammation, chronic intestinal pain as well as delayed bladder and bowel control [
7]. Modification of NLGN3 expression induces GI dysfunction in mice, as shown in both
Nlgn3 knockout (KO) and
Nlgn3R451C mice [
6,
7].
Nlgn3 KO mice exhibit distended colons and more rapidly propagating colonic muscle contractions [
6].
Nlgn3R451C mice display faster intestinal transit and increased numbers of myenteric neurons in the small intestine as well as GABA
A receptor-mediated colonic dysmotility [
7]. These findings suggest a role for NLGN3 in gut function.
The R451C mutation impacts
Nlgn3 mRNA and protein expression in the brain. In mouse whole brain samples,
Nlgn3 mRNA expression levels are unchanged by the
Nlgn3 R451C mutation but NLGN3 protein levels are reduced [
1]. Although initial studies of NLGN3 expression in the GI tract have been reported in rodents [
7,
14,
15] and humans [
14], the distribution of NLGN3 expression in different cell populations of the mouse enteric nervous system has not been profiled. Characterising
Nlgn3 expression in enteric cell subtypes is essential for identifying underlying mechanisms contributing to GI activity and to functional GI changes observed in patients and the
Nlgn3R451C mouse model of autism.
Localizing the NLGN3 protein in the mouse GI tract is challenging due to non-specific labelling of commercially available antibodies which commonly yield false-positive results in this tissue (Leembruggen et al., unpublished). Therefore, we used the enhanced in situ hybridization technique, RNAScope, to label Nlgn3 mRNA in enteric neurons. We combined RNAScope and immunofluorescence with high-resolution microscopy and 3D image analysis software to compare Nlgn3 mRNA expression in the wild type (WT) and Nlgn3R451C mouse gut.
Overall, we found that
Nlgn3 mRNA is expressed in the mouse enteric nervous system (including enteric glia) and that
Nlgn3R451C mice have reduced
Nlgn3 mRNA expression in subpopulations of submucosal and myenteric plexus neurons. The molecular identities of enteric neuronal populations have now been well described in mice based on scRNASeq combined with immunostaining verification [
16,
17]. These studies show the presence of 12 enteric neuronal populations with distinct neurochemical signatures and enteric glia. We therefore investigated the cellular expression of
Nlgn3 in the mouse enteric nervous system based on previously reported scRNASeq datasets and found that all enteric neuronal subtypes and glia express
Nlgn3 at varying levels. In mice, the R451C mutation reduces
Nlgn3 mRNA expression in submucosal cholinergic as well as myenteric nitrergic and calretinin neurons.
Nlgn3R451C mice also had reduced mRNA expression levels in myenteric (but not submucosal) glia. In contrast,
Nlgn3 mRNA expression was unchanged in VIPergic submucosal neurons in
Nlgn3R451C mice. Taken together, these findings suggest that NLGN3 is an important component of neuronal and neuron-glial communication in the gut, and that mutations in NLGN3 might play a role in GI dysfunction in individuals with autism.
4. Discussion
In this study, we showed that most submucosal and myenteric neurons in the mouse ileum express Nlgn3 mRNA. For the first time, we revealed that Nlgn3 mRNA is expressed in ileal submucosal and myenteric glia. In addition, we report that the autism-associated Nlgn3 R451C mutation reduces Nlgn3 mRNA expression in both submucosal and myenteric neurons as well as myenteric glia in the mouse ileum.
Most enteric neurons express Nlgn3 mRNA
Although there is ample evidence for the existence of prominent post-synaptic densities within the ENS [
21,
22,
23,
24], the molecular characterization remains to be fully characterized. In the rodent brain, neuroligin-3 is expressed at both excitatory and inhibitory synapses [
8] and closely associated with other PDZ-associated molecules to mediate postsynaptic signal transduction. There is evidence that PDZ domain proteins such as PSD95 [
25,
26], PSD93 [
26] as well as the cell adhesion molecules L1 [
27] and neuroligin-1 [
28] are expressed in the ENS.
Cholinergic transmission
It is well established that NLGN3 is involved in both excitatory and inhibitory synaptic transmission in the brain. However, relatively little is known about the distribution of post-synaptic proteins associated with excitatory (predominantly mediated via nicotinic acetylcholine receptors (nAChRs)) and inhibitory synapses in the ENS. We reveal that most cholinergic submucosal neurons also contain
Nlgn3 mRNA. Within the enteric neural circuitry, acetylcholine is the primary excitatory neurotransmitter utilised by cholinergic secretomotor neurons, excitatory muscle motor neurons, ascending interneurons, descending interneurons and intrinsic sensory neurons [
29,
30,
31,
32]. Since virtually all submucosal neurons receive synaptic inputs via cholinergic synapses [
33],
Nlgn3 is likely present in cholinergic synapses in the submucosal plexus. All submucosal neurons receive cholinergic fast excitatory post synaptic potentials hence can be expected to have post-synaptic densities with clusters of nicotinic receptors and NLGNs as a part of the synaptic structure. Although there is no available evidence of NLGN3 expression in the cholinergic system of the central or peripheral nervous systems, expression of other NLGN subtypes in cholinergic synapses has been reported. For example, NLGN2 is expressed at the postsynaptic membrane of cholinergic synapses in the mouse brain [
34] and NLGN1 is present in cholinergic synapses in the chick ciliary ganglion [
35]. Since NLGN3 and other NLGN isoforms can be colocalized in the same synapse [
8], NLGN3 might be co-expressed with NLGN1 and NLGN2 in cholinergic synapses in the submucosal plexus.
VIP-expressing neurons
We also revealed that
Nlgn3 mRNA is expressed in VIP-expressing submucosal neurons in the mouse ileum. In the submucosal plexus, VIP is a primary neurotransmitter of secretomotor neurons and stimulates intestinal secretion [
33,
36]. In the myenteric plexus, VIP is expressed in inhibitory muscle motor neurons and interneurons [
37,
38]. VIP also acts as a co-transmitter alongside ACh and NO in a subset of descending interneurons in the mouse and guinea pig ileum [
38,
39,
40]. Moreover, an excitatory role for the Vasoactive Intestinal Peptide 1 (VPAC1) receptor on cholinergic neurons has been identified in the myenteric plexus [
41]. These findings open a novel avenue for research in
Nlgn3R451C mice since the expression profiles of NLGN3 or other NLGN isoforms in VIP-containing synapses have not been reported to date in either the CNS or the peripheral nervous system.
NOS-expressing neurons
One of the major findings of this study is that most NO-containing enteric neurons express
Nlgn3 mRNA in the cell soma. Nitric oxide (NO) is the predominant inhibitory neurotransmitter to the smooth muscle in the enteric nervous system [
38,
39] and is also expressed in descending interneurons together with other neurotransmitters [
42]. Given that NLGN complexes anchor postsynaptic densities and PSD95 has at least one nNOS binding PDZ domain,
Nlgn3 may contribute to synaptic specialization of enteric NO neurons. However, the distributions of PSD95 and PSD93 have not been well characterized in ENS. It has been shown that PSD93 interacts with nNOS via a PDZ-PDZ domain interaction [
26]. Some nNOS expressing myenteric neurons express PSD93 [
43]. In agreement with this finding, our data indicating
Nlgn3 expression in this neuronal population suggests that Nlgn3 might be expressed at the postsynaptic membrane of nNOS immunoreactive myenteric neurons.
Calretinin-expressing neurons
We also show
Nlgn3 mRNA expression in calretinin-expressing myenteric neurons in the mouse ileum. In the enteric nervous system, calretinin is expressed in intrinsic primary afferent neurons, interneurons and excitatory motor neurons innervating the smooth muscle layer [
38,
39]. In line with these findings, scRNASeq data analysis confirmed expression of
Nlgn3 at varying levels in mouse myenteric neurons including putative motor neurons, interneurons and intrinsic primary afferent neurons in the mouse small intestine. It has been reported that the PSD93 protein is localized to calretinin neurons in mouse myenteric plexus [
43]. Therefore, it is possible that
Nlgn3 is associated with postsynaptic protein complexes within calretinin neurons in the myenteric plexus
Other signalling systems
Given that glutamate and GABA have active roles in the ENS [
44,
45] alongside the presence of postsynaptic proteins including PSD93 [
43], the wide distribution of NLGN3 we observed in the ENS suggests that NLGN3 could also be expressed at glutamatergic and GABAergic synapses within the ENS.
Differential expression of Neuroligin binding partners in the enteric nervous system
When examining scRNASeq data,
Nlgn2 and
3 expression profiles are almost identical in all classes of enteric neurons. In contrast with findings in the central nervous system [
8] reviewed by [
46], this suggests that NLGN3 and NLGN2 co-express in enteric neurons subtypes and we speculate that they may form heterodimers in the enteric nervous system. Expression patterns of the neurexin family of neuroligin binding partners, however, show that
Nrxn2 and
Nrxn3 are more strongly expressed within the enteric neuronal cell clusters compared with
Nrxn1. These findings suggest that
Nlgn3 may preferentially bind with
Nrxn2 and
3 at synapses in IPANs of the myenteric plexus. In the CNS, NLGNs are present at classical synapses receiving fast EPSPs but most functional evidence suggests that IPANs do not exhibit fast EPSPs. For example, findings from Hibberd and colleagues [
47] indicate that IPANs receive fast EPSPs, but in contrast, both [
48] and [
49] reported that AH neurons do not exhibit fast EPSPs. Nevertheless, it is unclear whether junctions producing slow EPSPs might express neuroligin complexes, therefore, further research is required to identify the role of NLGN3 in IPANs in the ENS.
A role for Nlgn3 in enteric neuronal-glial synapses
For the first time, we reveal that Nlgn3 mRNA is expressed in enteric glia. Intriguingly, scRNAseq analysis showed that most enteric glia express Nlgn3 mRNA, but negligible/low levels of Nlgn1 and Nlgn2. Therefore, we propose that NLGN3 acts as a main adhesion protein in glia for modulating glial-neuron synaptic activity in the enteric nervous system.
Enteric glia play a major role in enteric nervous system-mediated GI functions including mucosal secretion, intestinal permeability, mucosal sensation, GI motility and immune responses [
50,
51,
52] in concert with enteric neurons. Enteric neurons synapse onto enteric glia to regulate enteric nervous system-coordinated GI responses [
50,
53,
54]. Enteric glial-neuronal associations revealed by immunocytochemistry highlight that enteric glia are in close contact with nerve fibers and varicose release sites [
55]. Evidence for neuro-glia communication also comes from live imaging experiments reporting the presence of several signalling pathways. Ca
2+ imaging studies indicate that neuron-glia communication is regulated by neurally released purines which activate purinergic receptors on enteric glia [
54,
55,
56,
57]. In the submucosal plexus, neuron-glial transmission occurs via P2Y
1 and P2Y
4 receptors [
58]. Furthermore, neuronal ATP release via the pannexin-1 channel also influences neuro-glia interactions [
59,
60]. Based on this evidence, we propose that NLGN3 expressed by both enteric neurons and glia modulate neuro-glia signalling pathways. This hypothesis is well-established in the mouse brain where RNA sequencing of the transcriptome revealed that
Nlgn3 transcripts are enriched in glial cell types including astrocytes and oligodendrocytes [
61]. Interestingly, NLGNs expressed in astrocytes aid in neuron-astrocyte communication via bi- and tripartite synapses in mice in
C. elegans [
62] suggesting that a neuronal-glial communication role may not be specific to mice. Similarly, NLGN3 expressed in enteric glia could also be involved in establishing bi-partite and tripartite synapses in the enteric nervous system. Given this evidence, this study strongly suggests that NLGN3 might play a role in synapse formation and modifying synaptic function during neuron-glia communication in the enteric nervous system.
The R451C mutation reduces Nlgn3 mRNA expression in the enteric nervous system
We previously demonstrated that mice expressing the
Nlgn3 R451C mutation have faster small intestinal transit and increased numbers of myenteric neurons in the small intestine [
7]. How this mutation affects NLGN3 expression in the enteric nervous system, however, is not understood. Here we show that the R451C mutation decreases
Nlgn3 mRNA expression in both neurons and glia in the mouse ileal submucosal plexus. For example, cholinergic submucosal neurons in mutant mice contain fewer
Nlgn3 mRNA copies compared to WT. These changes could potentially alter cholinergic signalling in the submucosal plexus which could affect functions mediated by cholinergic neurons such as secretion, absorption and mucosal barrier functions. Although there are no reports available on changes to the cholinergic system of the enteric nervous system in ASD, CNS studies showed that altered cholinergic neurons are associated with the pathophysiology of autism [
63,
64]. Specifically, ASD patient basal forebrain tissues show altered cholinergic neuronal numbers, size and structure [
65]. In addition, a decreased plasma concentration of choline, a precursor for acetylcholine has been reported in ASD patients [
66,
67] and reduced levels of hippocampal cytosolic choline have been correlated with autism severity [
66]. In the human GI tract, NLGN3-mediated alterations to the cholinergic system could induce GI dysfunction, however further studies are required to determine effects of the R451C mutation on cholinergic signalling in the enteric nervous system.
In the myenteric plexus of the distal ileum, the R451C mutation reduces
Nlgn3 mRNA expression levels in neurons. Specifically,
Nlgn3 mRNA expression is substantially reduced in both calretinin and nNOS-immunoreactive neuronal populations. Although the contribution of calretinin neurons to ASD or ASD-related GI pathophysiology is unclear, the significance of calretinin neurons in other ENS-related diseases has been highlighted [
68]. As mentioned, an increased proportion of NOS1 expressing neurons has previously has previously been observed in
Nlgn3R451C mice [
7], and our current findings showing reduced
Nlgn3 expression in these cells might contribute to altered NO neuronal signalling in the enteric nervous system in these mice.
Finally, the impact of the R451C mutation on Nlgn3 expression in glia has not previously been reported. Here we show that in Nlgn3R451C mice, Nlgn3 mRNA expression is dramatically reduced in myenteric glia but unchanged in glia located within the submucosal plexus. Since glia play an important role in mediating intestinal functions such as mucosal barrier regulation, gut motility, immune responses and neurotransmission, such a reduction in NLGN3 levels could contribute to GI dysfunction in Nlgn3R451C mutant mice. Therefore, in addition to neuronal dysfunction, glial dysfunction could also contribute to GI pathology in individuals diagnosed with ASD.
Author Contributions
Conceptualization, M.H, E.H-Y, J.B and A.F.; methodology, M.H.; software, M.H.; validation, M.H., E.H-Y, J.B, E.C and A.F.; formal analysis, M.H.; investigation, E.H-Y, J.B and A.F; resources, M.H.; data curation, M.H.; writing - original draft preparation, M.H., E.H-Y, J.B, E.C and A.F; writing—review and editing, M.H.; visualization, E.H-Y, J.B and A.F.; supervision, E.H-Y, J.B and A.F.; project administration, E.H-Y, J.B and A.F.; funding acquisition, E.H-Y, J.B and A.F. All authors have read and agreed to the published version of the manuscript.
Figure 1.
The distribution of Nlgn3 mRNA in ileal submucosal neurons. Confocal images of A1: submucosal neurons labelled with Hu pan neuronal marker; A2: Nlgn3 mRNA expression. A3: merged image. A4: 3D reconstruction of submucosal neurons; A5: cellular Nlgn3 mRNA expression; A6: merged image. A7: Frequency distribution of Nlgn3 mRNA expression in submucosal neurons. Confocal images of (B1) submucosal ganglion (B2) Nlgn3 mRNA expression, B3: ChAT neuron, B4: merge. 3D structure of (B5) submucosal neurons; B6: Nlgn3 mRNA, B7: a cholinergic submucosal neuron (B8), merge. B9: The distribution of Nlgn3 mRNA expression in cholinergic neurons is similar to that in total neurons in the submucosal plexus. B10: Average number of Nlgn3 mRNA copies in cholinergic neuronal soma is similar to that in total neurons in the submucosal plexus. Confocal micrographs of: C1: submucosal neurons, C2: Nlgn3 mRNA expression, C3: VIP expression, C4: merge. 3D reconstruction of (C5) submucosal neurons, C6: Nlgn3 mRNA expression C7: VIP-expressing neurons; C8: merge. C9: Frequency distribution of Nlgn3 mRNA expression in VIP submucosal neurons is significantly different to total submucosal neurons. C10: On average, VIP submucosal neurons express similar numbers of Nlgn3 mRNA copies as observed in the total number of neurons in the submucosal plexus. Open arrowheads: Hu staining; arrows: Nlgn3 mRNA expression. Scale bar=10 µm.
Figure 1.
The distribution of Nlgn3 mRNA in ileal submucosal neurons. Confocal images of A1: submucosal neurons labelled with Hu pan neuronal marker; A2: Nlgn3 mRNA expression. A3: merged image. A4: 3D reconstruction of submucosal neurons; A5: cellular Nlgn3 mRNA expression; A6: merged image. A7: Frequency distribution of Nlgn3 mRNA expression in submucosal neurons. Confocal images of (B1) submucosal ganglion (B2) Nlgn3 mRNA expression, B3: ChAT neuron, B4: merge. 3D structure of (B5) submucosal neurons; B6: Nlgn3 mRNA, B7: a cholinergic submucosal neuron (B8), merge. B9: The distribution of Nlgn3 mRNA expression in cholinergic neurons is similar to that in total neurons in the submucosal plexus. B10: Average number of Nlgn3 mRNA copies in cholinergic neuronal soma is similar to that in total neurons in the submucosal plexus. Confocal micrographs of: C1: submucosal neurons, C2: Nlgn3 mRNA expression, C3: VIP expression, C4: merge. 3D reconstruction of (C5) submucosal neurons, C6: Nlgn3 mRNA expression C7: VIP-expressing neurons; C8: merge. C9: Frequency distribution of Nlgn3 mRNA expression in VIP submucosal neurons is significantly different to total submucosal neurons. C10: On average, VIP submucosal neurons express similar numbers of Nlgn3 mRNA copies as observed in the total number of neurons in the submucosal plexus. Open arrowheads: Hu staining; arrows: Nlgn3 mRNA expression. Scale bar=10 µm.
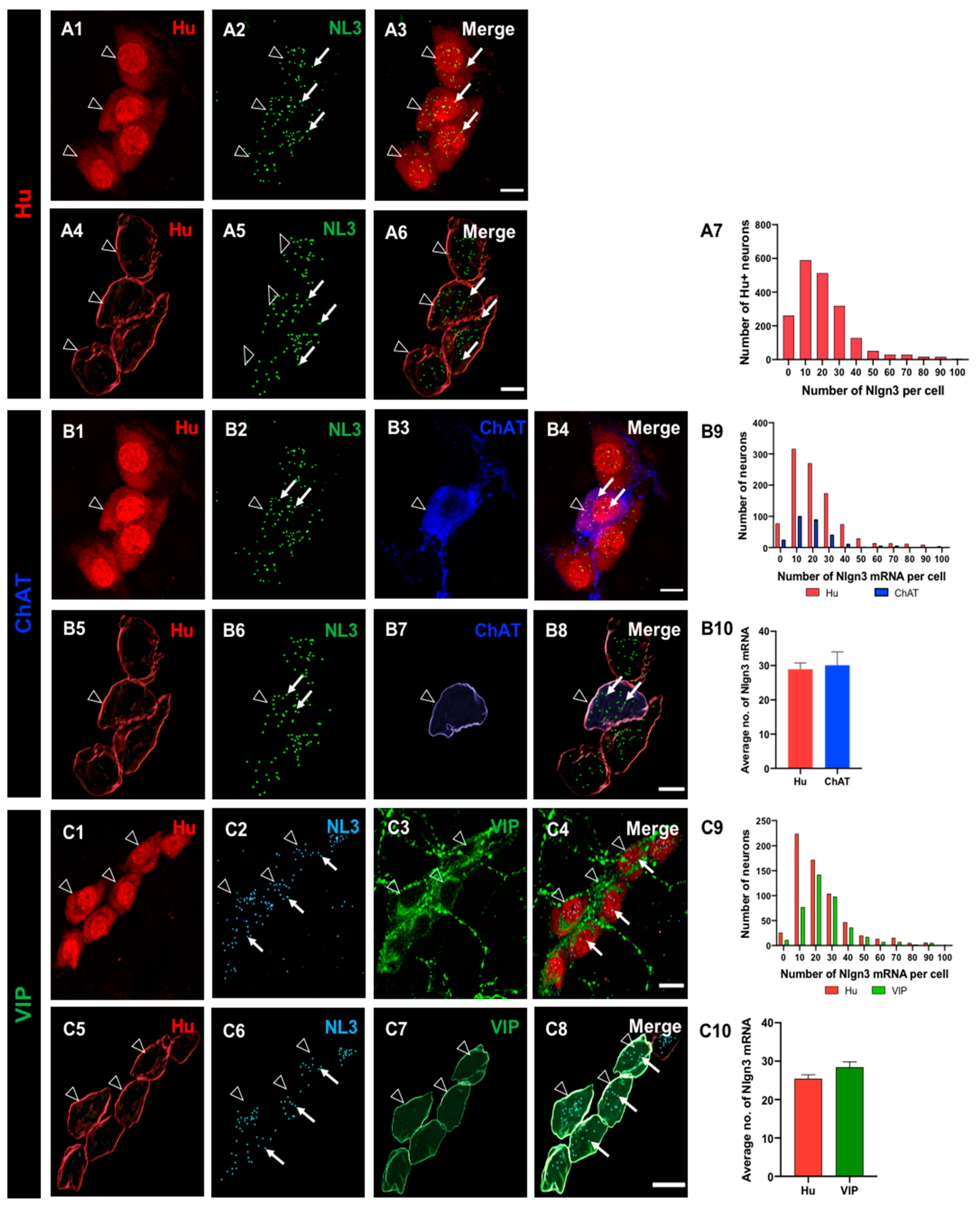
Figure 2.
Expression and the distribution of Nlgn3 mRNA in distal ileal myenteric neurons. A1: Myenteric neurons labelled using Hu immunofluorescence. A2: Nlgn3 mRNA expression (RNAScope); A3: merge. 3D structure of (A4) myenteric neurons, A5: Nlgn3 mRNA, A6: merge. A7: Frequency distribution of Nlgn3 expression in myenteric neurons. Confocal micrographs of (B1) myenteric neurons labelled with the pan-neuronal marker, Hu; B2: Nlgn3 mRNA, B3; calretinin. B4: Expression of Nlgn3 mRNA in calretinin neurons. 3D rendering of (B5) myenteric neurons, B6: Nlgn3 mRNA, B7: calretinin, B8: merge. B9: Frequency distribution of Nlgn3 mRNA expression in calretinin-positive myenteric neurons. B10: Calretinin-expressing neurons contain similar copy numbers of Nlgn3 mRNA to that of myenteric neurons overall. Triple labelling of (C1) myenteric neurons, C2: Nlgn3 mRNA and C3: nNOS expressing myenteric neurons. C4. Expression of Nlgn3 mRNA in nNOS containing myenteric neurons. 3D structure of (C5) myenteric neurons, C6: Nlgn3 mRNA, C7: NOS-expressing myenteric neurons, C8: merge. C9: Frequency distribution of Nlgn3 mRNA in nNOS containing neurons. C10: Average number of Nlgn3 mRNA copies in myenteric nNOS-containing compared to total myenteric neurons. Open arrowheads: Hu staining; arrows: Nlgn3 mRNA labelling. Scale bar=10 µm.
Figure 2.
Expression and the distribution of Nlgn3 mRNA in distal ileal myenteric neurons. A1: Myenteric neurons labelled using Hu immunofluorescence. A2: Nlgn3 mRNA expression (RNAScope); A3: merge. 3D structure of (A4) myenteric neurons, A5: Nlgn3 mRNA, A6: merge. A7: Frequency distribution of Nlgn3 expression in myenteric neurons. Confocal micrographs of (B1) myenteric neurons labelled with the pan-neuronal marker, Hu; B2: Nlgn3 mRNA, B3; calretinin. B4: Expression of Nlgn3 mRNA in calretinin neurons. 3D rendering of (B5) myenteric neurons, B6: Nlgn3 mRNA, B7: calretinin, B8: merge. B9: Frequency distribution of Nlgn3 mRNA expression in calretinin-positive myenteric neurons. B10: Calretinin-expressing neurons contain similar copy numbers of Nlgn3 mRNA to that of myenteric neurons overall. Triple labelling of (C1) myenteric neurons, C2: Nlgn3 mRNA and C3: nNOS expressing myenteric neurons. C4. Expression of Nlgn3 mRNA in nNOS containing myenteric neurons. 3D structure of (C5) myenteric neurons, C6: Nlgn3 mRNA, C7: NOS-expressing myenteric neurons, C8: merge. C9: Frequency distribution of Nlgn3 mRNA in nNOS containing neurons. C10: Average number of Nlgn3 mRNA copies in myenteric nNOS-containing compared to total myenteric neurons. Open arrowheads: Hu staining; arrows: Nlgn3 mRNA labelling. Scale bar=10 µm.
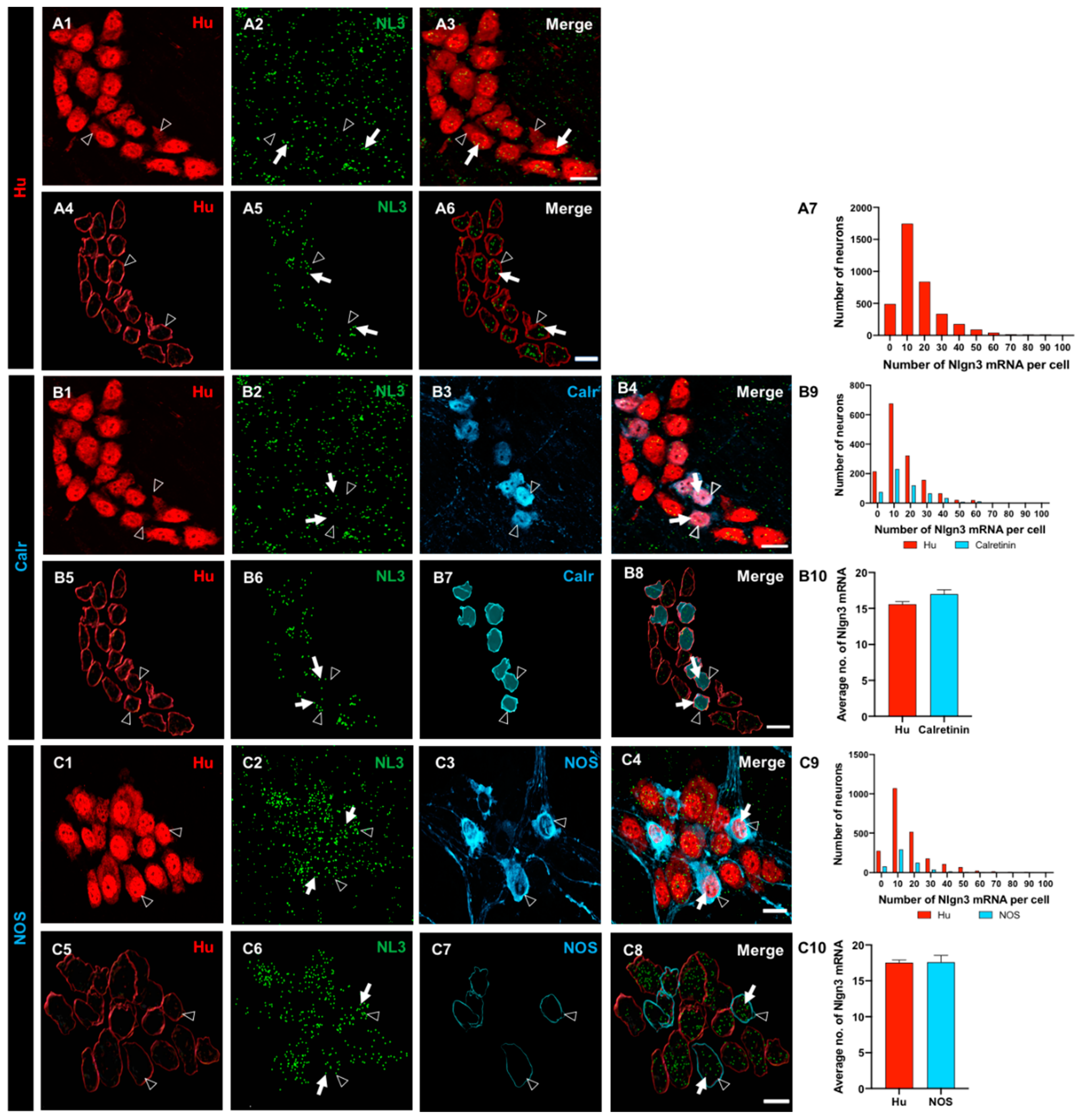
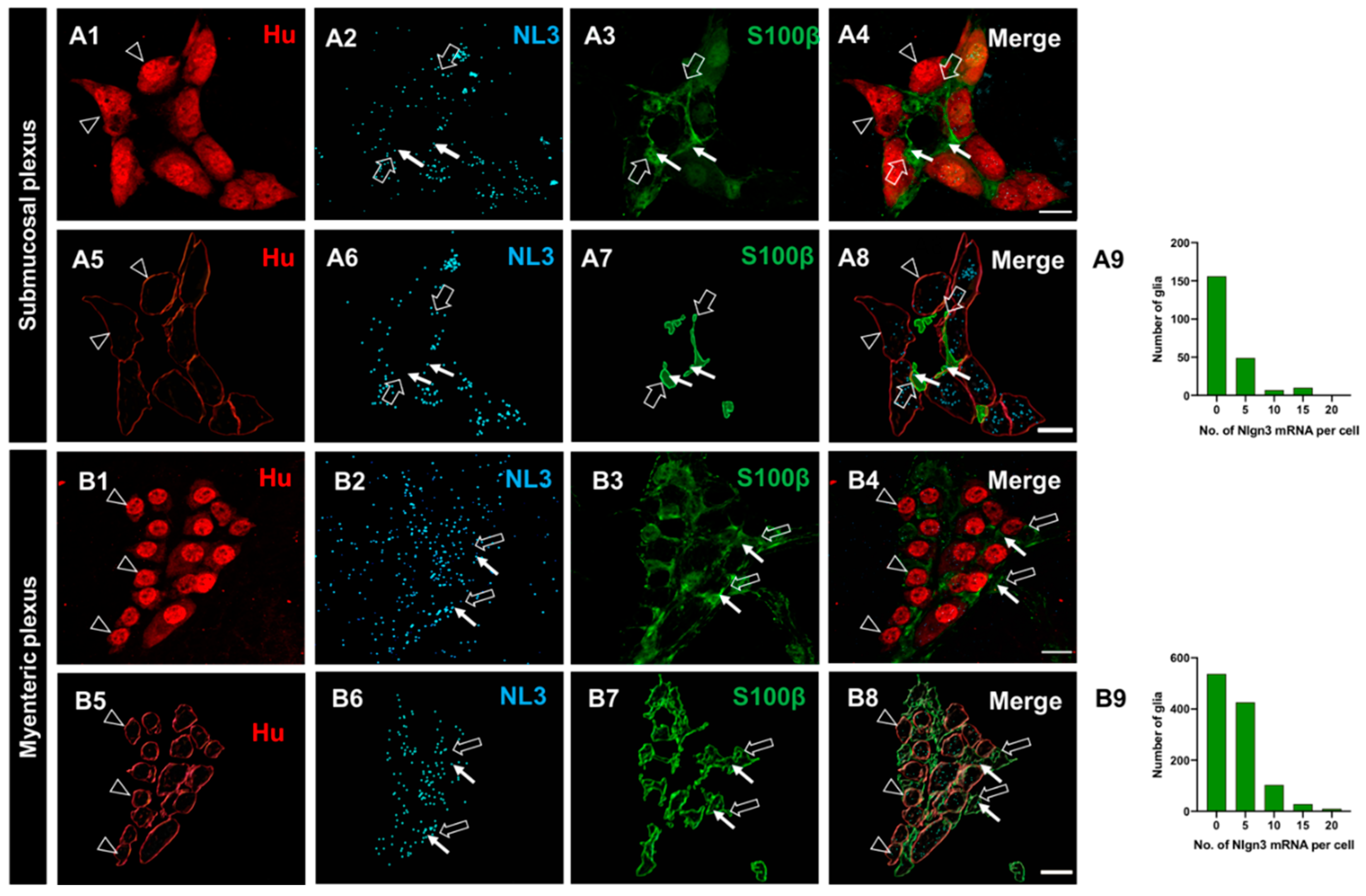
Figure 3.
The expression and distribution of Nlgn3 mRNA in enteric glia. Confocal micrographs of (A1) submucosal neurons labelled with the pan-neuronal marker Hu (A2) Nlgn3 mRNAexpression in the submucosal plexus (A3) submucosal glia labelled with S100β (A4) Nlgn3 mRNA in submucosal glia. Imaris-based 3D rendering of (A5) submucosal neurons; A6: Nlgn3 mRNA, A7: submucosal glia, A8: merge. A9: Frequency distribution analysis demonstrates that Nlgn3 mRNA is expressed in the majority of submucosal glial cells. Triple labelling of (B1) myenteric neurons using the pan-neuronal marker, Hu, B2: Nlgn3 mRNA, B3: myenteric glia (using S100β). B4: Expression of Nlgn3 mRNA in myenteric glia. 3D rendering of (B5) myenteric neurons, B6: Nlgn3 mRNA, B7: myenteric glia, B8: merge. B9: Frequency distribution analysis indicating that the vast majority of myenteric glia do not express Nlgn3 mRNA. Open arrowheads: Hu staining, filled arrows: Nlgn3 mRNA, open arrows: submucosal glia. Scale bar=10 µm.
Figure 3.
The expression and distribution of Nlgn3 mRNA in enteric glia. Confocal micrographs of (A1) submucosal neurons labelled with the pan-neuronal marker Hu (A2) Nlgn3 mRNAexpression in the submucosal plexus (A3) submucosal glia labelled with S100β (A4) Nlgn3 mRNA in submucosal glia. Imaris-based 3D rendering of (A5) submucosal neurons; A6: Nlgn3 mRNA, A7: submucosal glia, A8: merge. A9: Frequency distribution analysis demonstrates that Nlgn3 mRNA is expressed in the majority of submucosal glial cells. Triple labelling of (B1) myenteric neurons using the pan-neuronal marker, Hu, B2: Nlgn3 mRNA, B3: myenteric glia (using S100β). B4: Expression of Nlgn3 mRNA in myenteric glia. 3D rendering of (B5) myenteric neurons, B6: Nlgn3 mRNA, B7: myenteric glia, B8: merge. B9: Frequency distribution analysis indicating that the vast majority of myenteric glia do not express Nlgn3 mRNA. Open arrowheads: Hu staining, filled arrows: Nlgn3 mRNA, open arrows: submucosal glia. Scale bar=10 µm.
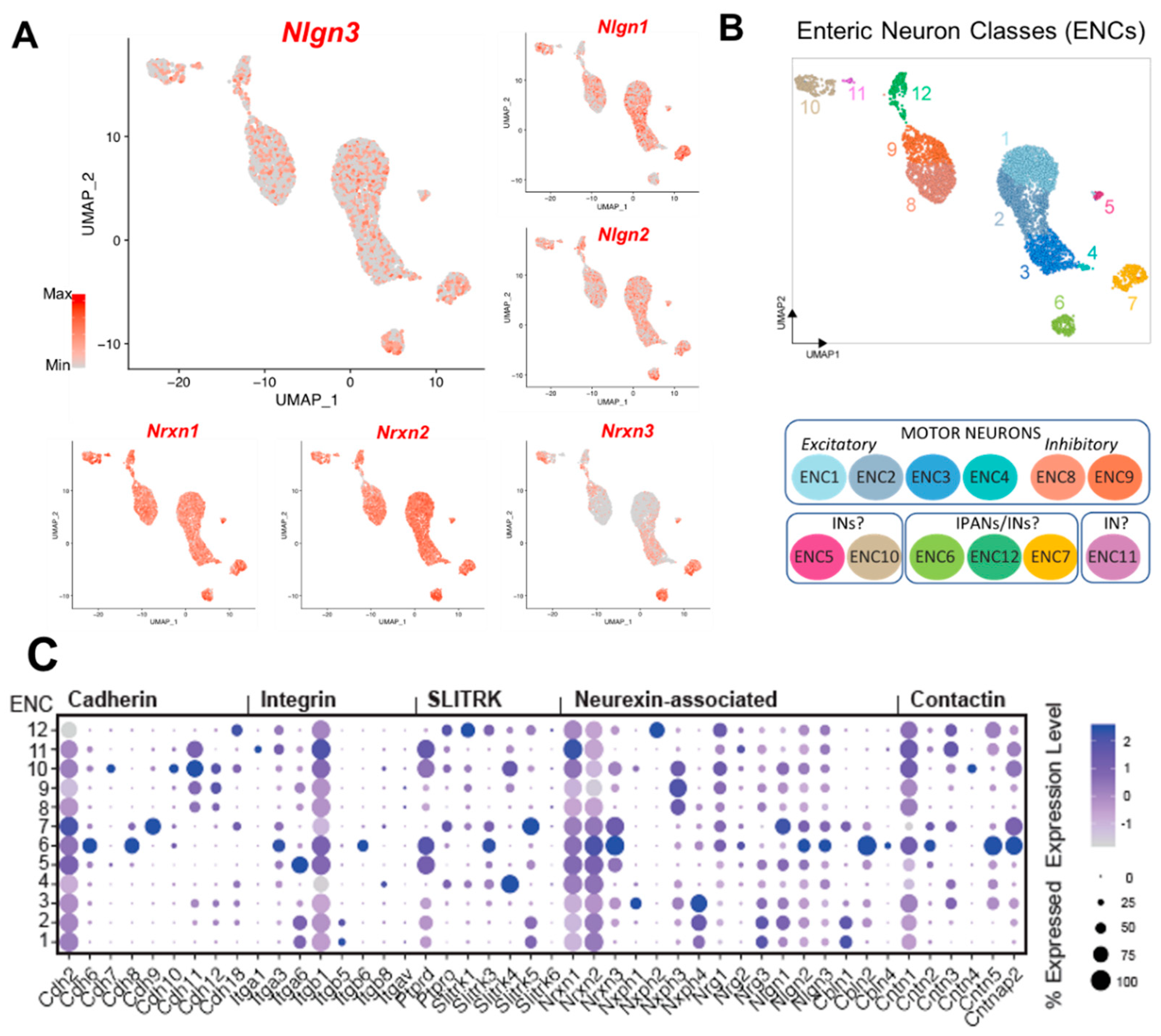
Figure 4.
Single cell RNA Sequencing data analysis reveal broad Nlgn3 expression in enteric neurons. A: UMAP of myenteric neuronal subtypes indicating expression of
Nlgn3 in addition to
Nlgn1 and
Nlgn2 at varying expression levels. Colour bar indicates relative expression level with maximum cut-off at the 90
th percentile.
B: ENC1-12 displayed on UMAP and schematic indicating plausible functional annotations (modified from [
17]).
C: Dotblots indicating relative expression of
Nrxn and
Nlgn genes across ENCs (modified version of figure in [
17]) UMAP: Uniform Manifold Approximation and Projection, IN: Interneuron; IPANS: Intrinsic primary afferent neurons. ENC: Enteric Neuron Class.
Figure 4.
Single cell RNA Sequencing data analysis reveal broad Nlgn3 expression in enteric neurons. A: UMAP of myenteric neuronal subtypes indicating expression of
Nlgn3 in addition to
Nlgn1 and
Nlgn2 at varying expression levels. Colour bar indicates relative expression level with maximum cut-off at the 90
th percentile.
B: ENC1-12 displayed on UMAP and schematic indicating plausible functional annotations (modified from [
17]).
C: Dotblots indicating relative expression of
Nrxn and
Nlgn genes across ENCs (modified version of figure in [
17]) UMAP: Uniform Manifold Approximation and Projection, IN: Interneuron; IPANS: Intrinsic primary afferent neurons. ENC: Enteric Neuron Class.
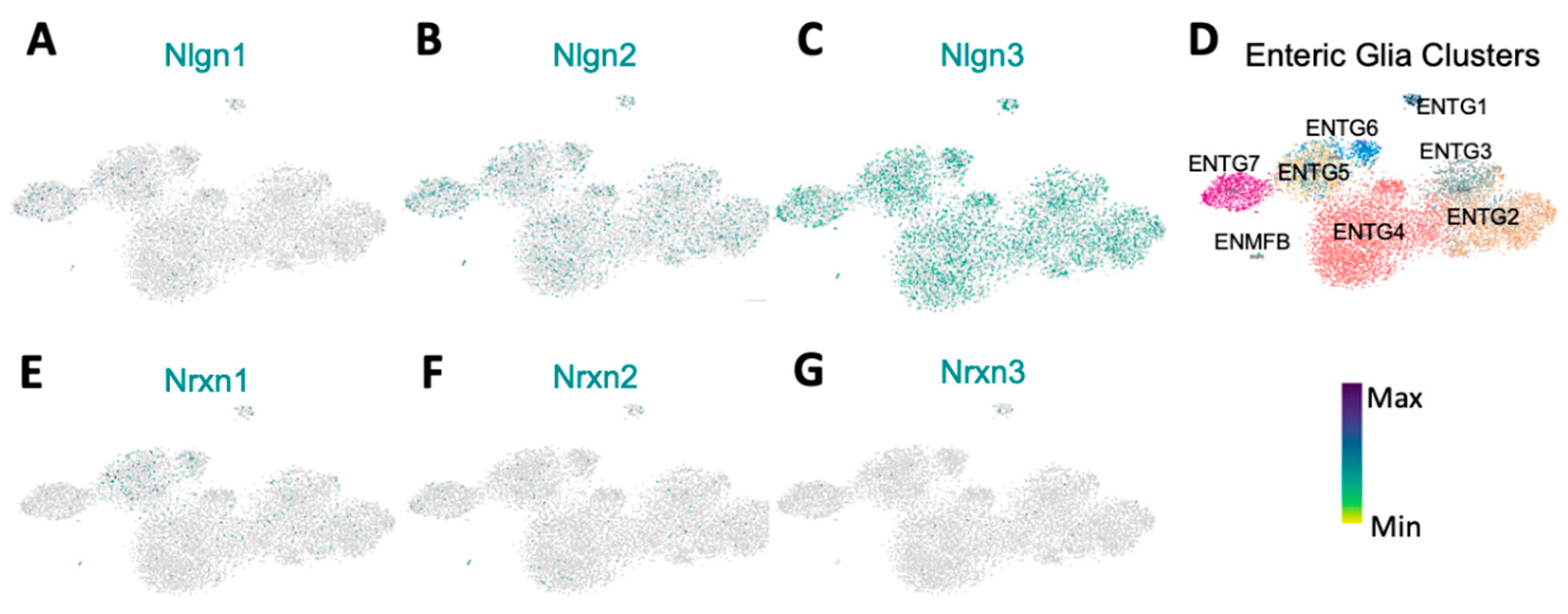
Figure 5.
Differential expression of Neuroligins and Neurexins in enteric glia. A: Nlgn1 is negligible expressed in EGCs B: Nlgn2 is expressed in a low proportion of all ENTGs. C: Nlgn3 is strongly expressed compared to Nlgn1 and Nlgn2 across all ENTGs. D: Representation of enteric glia clusters. E-G: Nrxn1, 2 and 3 are minimally expressed in enteric glia. Colour bar indicates relative gene expression. ENTG: enteric glia cluster; ENMFB: enteric mesothelial fibroblasts. Pseudocolored scale bar refers to maximum and minimum expression levels within ENTGs and ENMFB.
Figure 5.
Differential expression of Neuroligins and Neurexins in enteric glia. A: Nlgn1 is negligible expressed in EGCs B: Nlgn2 is expressed in a low proportion of all ENTGs. C: Nlgn3 is strongly expressed compared to Nlgn1 and Nlgn2 across all ENTGs. D: Representation of enteric glia clusters. E-G: Nrxn1, 2 and 3 are minimally expressed in enteric glia. Colour bar indicates relative gene expression. ENTG: enteric glia cluster; ENMFB: enteric mesothelial fibroblasts. Pseudocolored scale bar refers to maximum and minimum expression levels within ENTGs and ENMFB.
Figure 6.
Expression of Nlgn3 mRNA in the ileal submucosal plexus of Nlgn3R451C mutant mice. Confocal micrographs of submucosal neurons labeled with the pan-neuronal marker Hu (A1), Nlgn3 mRNA expression (A2); merge (A3). The same submucosal neurons shown following 3D reconstruction (A4), corresponding cellular Nlgn3 mRNA expression (A5); merge (A6). A7: The distribution of Nlgn3 mRNA in WT is significantly different compared to Nlgn3R451C mutant mice. A8: Nlgn3R451C mutant mice express fewer copies of Nlgn3 mRNA in submucosal neurons compared to WT. Submucosal neurons labelled with Hu pan-neuronal marker (B1) Nlgn3 mRNA (B2) and the cholinergic marker, ChAT (B3); merge (B4). 3D rendering of (B5) Hu neurons (B6) and corresponding Nlgn3 mRNA expression (B7) and ChAT labelling (B8). B9: Frequency distribution of Nlgn3 mRNA in cholinergic submucosal neurons. B10: Cholinergic neurons express significantly fewer copies of Nlgn3 mRNA in Nlgn3R451C mutants compared to WT. Confocal micrographs of submucosal neurons labelled with C1: the pan-neuronal marker, Hu; C2: Nlgn3 mRNA labelled using RNAScope and C3: non-cholinergic neurons labelled with VIP; C4: merge. 3D reconstruction of C5: submucosal neurons, C6: corresponding Nlgn3 mRNA puncta, C7: VIPergic neurons, C8; merge. C9: Frequency distribution of Nlgn3 mRNA in non-cholinergic submucosal neurons C10: Nlgn3R451C mice express similar numbers of Nlgn3 mRNA copies in non-cholinergic neurons compared to WT in the submucosal plexus. Nlgn3 mRNA is indicated by the filled arrow and submucosal glia are labelled with open arrows **p<0.01, ****p<0.0001, Scale bar=10 µm.
Figure 6.
Expression of Nlgn3 mRNA in the ileal submucosal plexus of Nlgn3R451C mutant mice. Confocal micrographs of submucosal neurons labeled with the pan-neuronal marker Hu (A1), Nlgn3 mRNA expression (A2); merge (A3). The same submucosal neurons shown following 3D reconstruction (A4), corresponding cellular Nlgn3 mRNA expression (A5); merge (A6). A7: The distribution of Nlgn3 mRNA in WT is significantly different compared to Nlgn3R451C mutant mice. A8: Nlgn3R451C mutant mice express fewer copies of Nlgn3 mRNA in submucosal neurons compared to WT. Submucosal neurons labelled with Hu pan-neuronal marker (B1) Nlgn3 mRNA (B2) and the cholinergic marker, ChAT (B3); merge (B4). 3D rendering of (B5) Hu neurons (B6) and corresponding Nlgn3 mRNA expression (B7) and ChAT labelling (B8). B9: Frequency distribution of Nlgn3 mRNA in cholinergic submucosal neurons. B10: Cholinergic neurons express significantly fewer copies of Nlgn3 mRNA in Nlgn3R451C mutants compared to WT. Confocal micrographs of submucosal neurons labelled with C1: the pan-neuronal marker, Hu; C2: Nlgn3 mRNA labelled using RNAScope and C3: non-cholinergic neurons labelled with VIP; C4: merge. 3D reconstruction of C5: submucosal neurons, C6: corresponding Nlgn3 mRNA puncta, C7: VIPergic neurons, C8; merge. C9: Frequency distribution of Nlgn3 mRNA in non-cholinergic submucosal neurons C10: Nlgn3R451C mice express similar numbers of Nlgn3 mRNA copies in non-cholinergic neurons compared to WT in the submucosal plexus. Nlgn3 mRNA is indicated by the filled arrow and submucosal glia are labelled with open arrows **p<0.01, ****p<0.0001, Scale bar=10 µm.
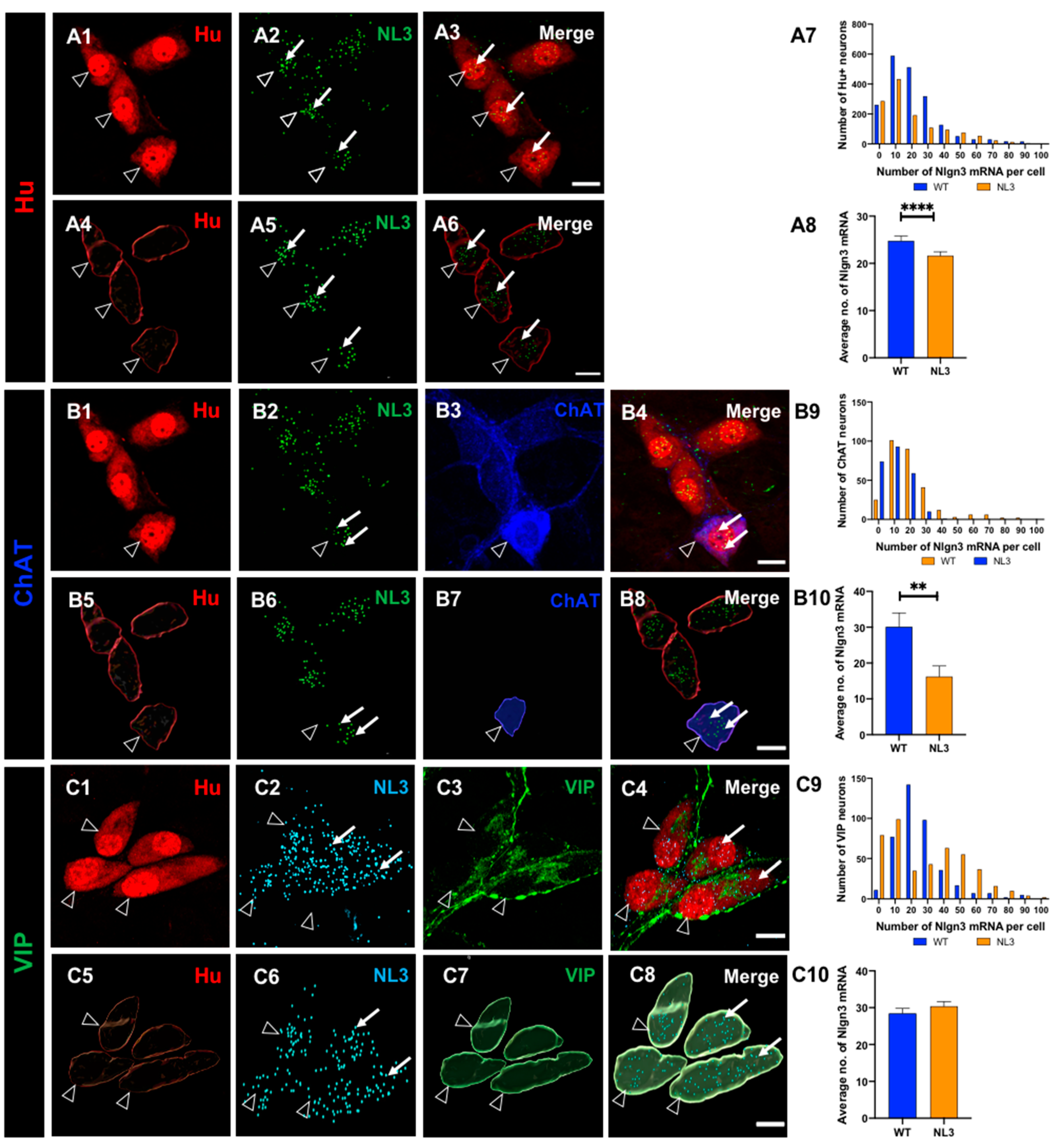
Figure 7.
Effects of the Nlgn3 R451C mutation on Nlgn3 mRNA expression in myenteric neurons. Confocal images of (A1) myenteric neurons (A2) Nlgn3 mRNA expression (A3) Nlgn3 mRNA expression in myenteric neurons. The 3D structure of (A4) myenteric neurons (A5) Nlgn3 mRNA (A6) Nlgn3 mRNA expression in myenteric neurons. A7. The frequency distribution of Nlgn3 mRNA in Nlgn3R451C mutant mice compared WT A8. In the Nlgn3R451C mouse ileum, myenteric neurons express fewer copies of Nlgn3 mRNA compared to WT. Triple labelling of (B1) myenteric neurons using B2: the pan-neuronal marker Hu, B3: Nlgn3 mRNA, B3: calretinin; B4: merge. 3D reconstruction of (B5) myenteric neurons, B6: Nlgn3 mRNA, B7: calretinin expressing neurons and B8: merge. B9. The frequency distribution of Nlgn3 mRNA in calretinin positive myenteric neurons in Nlgn3R451C mutant mice and WT. B10: The Nlgn3 R451C mutation reduces Nlgn3 mRNA expression in mutant mice compared to WT. Triple labelling of (C1) myenteric neurons, C2: Nlgn3 mRNA, C3: nNOS expressing neurons, C4: merge. 3D rendering of (C5) myenteric neurons; C6: nNOS positive neurons, C7: Nlgn3 mRNA and C8: merge. C9: Frequency distribution of Nlgn3 mRNA expression in NOS-positive neurons in Nlgn3R451C mutant mice. C10: In Nlgn3R451C mutant mice, NOS neurons contain fewer Nlgn3 mRNA copies compared to WT. Filled arrows: Nlgn3 mRNA. Open arrows: submucosal glia. ****p<0.0001, Scale bar=10 µm.
Figure 7.
Effects of the Nlgn3 R451C mutation on Nlgn3 mRNA expression in myenteric neurons. Confocal images of (A1) myenteric neurons (A2) Nlgn3 mRNA expression (A3) Nlgn3 mRNA expression in myenteric neurons. The 3D structure of (A4) myenteric neurons (A5) Nlgn3 mRNA (A6) Nlgn3 mRNA expression in myenteric neurons. A7. The frequency distribution of Nlgn3 mRNA in Nlgn3R451C mutant mice compared WT A8. In the Nlgn3R451C mouse ileum, myenteric neurons express fewer copies of Nlgn3 mRNA compared to WT. Triple labelling of (B1) myenteric neurons using B2: the pan-neuronal marker Hu, B3: Nlgn3 mRNA, B3: calretinin; B4: merge. 3D reconstruction of (B5) myenteric neurons, B6: Nlgn3 mRNA, B7: calretinin expressing neurons and B8: merge. B9. The frequency distribution of Nlgn3 mRNA in calretinin positive myenteric neurons in Nlgn3R451C mutant mice and WT. B10: The Nlgn3 R451C mutation reduces Nlgn3 mRNA expression in mutant mice compared to WT. Triple labelling of (C1) myenteric neurons, C2: Nlgn3 mRNA, C3: nNOS expressing neurons, C4: merge. 3D rendering of (C5) myenteric neurons; C6: nNOS positive neurons, C7: Nlgn3 mRNA and C8: merge. C9: Frequency distribution of Nlgn3 mRNA expression in NOS-positive neurons in Nlgn3R451C mutant mice. C10: In Nlgn3R451C mutant mice, NOS neurons contain fewer Nlgn3 mRNA copies compared to WT. Filled arrows: Nlgn3 mRNA. Open arrows: submucosal glia. ****p<0.0001, Scale bar=10 µm.
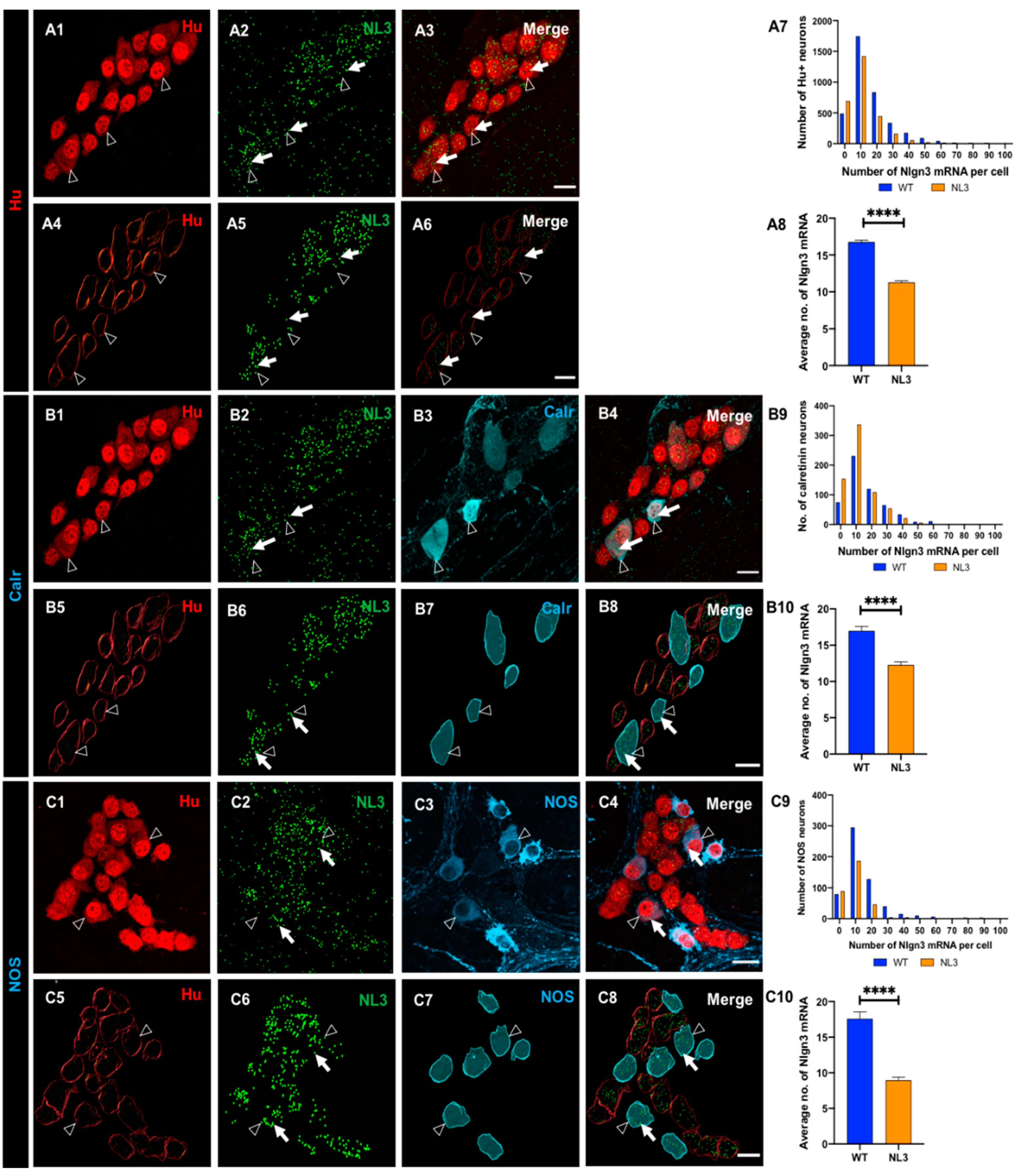
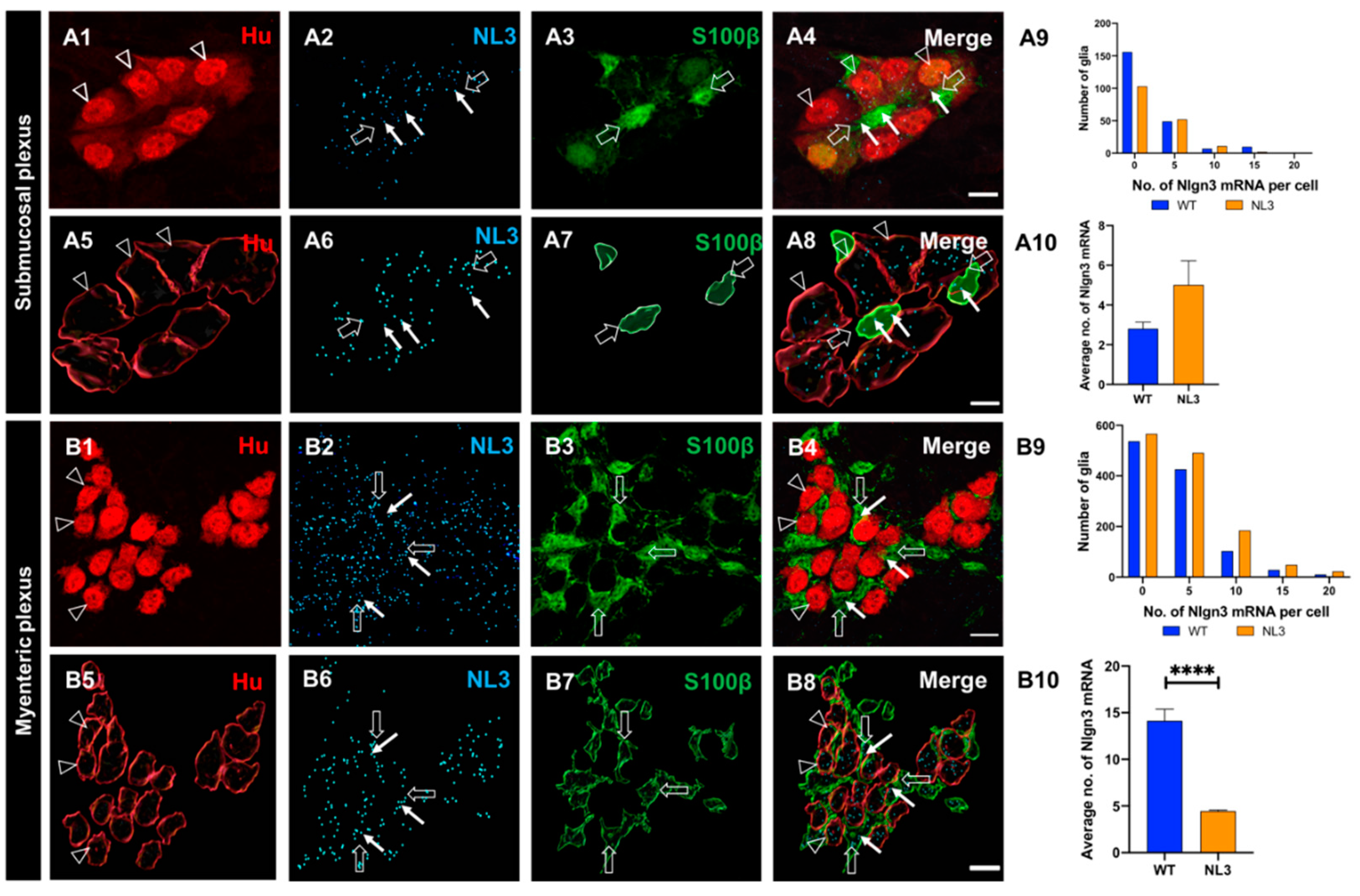
Figure 8.
Effects of Nlgn3 R451C mutation on Nlgn3 mRNA expression in enteric glia. Confocal images of (A1) submucosal neurons labelled with A2: Hu, A3: Nlgn3 mRNA, A4: merge. 3D structure of (A5) submucosal neurons; A6: Nlgn3 mRNA, A7: submucosal glia, A8: merge. A9: Frequency distribution of Nlgn3 mRNA in submucosal glia in Nlgn3R451C mutant mice compared to WT. A10: Nlgn3 mRNA copy numbers per cell in WT compared to Nlgn3R451C mutant mice. Immunofluorescent labelling of (B1) myenteric neurons (Hu), B2: Nlgn3 mRNA, B3: myenteric glia (S100β), B4: merge. 3D reconstruction of (B5) myenteric neurons; B6: Nlgn3 mRNA, B7: myenteric glia, B8: merge. B9: Frequency distribution of Nlgn3 mRNA expression in WT and Nlgn3R451C mutant mice. B10: Mutant mice express significantly fewer Nlgn3 mRNA copies in myenteric glia compared to WT. ****p<0.0001, Scale bar=10 µm.
Figure 8.
Effects of Nlgn3 R451C mutation on Nlgn3 mRNA expression in enteric glia. Confocal images of (A1) submucosal neurons labelled with A2: Hu, A3: Nlgn3 mRNA, A4: merge. 3D structure of (A5) submucosal neurons; A6: Nlgn3 mRNA, A7: submucosal glia, A8: merge. A9: Frequency distribution of Nlgn3 mRNA in submucosal glia in Nlgn3R451C mutant mice compared to WT. A10: Nlgn3 mRNA copy numbers per cell in WT compared to Nlgn3R451C mutant mice. Immunofluorescent labelling of (B1) myenteric neurons (Hu), B2: Nlgn3 mRNA, B3: myenteric glia (S100β), B4: merge. 3D reconstruction of (B5) myenteric neurons; B6: Nlgn3 mRNA, B7: myenteric glia, B8: merge. B9: Frequency distribution of Nlgn3 mRNA expression in WT and Nlgn3R451C mutant mice. B10: Mutant mice express significantly fewer Nlgn3 mRNA copies in myenteric glia compared to WT. ****p<0.0001, Scale bar=10 µm.
Table 1.
Primary antibodies used for immunocytochemistry.
Table 1.
Primary antibodies used for immunocytochemistry.
| Primary antisera |
Raised in |
Dilution |
Source |
| ANNA-1(anti HuC/D)*
|
Human |
1:5000 |
Gift from Dr. V. Lennon |
| VIP |
Rabbit |
1:1000 |
Merck Millipore |
| ChAT |
Goat |
1:100 |
Chemicon |
| nNOS |
Sheep |
1:1000 |
Gift from Dr P. Emson |
| Calretinin |
Goat |
1:1000 |
SWANT |
| S100β |
Rabbit |
1:1000 |
DAKO |
Table 2.
Secondary antibodies used for immunocytochemistry.
Table 2.
Secondary antibodies used for immunocytochemistry.
| Secondary antisera |
Raised in |
Dilution |
Source |
| Anti-human AF 594 |
Donkey |
1:500 |
Jackson Immuno Labs |
| Anti-sheep AF 647 |
Donkey |
1:500 |
Molecular Probes |
| Anti-rabbit AF 647 |
Donkey |
1:400 |
Molecular Probes |
| Anti-sheep AF 594 |
Donkey |
1:100 |
Molecular Probes |